Abstract
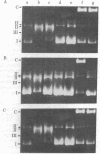
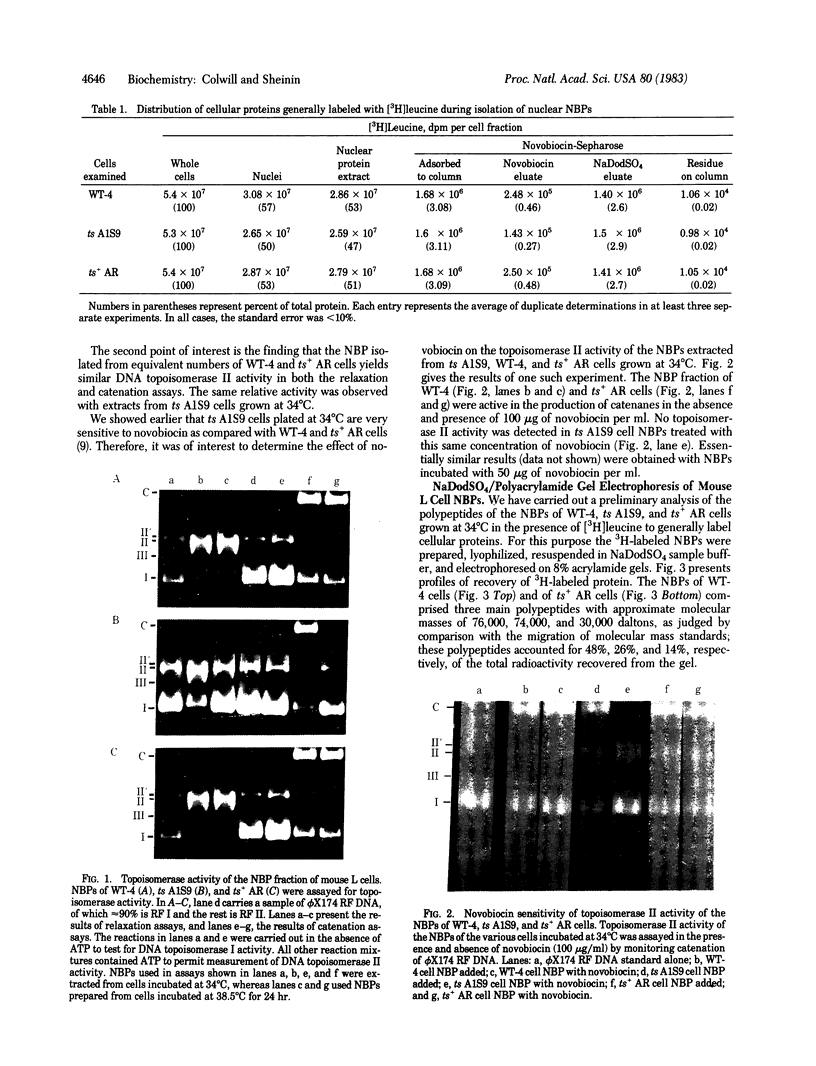
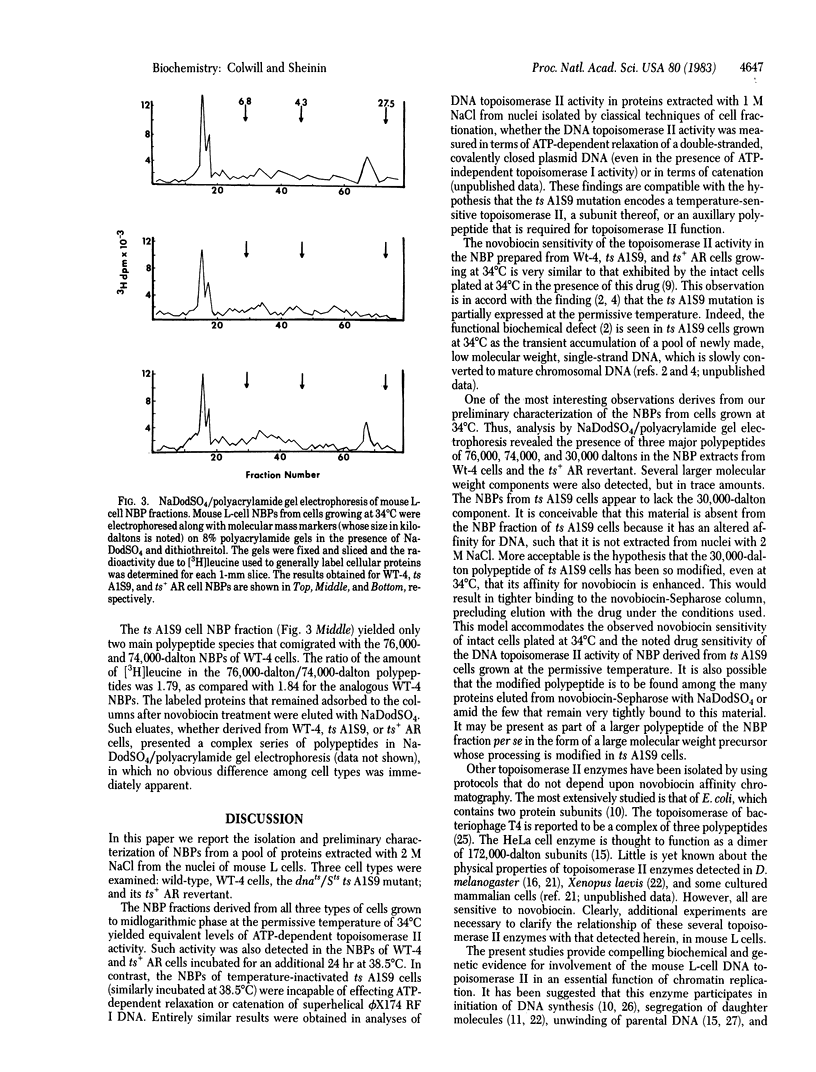
Nuclear novobiocin binding proteins (NBPs) from a set of mouse L cells have been extensively purified by affinity chromatography on novobiocin-Sepharose columns. The NBPs, specifically eluted with 100 micrograms of novobiocin per ml, exhibited equivalent DNA topoisomerase activities (measured as ATP-dependent relaxation or catenation of phi X174 replicative-form I DNA substrate) when extracted from equal numbers of wild-type (WT-4) mouse L cells growing logarithmically at 34 degrees C or at 38.5 degrees C, from ts A1S9 cells similarly cultivated at the low, permissive temperature or from revertant ts+ AR cells in exponential growth at either temperature. The NBPs isolated from similar numbers of ts A1S9 cells grown to midlogarithmic phase and then incubated for 24 hr at 38.5 degrees C (the nonpermissive temperature) showed no topoisomerase II activity. Preliminary NaDodSO4/polyacrylamide gel electrophoretic analysis of enzymatically active material revealed that the NBPs of WT-4 and ts+ AR cells grown at 34 degrees C comprised three major polypeptides of 76,000, 74,000, and 30,000 daltons and a number of larger molecular mass components present in trace amounts. The NBP of ts A1S9 cells grown at the permissive temperature was similar, except that the 30,000-kilodalton polypeptide was not detected. Such enzymatically active NBPs from WT-4 and ts+ AR cells were unaffected by 100 micrograms of novobiocin per ml, whereas the analogous preparation from ts A1S9 cells was totally inhibited. On the basis of these and other considerations, it is postulated that the ts A1S9 locus of mouse L cells encodes a temperature-sensitive polypeptide that is required for normal DNA topoisomerase II activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldi M. I., Benedetti P., Mattoccia E., Tocchini-Valentini G. P. In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase. Cell. 1980 Jun;20(2):461–467. doi: 10.1016/0092-8674(80)90632-7. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Proteins that affect DNA conformation. Annu Rev Biochem. 1978;47:449–479. doi: 10.1146/annurev.bi.47.070178.002313. [DOI] [PubMed] [Google Scholar]
