Abstract
With the emerging interest in personalized medicine, there is strong demand for new technologies for clinical sample interrogation. Exfoliated tumor cells in variety of pathological samples (e.g., blood, bone marrow, urine) could provide invaluable information for diagnosis and prognosis of cancers. Here we describe a detailed method for capture and isolation of tumor cells in medium, blood, or large-volume buffy coat using EpCAM-targeted buoyant microbubbles (MBs). Perflorohexane gas lipid shell MBs were prepared with emulsification method and conjugated with antibody as described by us before (Shi et al,. PLoS One, 2013). The binding of EpCAM-targeted MBs to A549 (human lung carcinoma) and 4T1 (mouse breast carcinoma) cells spiked into BSA/PBS or blood was more than 90%, which was comparable with commercial anti-EpCAM immunomagnetic beads (DynaBeads). Anti-EpCAM MBs efficiently (75–82%) isolated BxPC3 pancreatic tumor cells spiked into medium, blood or a buffy coat, within 15–30 min of incubation. We discuss MB parameters and experimental conditions critical to achieve efficient cells binding and isolation. In conclusion, MB-assisted cell isolation is a promising method for rapid enrichment of cells and biomarkers from biological samples.
1. INTRODUCTION
1.1. Tumor cell isolation methods
A key component of cancer progression is the shedding of malignant circulating tumor cells (CTCs) into blood (1–3). Isolation and analysis of CTCs could provide invaluable information for the diagnosis and prognosis of cancer patients (1, 4–8). Several isolation methods from blood are currently available. The crude enrichment (de-bulking) could be achieved by density-gradient centrifugation or Ficoll-Paque gradient (9, 10). These procedures eliminate most of the red blood cells in a sample, but lead to inevitable loss of the rare cells of interest. Another common strategy is to isolate CTCs directly from blood using immunomagnetic beads. Magnetic beads are nano- to micron-sized particles made of paramagnetic iron oxide core (i.e., become magnetized when placed into magnetic field) and are usually polymer-coated to improve solubility and biocompatibility (11). CellSearch Assay (Veridex) has recently received U.S. Food and Drug Administration clearance for detection of CTCs in metastatic breast cancer patient. In this technique, anti-epithelial cell adhesion molecule (EpCAM)-coated, micron-sized magnetic beads capture the CTCs in blood and then are trapped by an external magnetic field to wash away the unbound cells. The capturing efficiency of rare tumor cells with magnetic beads ranges between 60–90% (12, 13). The most significant limitations of the assay are its relatively long processing time, non-specific carryover and contamination with leukocytes (14–17). Recently, the field of CTC isolation witnessed a surge of technologies, including microfluidics and filtration. These state-of-the-art technologies allow to isolate, count and even to manipulate single CTCs (18–21). At the same time, there is a continuing interest in development and testing of cost-efficient, scalable and simple technologies for CTC isolation.
1.2. Microbubbles for cell isolation
Perfluorocarbon gas microbubbles (MBs) are very well described as ultrasound contrast agents (22, 23). For the lipidic MB preparation, a mixture of lipids is homogenized in the presence of gas. Perfluorocarbon gas is especially suitable due to its low solubility in water, which is necessary for maintaining the stability of MBs in the aqueous phase (22). Gas solubility and its partial pressure also determine the size of the MB (22, 24). Presence of perfluorocarbon gas makes the MBs buoyant. The use of MBs for the flotation separation of tumor cells could be an attractive alternative to immunomagnetic separation. We previously reported preparation and isolation of tumor cells using anti-EpCAM MBs (25). The principle of buoyancy separation is schematically shown in Fig. 1. Here we present a detailed method for preparation of MBs and testing the binding and separation of tumor cells in biological media.
Figure 1.
