Atrophin suppresses Hedgehog signaling by interacting with the transcriptional effector CiR and recruiting the histone deacetylase Rpd3 to the dpp locus to repress its transcription.
Abstract
The evolutionarily conserved Hedgehog (Hh) signaling pathway is transduced by the Cubitus interruptus (Ci)/Gli family of transcription factors that exist in two distinct repressor (CiR/GliR) and activator (CiA/GliA) forms. Aberrant activation of Hh signaling is associated with various human cancers, but the mechanism through which CiR/GliR properly represses target gene expression is poorly understood. Here, we used Drosophila melanogaster and zebrafish models to define a repressor function of Atrophin (Atro) in Hh signaling. Atro directly bound to Ci through its C terminus. The N terminus of Atro interacted with a histone deacetylase, Rpd3, to recruit it to a Ci-binding site at the decapentaplegic (dpp) locus and reduce dpp transcription through histone acetylation regulation. The repressor function of Atro in Hh signaling was dependent on Ci. Furthermore, Rerea, a homologue of Atro in zebrafish, repressed the expression of Hh-responsive genes. We propose that the Atro–Rpd3 complex plays a conserved role to function as a CiR corepressor.
Introduction
Hedgehog (Hh) is a secreted morphogen received by Patched (Ptc)-iHog coreceptors to relieve an inhibitory effect of Ptc on Smoothened (Smo). Activated Smo transduces a signal to the transcription factor Cubitus interruptus (Ci), which enters the nucleus and regulates the expression of target genes (Hooper and Scott, 2005; Chen et al., 2007; Jiang and Hui, 2008; Ingham et al., 2011). Transcriptional activity of Ci is finely regulated by a Hh gradient. In the absence of Hh, full-length Ci (Ci155) is phosphorylated and partially degraded to generate the Ci truncated form (Ci75), which primarily functions as a repressor (CiR; Aza-Blanc et al., 1997; Méthot and Basler, 1999). In the presence of Hh, Hh signaling stimulates the formation of the Ci active form (CiA), which binds to the specific promoter region of target genes and recruits coactivator dCBP (also known as nej; Akimaru et al., 1997) to activate downstream gene expression. Despite the fact that CiR and CiA transduce opposing signals, how a graded signal is achieved at the chromatin level remains unknown, especially through potential cofactors of CiR. From a yeast two-hybrid screen for Ci interacting proteins, we identified Atrophin (Atro) as a potential Ci regulator.
Atro (also known as Grunge) was first reported as a transcriptional corepressor in Drosophila melanogaster (Erkner et al., 2002; Zhang et al., 2002). It is involved in multiple developmental processes such as controlling planar polarity by interacting with Fat (Fanto et al., 2003), negative regulation of EGF receptor signaling (Charroux et al., 2006), and acting as corepressor of Tailless (Wang et al., 2006; Haecker et al., 2007). Atro is evolutionarily conserved with its homologous counterparts—arginine-glutamic acid dipeptide repeats a (Rerea) in zebrafish and atrophin1 and arginine-glutamic acid dipeptide repeats in human.
Here, we demonstrated that in D. melanogaster Atro acts as a corepressor of CiR to inhibit Hh target gene decapentaplegic (dpp) expression. Atro binds to Ci and recruits histone deacetylase Rpd3 to form a trimetric complex, which modifies the acetylation of histone H3 around the transcriptional start region of dpp, to achieve the final step of Ci repression. Furthermore, the zebrafish Atro homologue, Rerea, down-regulates different Hh-responsive genes. Together, our findings show a conserved role of Atro as a CiR corepressor in Hh signaling and reveal the mechanism of the Atro complex in Ci repression.
Results and discussion
Identification of Atro as a Ci interacting protein
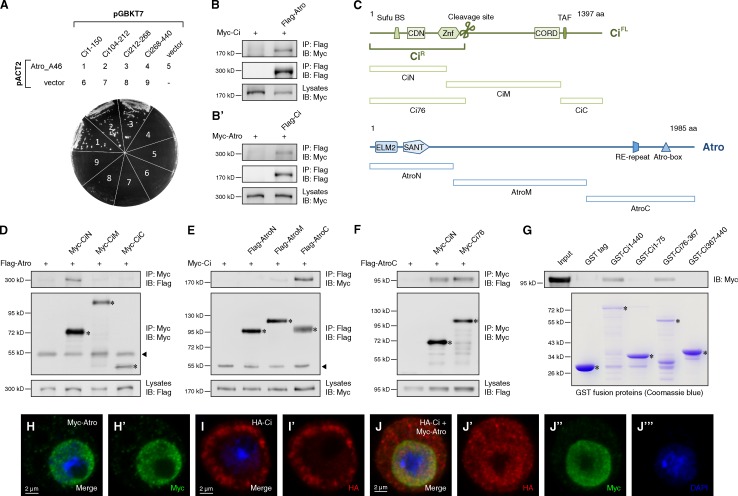
To identify new factors involved in Ci regulation, we performed a yeast two-hybrid screen using the N terminus (1–440 aa) of Ci (CiN) as bait and D. melanogaster embryonic cDNA library as prey. As a result, a positive clone termed A46 was identified as the C-terminal fragment (1727–1986 aa) of Atro. Further validation experiments showed that A46 interacted with different N-terminal fragments of Ci (Fig. 1 A). The interaction between Atro and Ci was then verified by coimmunoprecipitation (coIP) of overexpressed full-length Ci and Atro in S2 cells (Fig. 1, B and B′). A specific interaction was revealed between the C terminus of Atro (AtroC) and CiN using coIP in S2 cells (Fig. 1, C–F). A CiR mimic form, Ci76 (Aza-Blanc et al., 1997), interacted with AtroC in S2 cells (Fig. 1 F), indicating a possible role of Atro as a regulator of CiR. We identified that the region around 76–367 aa of CiN is essential to mediate the direct binding with AtroC by a GST pull-down assay (Fig. 1 G). When expressed alone, Atro localized in the nucleus (Fig. 1, H and H′), whereas Ci mainly localized in the cytoplasm (Fig. 1, I and I′). However, a portion of Ci translocated into the nucleus and colocalized with Atro when they were coexpressed (Fig. 1, J–J‴). Collectively, interaction and colocalization between Atro and Ci suggests a potential role of Atro in regulating Ci function.
Figure 1.
AtroC binds to the CiN. (A) Yeast two-hybrid assay between Atro_A46 fragment and indicated Ci fragments. (B and B′) Western blots of immunoprecipitates (top two panels) or lysates (bottom) from S2 cells expressing the indicated proteins. (C) Schematic representation of domains and motifs in Atro and Ci proteins and their fragments used in subsequent coIP assay. (D–F) Western blots of immunoprecipitates (top two panels) or lysates (bottom) from S2 cells expressing the indicated proteins. (G) GST pull-down between Myc-tagged AtroC and GST or GST-tagged Ci fragments. (H–J‴) S2 cells expressing the indicated proteins were immunostained with HA (red), Myc (green) antibodies, and DAPI (blue) to visualize nuclei. In all blots, asterisks indicate the target proteins and arrowheads indicate IgG.
