Abstract
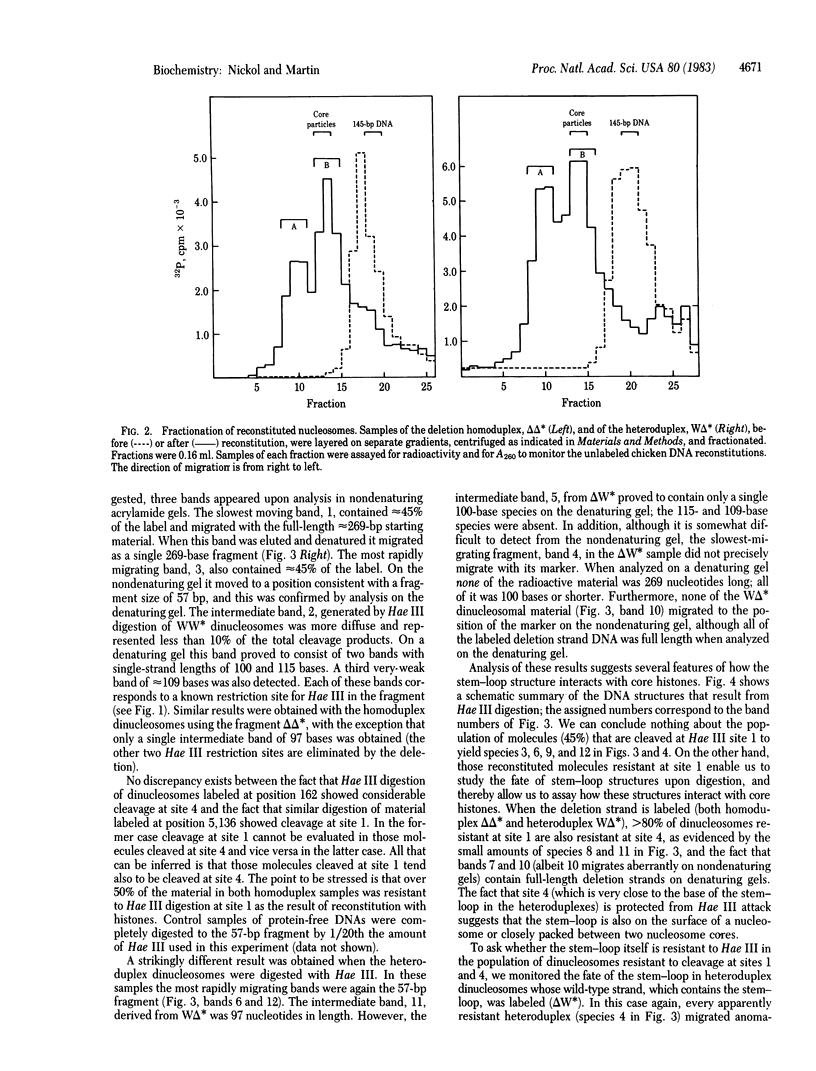
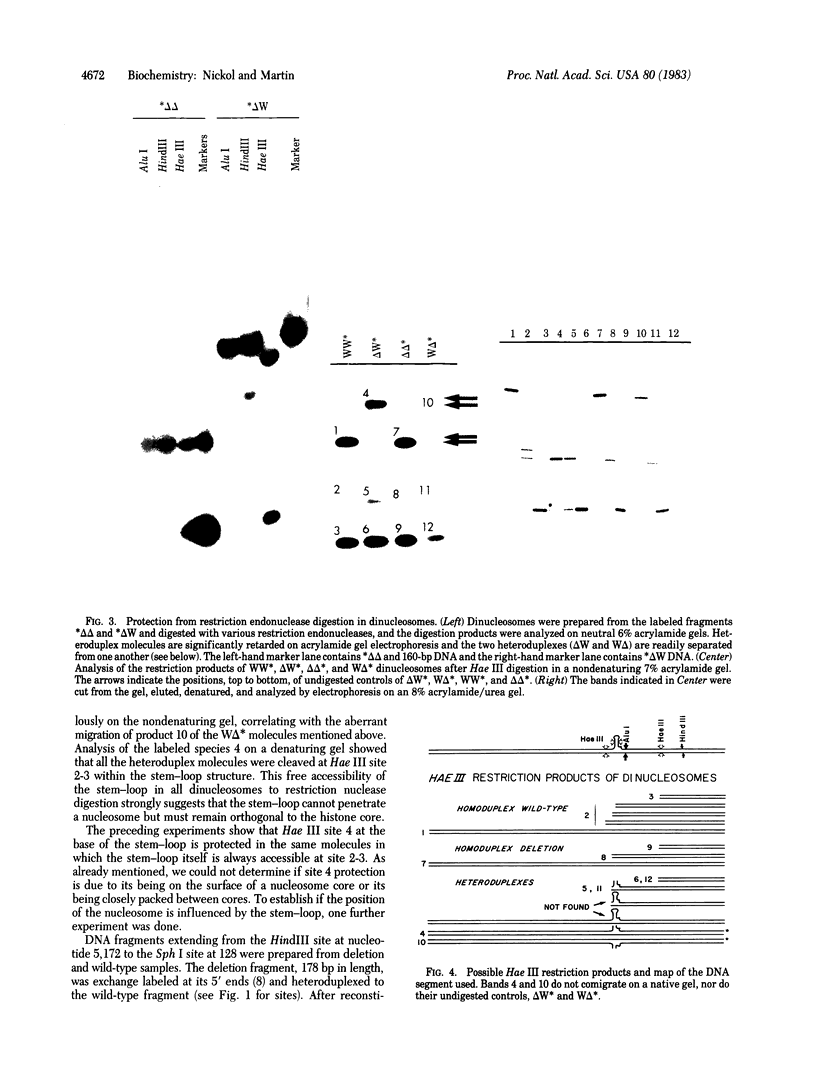
Heteroduplex DNA molecules were generated in which one of the two strands contained a 7-base-pair (double-stranded) stem and 3-base-pair (single-stranded) loop. The heteroduplexes and their corresponding homoduplex parental molecules, each of approximately 260 base pairs, were used for nucleosomes reconstitution. Protection from restriction endonuclease digestion was used to probe the structure of the resulting dinucleosomes. Although 50% or more of the potential cleavage sites in the homoduplex DNAs and in the linear portion of the heteroduplex DNAs were inaccessible to nuclease digestion, no protection of the stem-loop structures was observed. The results imply that a stem-loop structure preferentially occupies the spacer region between nucleosomes, but if it is part of a core particle the stem-loop is always pointed "outward" in such a way as to be accessible to nuclease digestion.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L., Eder G., Lurz R. In vitro core particle and nucleosome assembly at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3284–3288. doi: 10.1073/pnas.76.7.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. H., Camerini-Otero R. D., Felsenfeld G. An octamer of histones H3 and H4 forms a compact complex with DNA of nucleosome size. Nucleic Acids Res. 1978 Dec;5(12):4805–4818. doi: 10.1093/nar/5.12.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]