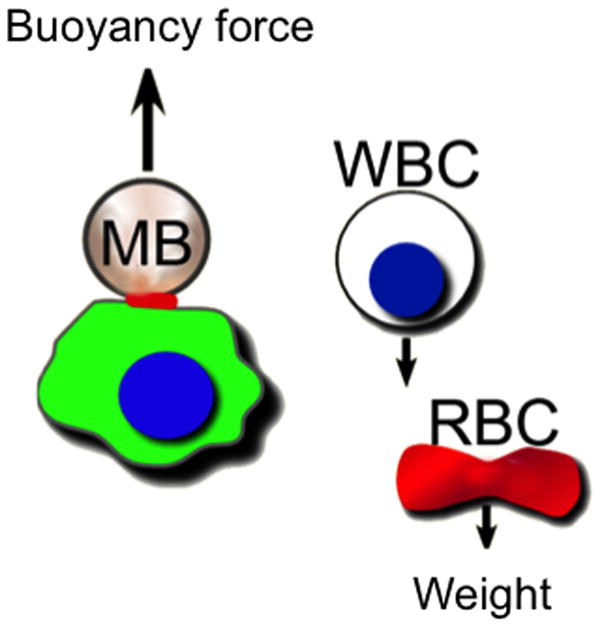
Overall concept of microbubble (MB) isolation of cancer cells, whereby attached MB drags the cell up due to buoyancy force and the blood cells (WBC=white blood cell, RBC=red blood cell) settle down due to weight (centrifugal force).
2. MICROBUBBLE PREPARATION
2.1 MB formation by emulsification
Solutions of DSPC (Avanti Polar lipids), DSPE-PEG-3400-maleimide (Laysan Bio, Inc.) and PEG40-stearate (Sigma) in chloroform are mixed at 10:1:1 mole ratio in a 2ml borosilicate glass vial (100–300 nmoles total lipid) and dried under argon stream.
The film is rehydrated in 1 ml phosphate-buffered saline (PBS, pH 7.4) at room temperature for 5 minutes.
The dispersion is sonicated under perflorohexane gas (Alfa Aesar) atmosphere by sonication (30-second cycle, 3–5 cycles total) using a MISONIX XL-2000 probe sonicator at power setting “1.”
Excess phospholipid membrane fragments and small MBs are removed by gentle centrifugation at 50g for 1 minute, repeated three times, and MBs are resuspended in 1ml PBS.
Following preparation, MBs are counted and sized using Countess™ cell counter (Invitrogen). Usually, the preparation results in MB concentration of 1×109/ml.
2.2. Conjugation of targeting antibodies
AffiniPure goat anti-rat or rabbit anti-mouse IgG, both Fc fragment specific (Jackson ImmunoResearch) are thiolated using Traut’s reagent (2-iminothiolane, Thermo Scientific) using 40:1 molar excess over antibody for 2h and washed with Zebra desalting column (Thermo Scientific).
Thiolated IgG is added to maleimide MBs at 40μg/ml and incubated with mixing at 10rpm for 1h.
MBs are washed 4 times and incubated with mouse anti-human CD326 (EpCAM) antibody (Bio Legend) or rat anti-mouse CD326 antibody (Bio Legend), 5μg total for 1h and then washed 3 times and resuspended in 1ml PBS. After conjugation and washing steps, MB concentration should be between 1×108/ml and 5×108/ml.
In order to verify antibody coating, the resulting MBs are stained with Alexa Fluor 488 secondary IgG (Invitrogen) for 10 min and washed 3 times.
2.3. MB characterization
MB preparation is shown schematically in Fig. 2A. As a result of washing steps, MBs should be enriched for large sized population, with average diameter between 3 and 5 μm (Fig. 2B). Microbubbles should float within a few minutes after resuspension (Fig. 2C). The MBs should be homogenously coated with anti-EpCAM antibody (Fig. 2D). At 7–10 mol% DSPE-PEG3400 maleimide, MBs have optimal coating with anti-EpCAM IgG.
Figure 2.