Atro functions as a repressor of Hh signaling in D. melanogaster
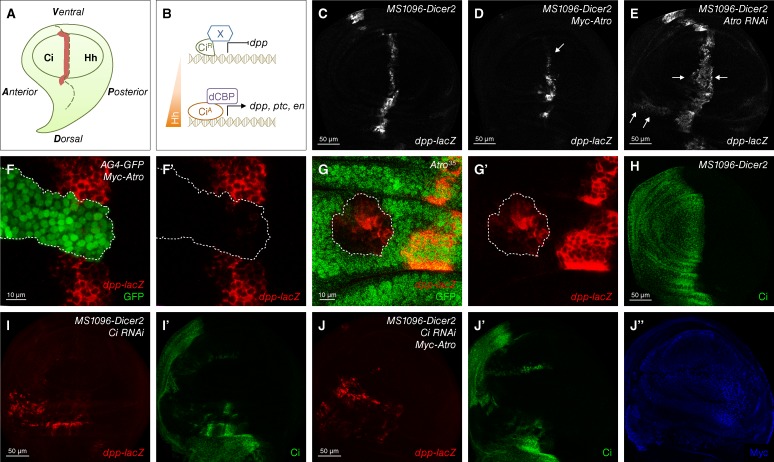
In the absence of Hh signaling, CiR represses dpp expression in the anterior compartment away from the anterior/posterior (A/P) compartment boundary (Fig. 2, A and B; Méthot and Basler, 1999; Müller and Basler, 2000). Hh signaling stimulates the formation of CiA to turn on different target genes around the A/P compartment boundary, including dpp, ptc, and engrailed (en), which are induced by low, medium, and high levels of Hh signaling, respectively (Fig. 2, A and B; Jiang and Hui, 2008). We first used dpp-lacZ staining to reveal dpp expression. Compared with control discs (Fig. 2 C), overexpression of Atro by MS1096-Dicer2 Gal4 down-regulated the level of dpp-lacZ (Fig. 2 D [arrow] and Fig. S1, A–A″). Knockdown of Atro (verified in Fig. S1, B and C) resulted in a notable increase of dpp-lacZ staining, which was expanded to the anterior compartment (Fig. 2 E, arrows). We confirmed these observations by inducing random clones. In the Atro overexpression clones marked by a GFP signal, dpp-lacZ staining was remarkably attenuated (Fig. 2, F and F′). Instead, ectopic expression of dpp-lacZ was observed in Atro35 (Erkner et al., 2002) mutant clones, which were identified by the absence of a GFP signal (Fig. 2, G and G′). We further examined the expression of ptc, a downstream gene of Hh signaling activated by CiA (Hepker et al., 1997), by ptc-lacZ. However, neither overexpression nor knockdown of Atro appeared to affect the staining of ptc-lacZ (Fig. S1, E–G). Moreover, knockdown of Ci blocked CiA-induced dpp-lacZ expression in the A/P compartment boundary and induced ectopic CiR-repressed dpp-lacZ expression in the anterior compartment. Overexpressed Atro did not repress ectopic dpp-lacZ expression in the absence of Ci (Fig. 2, H–J″). Collectively, these findings indicate that Atro specifically represses Hh signaling in a Ci-dependent manner in D. melanogaster.
Figure 2.
Atro is a repressor of Hh signaling pathway in D. melanogaster. (A) A cartoon of the wing discs from third-instar larva. The dashed line indicates the A/P compartment boundary. dpp expression region is shown in red. (B) Schematic shows that Hh gradient induces different target gene expression through transcriptional factor Ci. (C–E) A wild-type wing disc (C) or wing discs expressing Myc-tagged Atro (Myc-Atro; D) or Atro RNAi (E) with MS1096-Dicer2 Gal4 were immunostained to show the expression of dpp-lacZ. dpp-lacZ down-regulated in D (arrow) and up-regulated in E (arrows). (F and F′) A wing disc–expressing Myc-Atro with AG4-GFP was immunostained to show the expression of dpp-lacZ (red) and GFP (green). Atro-expressing clones (dashed lines) were marked by GFP-positive cells. (G and G′) A wing disc carrying Atro35 mutant clones (marked by GFP-positive cells in dashed lines) was immunostained to show the expression of dpp-lacZ (red) and GFP (green). (H) A MS1096-Dicer2 Gal4 wing disc was immunostained to show wild-type Ci staining. (I–J″) Wing discs expressing Ci RNAi alone or Ci RNAi and Myc-Atro together with MS1096-Dicer2 Gal4 were immunostained to show the expression of dpp-lacZ (red), Ci (green), and Myc-tagged Atro (blue). All of the phenotypes shown are stable and representative under each of the indicated genetic conditions.