- DULBECCO R., VOGT M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J Exp Med. 1954 Feb;99(2):167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980 Aug;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- Humbert J., Sheinin R. Properties of an invitro system for studying temperature-sensitive DNA synthesis in ts A1S9 mouse L-cells. Can J Biochem. 1978 Jun;56(6):444–451. doi: 10.1139/o78-069. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979 Oct 11;281(5731):456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- Mattern M. R., Painter R. B. Dependence of mammalian DNA replication on DNA supercoiling. II. Effects of novobiocin on DNA synthesis in Chinese hamster ovary cells. Biochim Biophys Acta. 1979 Jul 26;563(2):306–312. doi: 10.1016/0005-2787(79)90049-2. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Liu L. F., Englund P. T. A homogeneous type II DNA topoisomerase from HeLa cell nuclei. J Biol Chem. 1981 Sep 10;256(17):9334–9339. [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Ellison M. J. Purification and properties of type 1 topoisomerase from chicken erythrocytes: mechanism of eukaryotic topoisomerase action. Biochemistry. 1982 Mar 16;21(6):1155–1161. doi: 10.1021/bi00535a008. [DOI] [PubMed] [Google Scholar]
- Setterfield G., Sheinin R., Dardick I., Kiss G., Dubsky M. Structure of interphase nuclei in relation to the cell cycle. Chromatin organization in mouse L cells temperature-sensitive for DNA replication. J Cell Biol. 1978 Apr;77(1):246–263. doi: 10.1083/jcb.77.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin Polyoma and cell DNA synthesis in mouse L cells temperature sensitive for the replication of cell DNA. J Virol. 1976 Mar;17(3):692–704. doi: 10.1128/jvi.17.3.692-704.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin R., Darragh P., Dubsky M. Mitochondrial DNA synthesis in mouse L cells temperature sensitive in nuclear DNA replication. Can J Biochem. 1977 May;55(5):543–547. doi: 10.1139/o77-077. [DOI] [PubMed] [Google Scholar]
- Sheinin R. Preliminary characterization of the temperature-sensitive defect in DNA replication in a mutant mouse L cell. Cell. 1976 Jan;7(1):49–57. doi: 10.1016/0092-8674(76)90254-3. [DOI] [PubMed] [Google Scholar]
- Shishido K. The use of Haemophilus gallinarum DNA-relaxing enzyme to investigate the relationship between the number of superhelical turns and the molecular weight in a negatively twisted DNA. J Biochem. 1979 Sep;86(3):711–717. doi: 10.1093/oxfordjournals.jbchem.a132575. [DOI] [PubMed] [Google Scholar]
- Sparkuhl J., Sheinin R. Protein synthesis and degradation during expression of the temperature-sensitive defect in ts A1S9 mouse L-cells. J Cell Physiol. 1980 Nov;105(2):247–258. doi: 10.1002/jcp.1041050208. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L. H., Mankovitz R., Baker R. M., Till J. E., Siminovitch L., Whitmore G. F. Isolation of temperature-sensitive mutants of L-cells. Proc Natl Acad Sci U S A. 1970 Jun;66(2):377–384. doi: 10.1073/pnas.66.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]