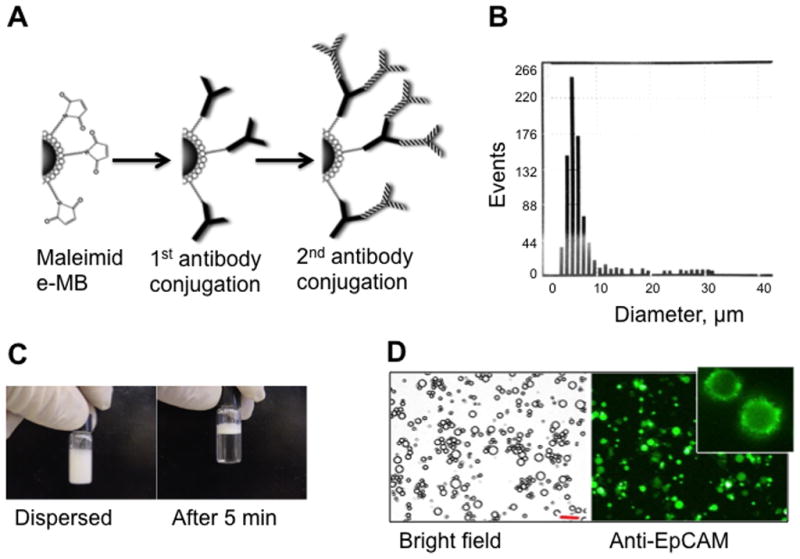
Preparation and characterization of MBs. (A) Maleimide-MBs are coated with anti-Fcspecific antibody and anti-EpCAM antibody in a two-step process. (B) Resulting MBs have a broad size distribution, as measured with Countess™ cell counter software. (C) MBs float soon after redispersion. (D) Microscope images of the resulting MBs after staining of anti-EpCAM IgG with Alexa 488-labeled secondary antibody. Inset shows cropped image of a MB with characteristic “dentate” fluorescence. Size bar 20 μm.
3. BINDING AND ISOLATION OF CELLS
3.1. MB–cell binding
In order to test the binding efficiency of anti-EpCAM MBs to tumor cells, EpCAM-positive human lung carcinoma A549 cells and mouse breast carcinoma 4T1 cells were used. The general method to test the binding efficiency of freshly prepared MBs is as follows:
Tumor cells are detached with trypsin, washed once in full medium, resuspended in buffer or cell medium at 1 × 106 cells/ml.
Human donors’ blood is diluted 1:3 with PBS and centrifuged at 1500g for 15 min. The washing step in PBS is repeated once more. The purpose of washings is to eliminate most of the plasma since MBs are less stable in whole blood.
Tumor cells are added to 1 ml of 1% BSA/PBS buffer, medium or blood at 1 × 105 cells/ml, followed by EpCAM-targeted MBs or control (first antibody only) MBs at MB/cell ratio 1:100–1600);
The cells are mixed at 10–30 rpm on a vertical rotator for 15 min;
Twenty microliters of cell suspension is placed on a slide, covered with cover glass and the binding efficiency is determined as a percentage of cells having at least one MB attached.
Following incubation with 4T1 cells, anti-EpCAM MBs formed characteristic rosettes (Fig. 3A), whereas control MBs did not show significant binding (Fig. 3B). Additional strategy to quantify binding efficiency is to prepare MBs with 0.1% DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, Biotium). Following incubation with DiI labeled MBs, cells were analyzed with flow cytometry. 4T1 cells coated with EpCAM MBs became fluorescent, while control MBs showed minimal shift in cell fluorescence (Fig. 3C). The binding efficiency of 4T1 cells was over 80% (Fig. 3D). Anti-EpCAM MBs also bound to A549 cells in 1 ml of 1% BSA in PBS or in human blood with over 90% efficiency (Fig. 4A,B). Commercial anti-EpCAM-coated magnetic beads (DynaBeads® Epithelial Enrich, Invitrogen) showed similar binding efficiency (>90%) in BSA/PBS (Fig. 4A) and blood (Fig. 4B).
Figure 3.