Atro/Rerea plays a conserved role in repressing Hh signaling in zebrafish
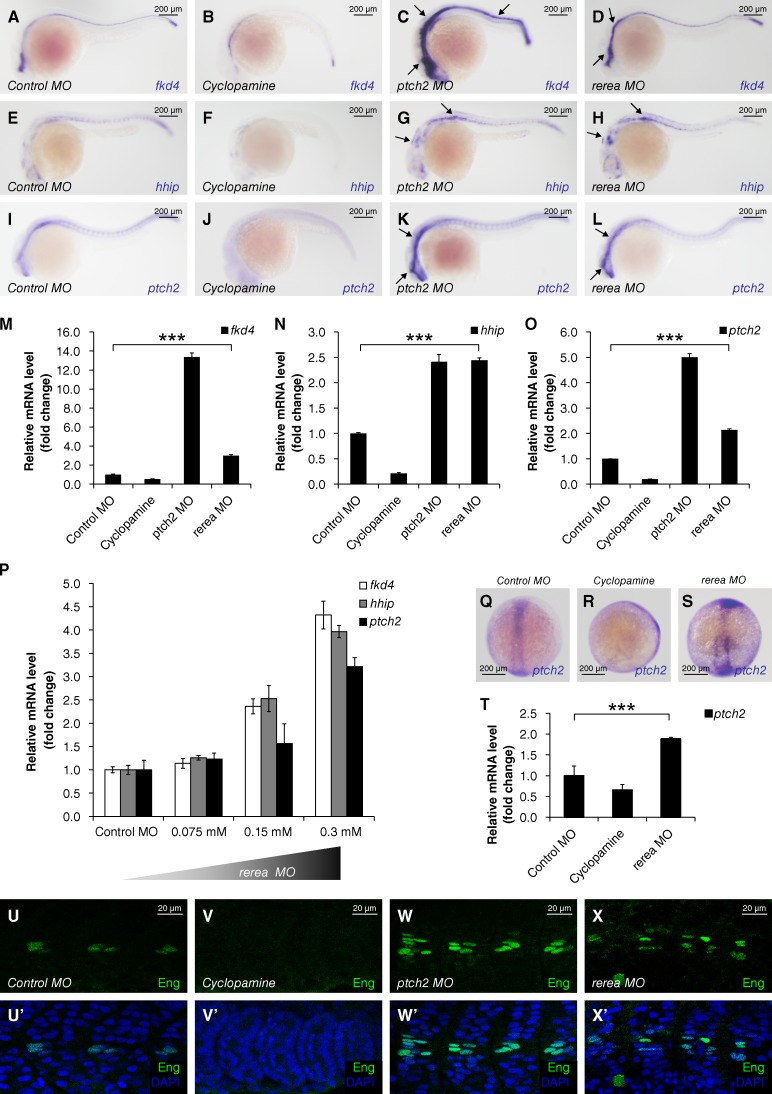
Given that Hh signaling is highly conserved from invertebrates to vertebrates (Jiang and Hui, 2008; Ingham et al., 2011), we wanted to determine whether Atro plays a conserved role in Hh signaling repression. Therefore, we checked the expression levels of several Hh-responsive genes in zebrafish embryos, including forkhead4 (fkd4; Tay et al., 2005; Tyurina et al., 2005), hedgehog interacting protein (hhip; Ochi et al., 2006), patched2 (ptch2; Concordet et al., 1996), NK2 homeobox 1b (nkx2.1b; Rohr et al., 2001), forkhead box A2 (foxa2; Tyurina et al., 2005), and GLI-Kruppel family member 1 (gli1; Karlstrom et al., 2003), 24 h postfertilization (hpf) by in situ hybridization and/or real-time PCR. Expression levels of Hh target genes were down-regulated in zebrafish embryos treated with cyclopamine (Wolff et al., 2003; Fig. 3, B, F, J, and M–O; and Fig. S1, H–J), which antagonized the Hh effector Smo (Chen et al., 2002), whereas knockdown of ptch2 by Morpholino (MO) up-regulated the expression of Hh target genes (Fig. 3, C, G, K, M-O; and Fig. S1, H–J). Compared with control morphants (Fig. 3, A, E, I, and M–O; and Fig. S1, H–J), Hh target gene expression was notably up-regulated in rerea morphants (Fig. 3 D, H, L, and M–O; and Fig. S1, H–J), which is the homologue of Atro in zebrafish. Most notably, Hh target gene expression was up-regulated in a rerea MO dosage-dependent manner (Fig. 3 P). We further examined ptch2 expression in bud stage (10 hpf) embryos (Fig. 3, Q–T). The expression of ptch2 was diminished in cyclopamine-treated embryos, whereas rerea morphants exhibited an elevated expression of ptch2. In a previous study, Wolff et al. (2003) showed that Hh signaling specifies cell fate in the zebrafish myotome in a dosage-dependent manner. Thus, we explored the impact of rerea MO-induced up-regulation of Hh signaling activity in muscle cells. Immunostaining revealed a high expression of Engrailed (Eng) in muscle pioneers (Fig. 3, U and U′). Cyclopamine treatment diminished Eng staining (Fig. 3, V and V′). In contrast, the number of Eng-positive cells increased in rerea morphants (Fig. 3, X and X′), similarly as observed in ptch2 morphants (Fig. 3, W and W′). Collectively, these data indicate that Rerea plays an evolutionarily conserved role in Hh signaling repression in zebrafish.
Figure 3.
The repressor function of Atro in Hh signaling is evolutionally conserved in zebrafish. (A–L) Expression of Hh-responsive genes in zebrafish embryos that were injected with the indicated MOs or treated with cyclopamine (10 µM) at 24 hpf. Up-regulated in situ stainings are marked by arrows. (M–O) Relative mRNA levels of fkd4, hhip, and ptch2 from 24 hpf zebrafish embryos indicated in A–L were revealed by real-time PCR (mean ± SD; n ≥ 3). (P) Relative mRNA levels of fkd4, hhip, and ptch2 from 24 hpf zebrafish embryos, which were injected with gradient concentrations of rerea MO. (Q–S) Expression of ptch2 in zebrafish embryos injected with the indicated MOs or treated with cyclopamine (100 µM) at bud stage (10 hpf). (T) Relative mRNA levels of ptch2 from bud stage embryos indicated in Q–S were revealed by real-time PCR (mean ± SD; n ≥ 3). (U–X’) Zebrafish embryos injected with the indicated MOs or treated with cyclopamine (10 µM) at 24 hpf were immunostained with Eng antibody (green) and DAPI (blue) to visualize nuclei. P-values in this figure were obtained by student’s t test between two groups (***, P < 0.001).
Atro cooperates with Rpd3 in Hh signaling repression
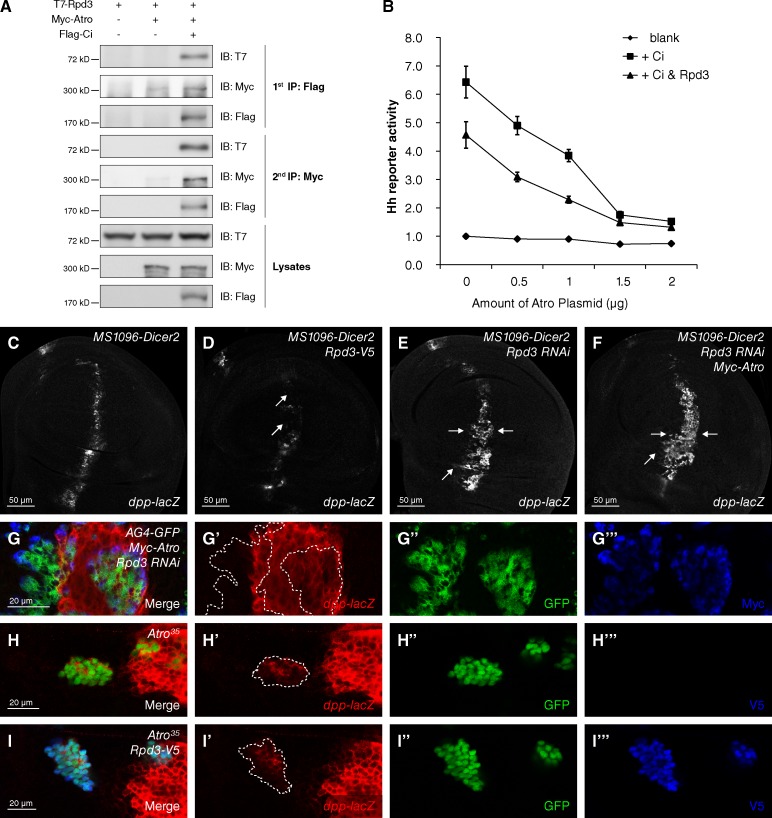
A previous study suggested that Atro family proteins associate with histone deacetylase 1 and 2 to function as nuclear receptor corepressors (Wang et al., 2006). We further investigated whether the D. melanogaster homologue of histone deacetylase 1 and 2 (Wang et al., 2008), Rpd3, was involved in Atro-mediated repression of Hh signaling. Results from a coIP experiment showed that Rpd3 preferentially bound to the N terminus of Atro (Fig. S2, A and B) rather than AtroC, which is responsible for the Ci interaction (Fig. 1 E). To investigate whether Atro–Ci and Atro–Rpd3 exist in two different complexes or whether they form a trimeric complex, we performed a two-step immunopreci experiment. As shown in Fig. 4 A, T7-Rpd3 was found in both Flag-Ci and Myc-Atro immunoprecipitates, indicating that Atro, Rpd3, and Ci are able to form a larger complex. Results from a reporter assay in S2 cells demonstrated the cooperation between Rpd3 and Atro in Hh signaling repression (Fig. 4 B). An increased amount of Atro down-regulated Hh reporter activity gradually, which was further decreased by coexpression with Rpd3 (Fig. 4 B).