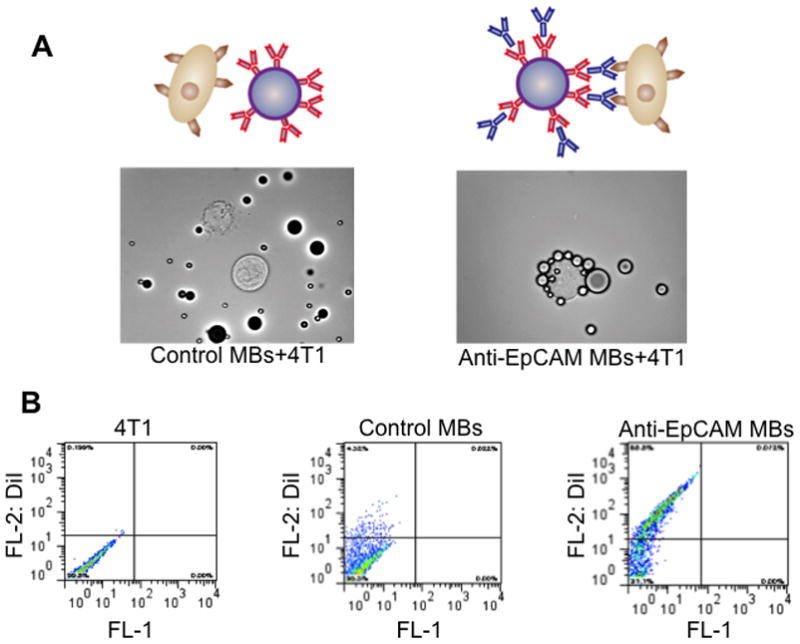
Measuring binding efficiency of MBs to 4T1 cells. (A) Anti-EpCAM MBs and control MBs were added to breast cancer 4T1 cells in 1% BSA/PBS. Targeted MBs but not control (first Ab only) MBs bind to the cells and form rosettes as observed under microscope. (B) Binding efficiency of DiI labeled anti-EpCAM MBs to 4T1 cells can be quantified with flow cytometry. Cell population becomes shifted on the dot plot, due to the binding of DiI MBs. In this experiment, 69% of the cells became coated with fluorescent anti-EpCAM MBs.
Figure 4.
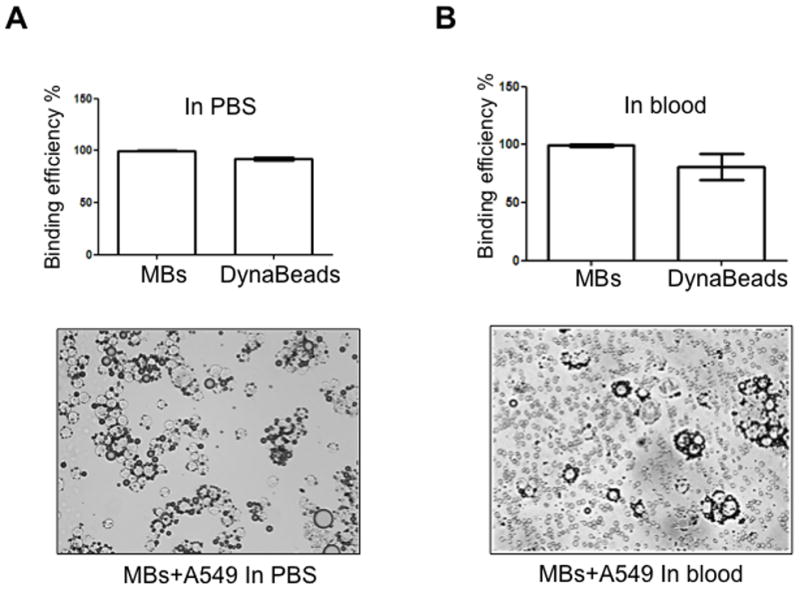
Binding of anti-human EpCAM MBs and DynaBeads to A549 lung carcinoma cells. (A) Efficiency of binding in PBS. Image shows A549 cells coated with targeted MBs. (B) Efficiency of binding in human blood. Image below shows A549 cells coated with targeted MBs among erythrocytes. MBs and DynaBeads demonstrate similar binding efficiency.
3.2. Cell isolation
3.2.1. Isolation of large numbers of tumor cells with MBs and DynaBeads
Isolation of GFP-BxPC3 pancreatic carcinoma cells by MBs and beads from PBS or heparinized donor blood was performed according to the following general protocol:
Tumor cells at 1×105/ml are added to 1 ml BSA/PBS buffer or plasma-depleted human blood (3.1) in eppendorf tube, followed by anti-EpCAM MBs (1×107) and incubated with mixing as described in 3.1. Anti-EpCAM DynaBeads are prepared according to the manufacturer’s instruction and added to cells at 1×107 beads/ml.
After 30 min incubation step, tube with MBs is inverted and centrifuged at 100g or 1,000g for 2 min. The inverted conical bottom part of the tube is cut off and 500–700 μl PBS is added to bring the level of MBs up. MB layer is carefully harvested into a new eppendorf tube for counting. DynaBeads are separated with external magnet (StemCell Technologies), washed 3 times with 1 ml of cold buffer (PBS w/0.1% BSA and 0.6% sodium citrate) and collected into a new eppendorf tube for counting.
For counting of nucleated cells, the isolated fraction is stained with 1 μg/ml Hoechst 33342 dye (Invitrogen). MBs can be briefly (1s) bath sonicated and destroyed prior to the counting. The cell recovery is determined by counting GFP-positive cells under microscope with hemocytometer.
According to Fig. 5A, MBs and magnetic beads show diametrically different principles of cell separation. As a result of a separation, MBs form a foamy layer on top of the tube, whereas magnetic beads form slurry on the tube. The GFP+/Hoechst+ tumor cells isolated with either method should be readily distinguishable under microscope from GFP−/Hoechst+ leukocytes (Fig. 5B). According to Fig. 5C, MBs efficiently isolated the cells from BSA/PBS or blood (75%), which is similar to DynaBeads, but the isolation efficiency decreased to 45% when MBs are centrifuged at 1,000g. According to our earlier theoretical calculations (25), MBs can detach from tumor cells at high centrifugation speeds.
Figure 5.
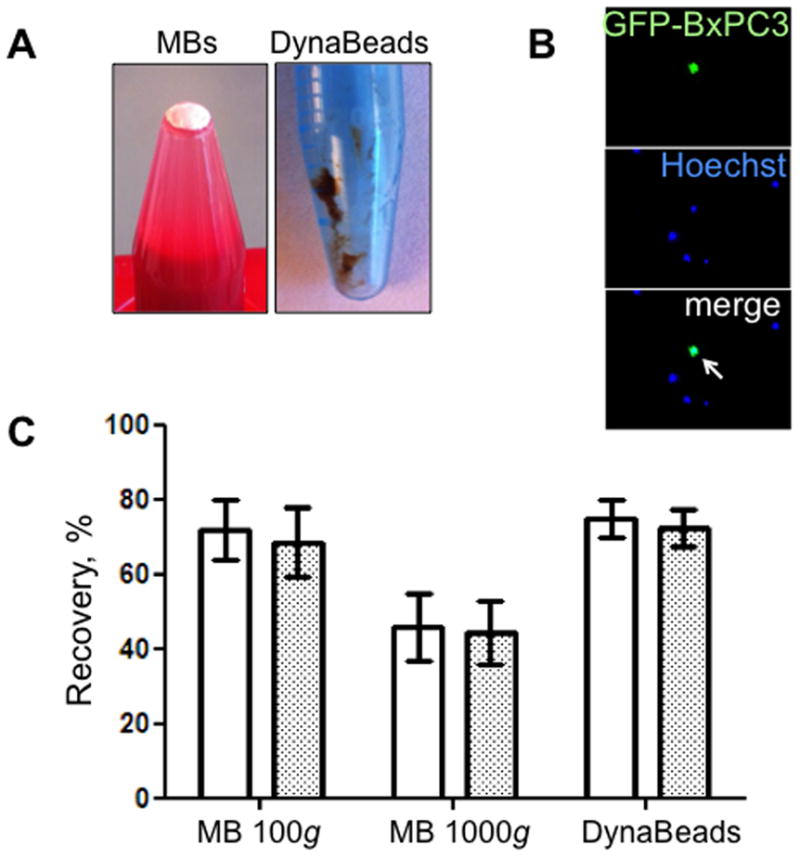
Isolation of GFP-positive BxPC3 pancreatic carcinoma cells from 1 ml blood with anti-EpCAM MBs and magnetic beads. (A) MBs after centrifugation form white pellet on top of the liquid. DynaBeads form slurry on the side of the tube after washing with external magnet. (B) Tumor cells are distinguished from leukocytes due to GFP/Hoechst label (arrow). (C) Effect of centrifugation speed on the recovery of BxPC3 cells from BSA/PBS (white bars) or blood (grey bars). Isolation efficiency in the MB layer decreased with 1000g force, possibly due to the tumor cell detachment from MBs. At 100g, the isolation efficiency was similar to DynaBeads.