Figure 4.
Atro recruits Rpd3 to repress Hh signaling. (A) S2 cells expressing the indicated proteins were harvested for the two-step immunoprecipitation and analyzed by Western blotting. (B) Rpd3 cooperates with Atro to repress Hh signaling in reporter assay in S2 cells (mean ± SD; n = 3). The whole DNA amount transfected in each group was normalized equal with blank vectors. (C–F) A wild-type wing disc (C) or wing discs expressing Rpd3-V5 (D), Rpd3 RNAi (E), or Rpd3 RNAi plus Myc-Atro (F) with MS1096-Dicer2 Gal4 were immunostained to show the expression of dpp-lacZ. The level of dpp-lacZ was reduced in Rpd3 overexpressing disc (D, arrows) and increased in Rpd3 knockdown disc (E, arrows). Atro overexpression failed to block the up-regulated expression of dpp-lacZ by Rpd3 knockdown (F, arrows). (G–G‴) A wing disc expressing Myc-Atro plus Rpd3 RNAi with AG4-GFP was immunostained to show the expression of dpp-lacZ– (red), GFP- (green), and Myc-tagged Atro (blue). (H–I‴) Wing discs carrying Atro35 clones or Atro35 plus Rpd3 overexpression were immunostained to show the expression of dpp-lacZ (red) and GFP (green). From G to I‴, clones were marked by GFP-positive cells (dashed lines).
In accordance with the Atro-overexpressing phenotype (Fig. 2, D–F), overexpression of Rpd3 down-regulated the dpp-lacZ staining in vivo (Fig. 4, compare D [arrows] with C). Overexpressed Rpd3 failed to repress dpp expression when Ci was knocked down (Fig. S2, C–F‴), indicating a similar requirement of Ci for the repressor role of Rpd3 in Hh signaling. We performed additional experiments to test whether Atro and Rpd3 cooperation is essential for Hh signaling repression. Knockdown of Rpd3 (verified in Fig. S2, G–G″) up-regulated dpp-lacZ staining (Fig. 4, compare E [arrows] with C), whereas coexpression with Atro failed to repress dpp expression (Fig. 4 F, arrows). A clonal assay with AG4-GFP further verified this phenotype. Coexpression of Rpd3 RNAi and Myc-Atro failed to block dpp-lacZ staining (Fig. 4, G–G‴), whereas Myc-Atro expressed alone remarkably reduced the dpp-lacZ signal (Fig. 2, F and F′). We next investigated the requirement of Atro in Rpd3-mediated Hh signaling repression. In a mosaic analysis with a repressible cell marker assay (Lee and Luo, 1999), Atro35 mutant clones showed increased dpp-lacZ staining (Fig. 4, I–I‴), which was not repressed by coexpression with Rpd3 in clones (Fig. 4, J–J‴). Collectively, these results suggest that Atro and Rpd3 cooperate to repress Hh signaling.
Hh signaling regulates the association of Atro–Rpd3 complex on the Ci regulatory region of the dpp locus
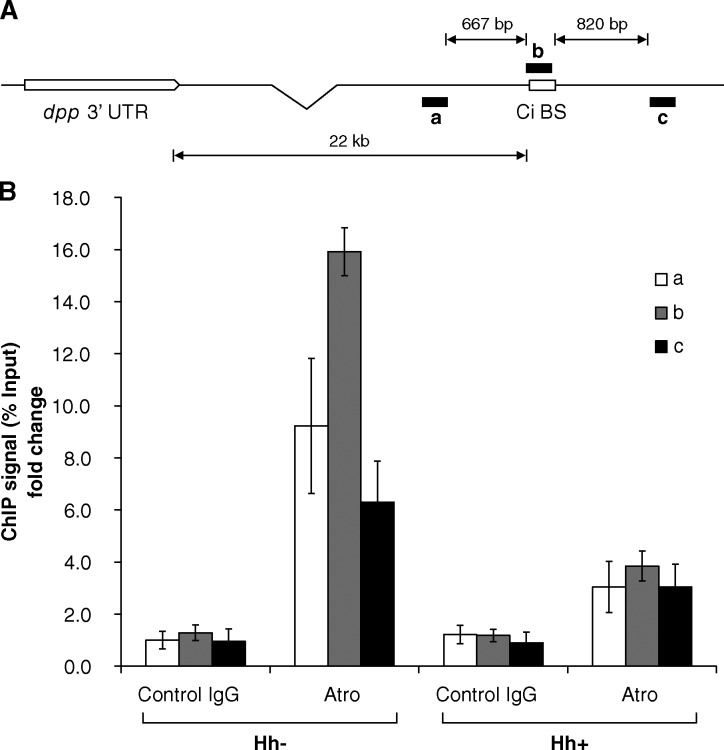
The dpp locus consists of a regulatory region >25 kb downstream of 3′ UTR (St Johnston et al., 1990), in which a 100-bp Ci binding site was mapped ∼22 kb away from a 3′ UTR (Müller and Basler, 2000; Fig. 5 A). Because we demonstrated that the Atro–Rpd3 complex acts together with CiR to repress dpp expression, Atro is likely recruited to the Ci binding site around the dpp locus. To test this possibility, we performed chromatin immunoprecipitation (ChIP) assays in wing disc–derived Cl.8 cells using an Atro antibody and control IgG. Our ChIP assays revealed that the Atro antibody, but not the control IgG, specifically precipitated chromatin fragments harboring the regions a, b, and c. Among these, the highest ChIP signal was located in region b covering the Ci binding site (Fig. 5 B, Hh−). Moreover, the intensity of the ChIP signal from the Atro group decreased in Cl.8 cells treated with Hh (Fig. 5 B, Hh+). Hence, Hh signaling activity regulates the association of Atro on the Ci regulatory region of the dpp locus. These data are consistent with our previous demonstration showing that Atro functions in the absence of Hh signaling as a corepressor of CiR.
Figure 5.
Atro–Rpd3 complex associates with a Ci-binding site at the dpp locus in the absence of Hh signaling. (A) Schematic diagram of dpp locus and regions amplified with corresponding PCR primers to detect ChIP products. (B) ChIP for Atro around Ci binding site in dpp locus in Cl.8 cells with/without Hh treatment. Data of ChIP signals were normalized to 1/10 of input and showed as the fold change to the first group (mean ± SD; n = 3).
The increased histone H3 acetylation correlates well with loss of Atro–Rpd3 complex and Hh activation
Rpd3 is a histone deacetylase that deacetylates acetylated histone H3 to turn off gene transcription (Foglietti et al., 2006). Thus, we speculated that Rpd3 links the repressor function of the Atro–Rpd3 complex to histone H3 deacetylation, whereas Hh activation relieves the association of Atro–Rpd3 with the corresponding dpp locus and thus increases histone H3 acetylation to turn on dpp transcription. We then performed ChIP assays to assess the alteration of histone H3 acetylation at the dpp transcription start site upon Hh activation. Among the reported lysine sites (K9, K11, K18, K23, and K27; Shahbazian and Grunstein, 2007) on the N terminus of histone H3, Hh activation increased the ChIP signal of H3K23 acetylation (H3K23Ac) and H3K27 acetylation (H3K27Ac) approximately three- and sevenfold, respectively (Fig. S3, A and B). Because the level of H3K27Ac exhibited a distinct increment upon Hh activation, we chose this site for in vivo verification. Both knockdown of Rpd3 (Fig. S3, C and D) and knockout of Atro (Fig. S3, E and E′) in wing discs increased the H3K27Ac level. To further investigate the correlation between histone acetylation and Hh activation, we used trichostatin A (TSA; Yoshida et al., 1995), which inhibits a wide range of histone deacetylases. TSA treatment accumulated H3K27Ac and up-regulated Hh reporter in S2 cells (Fig. S3, F–G′) and dpp-lacZ staining in wing discs (Fig. S3, H–I′). When flies were fed 40 µM TSA from the fertilized egg stage, the fused phenotype of c765-SmoPKA12 (Fig. S3 K, arrow shows the fusion of veins L3 and L4) was partially rescued (compare arrows in Fig. S3, K and M), suggesting that Hh activity increased upon TSA treatment. Furthermore, zebrafish embryos cultured at different TSA concentrations showed a graded up-regulation of Hh-responsive genes (Fig. S3 N) and H3K27Ac levels (Fig. S3, O and O’). Collectively, these data suggest an evolutionarily conserved association between histone acetylation, Atro–Rpd3 complex, and Hh signaling activity. In addition to Atro–Rpd3, other proteins may modify histone H3 or other histones in Hh activation, which involves both derepression of CiR and activation of CiA.
Activation of Gli proteins is related to the formation of various cancers (Hui and Angers, 2011), suggesting a critical role of Gli repression. In this study, we demonstrated how Ci/Gli family proteins repress target gene expression. An Atro C-terminal fragment was identified in a yeast two-hybrid screening with the N-terminal region of Ci as bait. Several assays verified their interaction both in S2 cells and in vitro. Atro repressed dpp expression in D. melanogaster and down-regulated Hh-responsive genes in zebrafish. Furthermore, our findings illustrate that Atro associates with Rpd3 to deacetylate histone during Hh target gene expression repression. Studies in mammalian systems suggested that a Ski corepressor complex regulates both full-length Gli3 and Gli3 repressor (Dai et al., 2002); testing how the Atro–Rpd3 and Ski complexes coordinate with each other in repressing Hh signaling or whether these two corepressors function at different developmental stages or in different tissues would be interesting.
Materials and methods
Fly mutants and transgenes
Atro35 (a gift from S. Kerridge, Université de la Méditerranée, Aix-en-Provence, France; Erkner et al., 2002) is a null allele. MS1096 Gal4 drives transgenes expression in the wing pouch, with a higher level in the dorsal half compared with the ventral half. UAS-Dicer2 was used to increase RNAi efficiency (Flybase). act>CD2>Gal4 UAS-GFP (AG4-GFP) induces random Gal4 expression by heat shock–induced “jump out” of CD2, so that Gal4-expressing clones were marked as CD2 minus and GFP positive region (Flybase). C765-SmoPKA12 is a dominant-negative form of Smo induced by a wing Gal4, C765-Gal4. It is a widely used tool to screen for genes that might affect Hh signaling activity (Jia et al., 2004). dpp-lacZ and ptc-lacZ have been described previously (Zhao et al., 2007; Zhang et al., 2013b). Ci RNAi (#2125R-1; Zhang et al., 2013a), Rpd3 RNAi (#46930 and #30599), and UAS-Rpd3-V5 (#32241) were obtained from the National Institute of Genetics, Vienna Drosophila RNAi Center, and Bloomington Drosophila Stock Center, respectively. We obtained full-length hs-Atro plasmid (Zhang et al., 2002) from T. Xu (Yale University, New Haven, CT). pUAST vector contains upstream activating sequence to induce gene expression by Gal4, P-element to make transgenic fly, “mini-white” marker to identify transgene by red eye, and “ampicillin-resistant” element for plasmid amplification. To construct UAS-Atro-RNAi (two constructs named R3 and R4), cDNA fragments with the coding sequence for Atro nucleotide 259–509 (R3) or 4902–5101 (R4) were cloned into pUAST with the corresponding genomic DNA fragment inserted into a reverse orientation, respectively. To construct UAS-Myc-Atro, full-length Atro was cloned into pUAST-Myc vector. All transgenic flies were generated by standard P-element–mediated transformation. Multiple independent transgenic lines were tested for activity. To achieve a better knockdown efficiency of Atro RNAi, lines R3 and R4 were recombined for all the experiment. Genotypes for generating clones are as follows: y hsflp/yw or Y; dpp-lacZ; UAS-GFP FRT2A/Atro35 FRT2A and y hsflp tub-Gal4 UAS-GFP/yw or Y; UAS-Rpd3-V5/dpp-lacZ Gal80 FRT2A/Atro35 FRT2A.
Immunostaining of imaginal discs
Third-instar larvae were dissected for immunostaining of imaginal discs with standard protocol as previously described (Zhang et al., 2013b). In brief, samples were fixed in 4% formaldehyde in PBS buffer, washed and incubated with primary and secondary antibody in PBTA (PBS, 0.1% Triton X-100, and 1% BSA) buffer, and mounted in 40% glycerol. For experiments involving comparisons between different genotypes, all samples were stained (e.g., dilute the antibody and aliquot together to add into different groups), processed (e.g., do the dissection at the same time), and imaged (e.g., scan the confocal pictures at the same time) in parallel to reduce the variables. Images of imaginal discs were acquired under a confocal microscope (LAS SP5; Leica) using a 40×/1.25 NA oil objective (Leica) or 63×/1.4 NA oil objective (Leica) at room temperature. Images shown were processed in Adobe Photoshop to convert to grayscale and to enhance visibility by adjusting brightness and contrast and pseudocoloring where appropriate. Primary antibodies used in this study were as follows: mouse anti-V5 (Invitrogen), anti-H3K27Ac (EMD Millipore), rabbit anti-Rpd3 (a gift from J.T. Kadonaga, University of California, San Diego, La Jolla, CA), anti–β-galactosidase (Cappel), and rat anti-Ci (Developmental Studies Hybridoma Bank). Rabbit anti-Atro antibody was generated in rabbit with 32–226 aa of Atro as antigen (ABclonal Technology).
Cell culture, transfection, immunostaining, luciferase reporter assay, immunoprecipitation, and Western blot analysis
S2 cells were cultured at 25°C in Schneider’s Drosophila Medium (Invitrogen) with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 µg/ml of streptomycin. Cell transfection was performed using the Calcium Phosphate Transfection kit (Specialty Media) according to the manufacturer’s instructions (Zhang et al., 2011). Cell immunostaining was performed as previously described (Yang et al., 2013). Cells were washed with PBS and fixed in freshly made 4% formaldehyde in PBS buffer at room temperature for 10 min, then washed again with PBS and treated with PBST (PBS and 0.25% Triton X-100) for permeabilization, and blocked with PBSA (PBS and 1% BSA) for 15 min. Cells were incubated with primary antibody diluted in PBSA for 1 h at room temperature, then washed with PBS and incubated with secondary antibody diluted in PBSA for 30 min at room temperature, and finally mounted in 40% glycerol. Images of cells were acquired under a confocal microscope (LAS SP5) using a 63×/1.4 NA oil objective (Leica) at room temperature. Images shown were processed in Adobe Photoshop to convert to grayscale and to enhance visibility by adjusting brightness and contrast and pseudocoloring where appropriate. Hh reporter activity was performed as previously described (Shi et al., 2013). In brief, Hh reporter firefly expression plasmid and constitutive Renilla expression plasmid were transfected equally in all experimental groups. Cells were harvested 48 h after transfection and lysed in passive lysis buffer (Promega) for Dual-luciferase assay according to the manufacturer’s instructions. Standard or two-step immunoprecipitation and Western blot were performed according to previously described methods (Zhang et al., 2013b). In brief, immunoprecipitation was performed at 4°C. Cells were lysed in standard NP-40 buffer; clarified lysates were first incubated with 2 µg of the indicated antibodies for 2 h and then with 20 µl protein A/G PLUS agarose (Santa Cruz Biotechnology, Inc.) for 1 h. Beads were washed three times and boiled in SDS loading buffer. Primary antibodies used in this study were mouse anti-HA, anti-Myc, anti-Flag (Sigma-Aldrich), anti-T7 (EMD Millipore), anti-H3K27Ac (EMD Millipore), rabbit anti-Myc (Sigma-Aldrich), and anti-H3 (Abcam).
Yeast two-hybrid assay and GST fusion protein pull-down assay
Matchmaker GAL4 Two-Hybrid System (Takara Bio Inc.) was used to screen for Ci interacting proteins and to verify the interactions according to the manufacturer’s instructions. N-terminal 1–440 aa of Ci were used as bait and D. melanogaster embryonic cDNA library (provided by Dahua Chen, Institute of Zoology, Chinese Academy of Sciences, Beijing, China) was used as prey. GST fusion proteins were produced in Escherichia coli BL21 and purified with glutathione agarose beads (GE Healthcare). GST fusion protein-loaded beads were incubated with S2 cell lysates expressing indicated proteins in GST pull-down lysis buffer (20 mM Tris-Cl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% NP-40, and PMSF) at 4°C for 1 h. The beads were washed three times with lysis buffer, followed by Western blot analysis.
In situ hybridization and immunostaining of zebrafish embryos
Zebrafish (Danio rerio) embryos were obtained by natural spawning of adult AB strain zebrafish. Embryos were maintained at 28.5°C on a 14-h light/10-h dark cycle. Whole mount in situ hybridization of zebrafish embryos was performed according to standard protocols (Thisse and Thisse, 2008). To avoid variability of in situ hybridization to the utmost extent, we injected the MO, collected the fish, performed the in situ, and took the picture at the same time under the same conditions. The RNA probe used was labeled with digoxigenin. The hhip probe encompassed the 1,193–2,112-bp regions (start from ATG) of mRNA. Other probes used were fkd4 (Tay et al., 2005) and ptch2 (also known as ptc1; Concordet et al., 1996). Immunostaining of zebrafish embryos was performed as previously described (Wang et al., 2013). In brief, zebrafish embryos were fixed in formaldehyde and stored in methanol at −20°C overnight. On the second day, embryos were washed out of methanol by serial washes in ethanol/PBStw (PBS + 0.1% Tween-20) and blocked in PBStwA (PBStw + 1%BSA). Embryos were incubated with primary antibody diluted in PBStwA at 4°C overnight and washed with PBStw, and then incubated with secondary antibody diluted in PBStwA for 2 h at room temperature. Images of zebrafish embryos were acquired under a confocal microscope (LAS SP5) using a 20×/0.7 NA objective (Leica) at room temperature. Images shown were processed in Adobe Photoshop to convert to grayscale and to enhance visibility by adjusting brightness and contrast and pseudocoloring where appropriate. Primary antibodies used in this study were mouse anti-Eng (Developmental Studies Hybridoma Bank).
MO knockdown
Antisense MOs (Gene Tools) were microinjected into one to four cell stage embryos according to standard protocols. A 4-nl volume of MOs was injected at the concentration of 0.15 mM for all experiments unless indicated. MO sequences used were rerea-MO (5′-TCCTTGGAGGCTGTAAACACAAATT-3′; Asai et al., 2006), smo-MO (5′-CGCTTGGAGGACATCTTGGAGACGC-3′; Robu et al., 2007), ptch2-MO (5′-CATAGTCCAAACGGGAGGCAGAAGA-3′; Wolff et al., 2003), and standard control-MO (Gene Tools).
RNA isolation, reverse transcription, and real-time PCR
About 20 zebrafish embryos at indicated stages were lysed in TRIzol (Invitrogen) for RNA isolation following standard protocol. 1 µg RNA was used for reverse transcription by Maxima First Strand cDNA Synthesis kit (Thermo Fisher Scientific). Real-time PCR was performed on ABI Fast7500 with Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific). 2-ΔΔCt method was used for relative quantification. The primer pairs used were as follows: fkd4, 5′-GCTTCACTGAACCATTTCGCA-3′ (forward) and 5′-CTGAGCCATAATACATCTCGCTG-3′ (reverse); hhip, 5′-CTTACGAGCCAAGTGTGAACTG-3′ (forward) and 5′-TGCTGTCTTTCTCACCGTCC-3′ (reverse); ptch2, 5′-TCCTCCTTATGAGTCCCAAACAG-3′ (forward) and 5′-CATGAACAACCTCAACAAACTTCC-3′ (reverse); gapdh, 5′-CATCACAGCAACACAGAAGACC-3′ (forward) and 5′-ACCAGTAAGCTTGCCATTGAG-3′ (reverse); nkx2.1b, 5′-CCCACGCTTCTCTACAATCTCT-3′ (forward) and 5′-ACCCATGCCTTTACCAACATC-3′ (reverse); foxa2, 5′-AATACAGGCATTTCTCGTGTGG-3′ (forward) and 5′-AGGTGTAACACTCAGGCTCTC-3′ (reverse); gli1, 5′-TTCTTGGTTTACTTGAAGGCAGAG-3′ (forward) and 5′-GCTCATTATTGATGTGATGCACC-3′ (reverse).
ChIP assay
D. melanogaster wing disc–derived Cl.8 cell was used for ChIP assay with endogenous antibodies. Cells were fixed in freshly made 1% formaldehyde in PBS buffer at room temperature for 15 min, added with 2.5 M glycine to a final concentration of 125 mM for 5 min incubation, and washed with cold PBS. Cells were lysed in ChIP lysis buffer (50 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and cocktail) and sonicated into small chromatin fragments. After centrifugation, lysates were incubated with 2 µg of the indicated antibodies for 5 h (or overnight) at 4°C. Samples were combined with 20 µl protein A/G PLUS agarose (Santa Cruz Biotechnology, Inc.) and incubated for 5 h (or overnight) on a rotator at 4°C. Beads were washed three times with ChIP wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 150 mM NaCl, and 20 mM Tris-Cl, pH 8.0) and finally washed with ChIP final wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 500 mM NaCl, and 20 mM Tris-Cl, pH 8.0). Beads were incubated with ChIP elution buffer (1% SDS and 100 mM NaHCO3) at 65°C for 30 min, and then added with 4 M NaCl to a final concentration of 200 mM for further incubation at 65°C for 4 h. After this, beads were mixed with 5 mM EDTA and 0.25 mg/ml proteinase K for incubation at 55°C for 2 h. The DNA on the beads was purified with DNA purification kit (QIAGEN) and sent for real-time PCR assay. Antibodies used in this study are mouse anti-H3K27Ac (EMD Millipore) and anti-H3K9Ac (Abcam) and rabbit anti-H3K14Ac, anti-H3K18Ac, anti-H3K23Ac (provided by Degui Chen, Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China), anti-H3 (Abcam), and anti-Atro (generated in this study). The primer pairs used were as follows: dpp (around the transcriptional start site), 5′-ACTTAAGCAACTTGCCATCGT-3′ (forward) and 5′-AGCAACATCAAAGCAAAGCAC-3′ (reverse); dpp-a, 5′-GTGTGCGCTGTCTTTATAAAGG-3′ (forward) and 5′-TACCTACACCTAACATTCTTGCAG-3′ (reverse); dpp-b, 5′-AATCACATTTCGAGAAGAGACGAC-3′ (forward) and 5′-GGACCCGGTTGGTTAATCAG-3′ (reverse); dpp-c, 5′-GTCCACACCGTTCATCACCA-3′ (forward) and 5′-GGCTACCCATCCACATCGTC-3′ (reverse).
Online supplemental material
Fig. S1 shows the verification of Atro RNAi and Atro35 mutant, the effect of Atro to ptc expression in D. melanogaster, and that Rerea represses additional Hh-responsive genes in zebrafish. Fig. S2 shows the interaction between Atro and Rpd3, the verification of Rpd3 RNAi, and the repression of Rpd3 to dpp expression, which is dependent on Ci. Fig. S3 shows that histone acetylation correlates well with Atro–Rpd3 and Hh signaling activity. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306012/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Dahua Chen, Tian Xu, Stephen Kerridge, Degui Chen, Guoliang Xu, James T. Kadonaga, Thomas B. Kornberg, Jing Jiang, Fengwei Yu, and Steve Wilson and the Developmental Studies Hybridoma Bank, Vienna Drosophila RNAi Center, National Institute of Genetics, and Bloomington Drosophila Stock Center for fly stocks and reagents. We thank Dr. Jinqiu Zhou for useful discussions on the manuscript.
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010405, XDA01010406, and XDA01010110) and also by grants from the National Basic Research Program of China (2010CB912101, 2011CB943900, 2012CB945001, and 2010CB945300), from National Natural Science Foundation of China (31171414, 31171394, and 31371492), and from the Science and Technology Commission of Shanghai Municipality (11140900100). L. Zhang is the scholar of the Hundred Talents Program of the Chinese Academy of Sciences.
Footnotes
Abbreviations used in this paper:
- A/P
- anterior/posterior
- Atro
- Atrophin
- AtroC
- C terminus of Atro
- ChIP
- chromatin immunoprecipitation
- Ci
- Cubitus interruptus
- CiN
- N terminus of Ci
- coIP
- coimmunoprecipitation
- dpp
- decapentaplegic
- Eng
- Engrailed
- H3K23Ac
- H3K23 acetylation
- Hh
- Hedgehog
- hpf
- hours postfertilization
- MO
- Morpholino
- Ptc
- Patched
- Smo
- Smoothened
- TSA
- trichostatin A
References
- Akimaru H., Chen Y., Dai P., Hou D.X., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S. 1997. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 386:735–738 10.1038/386735a0 [DOI] [PubMed] [Google Scholar]
- Asai Y., Chan D.K., Starr C.J., Kappler J.A., Kollmar R., Hudspeth A.J. 2006. Mutation of the atrophin2 gene in the zebrafish disrupts signaling by fibroblast growth factor during development of the inner ear. Proc. Natl. Acad. Sci. USA. 103:9069–9074 10.1073/pnas.0603453103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramírez-Weber F.A., Laget M.P., Schwartz C., Kornberg T.B. 1997. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 89:1043–1053 10.1016/S0092-8674(00)80292-5 [DOI] [PubMed] [Google Scholar]
- Charroux B., Freeman M., Kerridge S., Baonza A. 2006. Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Dev. Biol. 291:278–290 10.1016/j.ydbio.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Chen J.K., Taipale J., Cooper M.K., Beachy P.A. 2002. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16:2743–2748 10.1101/gad.1025302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.H., Wilson C.W., Chuang P.T. 2007. SnapShot: hedgehog signaling pathway. Cell. 130:386 10.1016/j.cell.2007.07.017 [DOI] [PubMed] [Google Scholar]
- Concordet J.P., Lewis K.E., Moore J.W., Goodrich L.V., Johnson R.L., Scott M.P., Ingham P.W. 1996. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 122:2835–2846 [DOI] [PubMed] [Google Scholar]
- Dai P., Shinagawa T., Nomura T., Harada J., Kaul S.C., Wadhwa R., Khan M.M., Akimaru H., Sasaki H., Colmenares C., Ishii S. 2002. Ski is involved in transcriptional regulation by the repressor and full-length forms of Gli3. Genes Dev. 16:2843–2848 10.1101/gad.1017302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkner A., Roure A., Charroux B., Delaage M., Holway N., Coré N., Vola C., Angelats C., Pagès F., Fasano L., Kerridge S. 2002. Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development. 129:1119–1129 [DOI] [PubMed] [Google Scholar]
- Fanto M., Clayton L., Meredith J., Hardiman K., Charroux B., Kerridge S., McNeill H. 2003. The tumor-suppressor and cell adhesion molecule Fat controls planar polarity via physical interactions with Atrophin, a transcriptional co-repressor. Development. 130:763–774 10.1242/dev.00304 [DOI] [PubMed] [Google Scholar]
- Foglietti C., Filocamo G., Cundari E., De Rinaldis E., Lahm A., Cortese R., Steinkühler C. 2006. Dissecting the biological functions of Drosophila histone deacetylases by RNA interference and transcriptional profiling. J. Biol. Chem. 281:17968–17976 10.1074/jbc.M511945200 [DOI] [PubMed] [Google Scholar]
- Haecker A., Qi D., Lilja T., Moussian B., Andrioli L.P., Luschnig S., Mannervik M. 2007. Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos. PLoS Biol. 5:e145 10.1371/journal.pbio.0050145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepker J., Wang Q.T., Motzny C.K., Holmgren R., Orenic T.V. 1997. Drosophila cubitus interruptus forms a negative feedback loop with patched and regulates expression of Hedgehog target genes. Development. 124:549–558 [DOI] [PubMed] [Google Scholar]
- Hooper J.E., Scott M.P. 2005. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6:306–317 10.1038/nrm1622 [DOI] [PubMed] [Google Scholar]
- Hui C.C., Angers S. 2011. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27:513–537 10.1146/annurev-cellbio-092910-154048 [DOI] [PubMed] [Google Scholar]
- Ingham P.W., Nakano Y., Seger C. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12:393–406 10.1038/nrg2984 [DOI] [PubMed] [Google Scholar]
- Jia J., Tong C., Wang B., Luo L., Jiang J. 2004. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 432:1045–1050 10.1038/nature03179 [DOI] [PubMed] [Google Scholar]
- Jiang J., Hui C.C. 2008. Hedgehog signaling in development and cancer. Dev. Cell. 15:801–812 10.1016/j.devcel.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstrom R.O., Tyurina O.V., Kawakami A., Nishioka N., Talbot W.S., Sasaki H., Schier A.F. 2003. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development. 130:1549–1564 10.1242/dev.00364 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 96:819–831 10.1016/S0092-8674(00)80592-9 [DOI] [PubMed] [Google Scholar]
- Müller B., Basler K. 2000. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic gli-binding sites. Development. 127:2999–3007 [DOI] [PubMed] [Google Scholar]
- Ochi H., Pearson B.J., Chuang P.T., Hammerschmidt M., Westerfield M. 2006. Hhip regulates zebrafish muscle development by both sequestering Hedgehog and modulating localization of Smoothened. Dev. Biol. 297:127–140 10.1016/j.ydbio.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. 2007. p53 activation by knockdown technologies. PLoS Genet. 3:e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr K.B., Barth K.A., Varga Z.M., Wilson S.W. 2001. The nodal pathway acts upstream of hedgehog signaling to specify ventral telencephalic identity. Neuron. 29:341–351 10.1016/S0896-6273(01)00210-0 [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100 10.1146/annurev.biochem.76.052705.162114 [DOI] [PubMed] [Google Scholar]
- Shi D., Lv X., Zhang Z., Yang X., Zhou Z., Zhang L., Zhao Y. 2013. Smoothened oligomerization/higher order clustering in lipid rafts is essential for high Hedgehog activity transduction. J. Biol. Chem. 288:12605–12614 10.1074/jbc.M112.399477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston R.D., Hoffmann F.M., Blackman R.K., Segal D., Grimaila R., Padgett R.W., Irick H.A., Gelbart W.M. 1990. Molecular organization of the decapentaplegic gene in Drosophila melanogaster. Genes Dev. 4:1114–1127 10.1101/gad.4.7.1114 [DOI] [PubMed] [Google Scholar]
- Tay S.Y., Ingham P.W., Roy S. 2005. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 132:625–634 10.1242/dev.01606 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3:59–69 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Tyurina O.V., Guner B., Popova E., Feng J., Schier A.F., Kohtz J.D., Karlstrom R.O. 2005. Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Dev. Biol. 277:537–556 10.1016/j.ydbio.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Wang L., Rajan H., Pitman J.L., McKeown M., Tsai C.C. 2006. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 20:525–530 10.1101/gad.1393506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Charroux B., Kerridge S., Tsai C.C. 2008. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Rep. 9:555–562 10.1038/embor.2008.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu T., Xu L., Gao Y., Wei Y., Duan C., Chen G.Q., Lin S., Patient R., Zhang B., et al. 2013. Fev regulates hematopoietic stem cell development via ERK signaling. Blood. 122:367–375 10.1182/blood-2012-10-462655 [DOI] [PubMed] [Google Scholar]
- Wolff C., Roy S., Ingham P.W. 2003. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13:1169–1181 10.1016/S0960-9822(03)00461-5 [DOI] [PubMed] [Google Scholar]
- Yang X., Mao F., Lv X., Zhang Z., Fu L., Lu Y., Wu W., Zhou Z., Zhang L., Zhao Y. 2013. Drosophila Vps36 regulates Smo trafficking in Hedgehog signaling. J. Cell Sci. 126:4230–4238 10.1242/jcs.128603 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Horinouchi S., Beppu T. 1995. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 17:423–430 10.1002/bies.950170510 [DOI] [PubMed] [Google Scholar]
- Zhang S., Xu L., Lee J., Xu T. 2002. Drosophila atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell. 108:45–56 10.1016/S0092-8674(01)00630-4 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mao F., Lu Y., Wu W., Zhang L., Zhao Y. 2011. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 21:1436–1451 10.1038/cr.2011.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lv X., Jiang J., Zhang L., Zhao Y. 2013a. Dual roles of Hh signaling in the regulation of somatic stem cell self-renewal and germline stem cell maintenance in Drosophila testis. Cell Res. 23:573–576 10.1038/cr.2013.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lv X., Yin W.C., Zhang X., Feng J., Wu W., Hui C.C., Zhang L., Zhao Y. 2013b. Ter94 ATPase complex targets k11-linked ubiquitinated ci to proteasomes for partial degradation. Dev. Cell. 25:636–644 10.1016/j.devcel.2013.05.006 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tong C., Jiang J. 2007. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 450:252–258 10.1038/nature06225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.