3.2.2. Isolation of rare cells from large volume of biological media
Tumor cells are present in blood at extremely low concentrations (12), therefore the method should be capable of isolating single cells. Recently, we described the isolation of rare cells from 1–7 ml blood (25). Below, we describe isolation of rare BxPC-3 cells from large volume of cell medium or buffy coats. The cell isolation setup was similar to the one described in 3.2.1, but instead of collecting MBs into an eppendorf tube, they were collected on a nitrocellulose membrane in order to immobilize the cells and facilitate staining for cytokeratin (cells were not-GFP labeled in this experiment).
Buffy coats derived from donor blood (22 ml from 125 ml whole blood, San Diego blood bank) are washed twice in PBS by centrifugation at 1500g for 15 min and resuspended in the original volume with PBS.
Tumor cells are added to 22 ml buffy coats in 50ml polypropylene tube. In another tube, the same number of cells is added to 22ml of FBS-supplemented cell medium (RPMI).
Anti-human EpCAM MBs are added (0.5×108 total amount) to tubes with buffy coat and medium and mixed at 30rpm for 15 min.
Tubes are inverted and centrifuged at 100g for 2 min.
The conical bottom of the tube (while the tube is in the inverted upright position) is cut off, 30 ml PBS are added and the tubes are centrifuged again. Alternatively, the bottom of the tube may be cut off prior to the experiment and sealed with Parafilm during the incubation.
MB layer on top of the tube (Fig. 6A) is carefully transferred to 1cm × 1cm nitrocellulose membrane. The same amount of cells that was spiked in the buffy coats is applied on another piece of membrane.
Membranes are washed 3 times with 0.5ml ice-cold isopropanol (to destroy MBs), blocked with 10% mouse serum for 1h, stained with anti-Pan-cytokeratin antibody (eBioscience, cat. 53-9003-80) and mounted with Hoechst on a slide.
CK+/Hoechst+ tumor cells in “applied” sample and “recovered” samples are counted under microscope.
Figure 6.
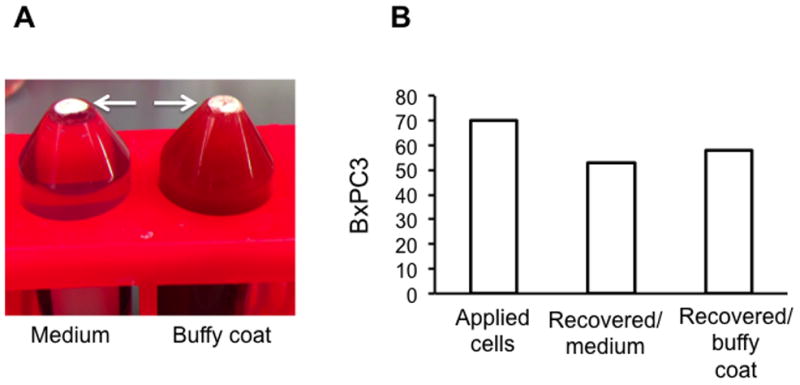
Isolation of spiked tumor cells from large volume of buffy coat or medium. (A) BxPC3 cells (non-GFP) were spiked into 22 ml medium (left tube) or 22 ml plasma-depleted buffy coats (right tube) and isolated with MBs. Arrows point to the MB layer. (B) Isolation (recovery) efficiency was determined after staining with cytokeratin (CK) antibody and Hoechst and counting CK+/Hoechst+ cells with fluorescent microscope.
According to the results of a representative experiment (Fig. 6B), out of 70 cells applied, MBs recovered 58 cells from buffy coat, and 52 cells from medium.
4. CRITICAL MB PARAMETERS FOR EFFICIENT ISOLATION
According to our previous theoretical calculations (25), MB size is the most important parameter in order to achieve cell isolation, since it is tied to buoyancy (the larger the diameter, the faster the separation). We increased the average MB size by washing away small MBs. Microfluidic manufacturing is another method for preparation of large-size uniform MBs (26).
Another important parameter is antibody density on MB surface. At 100g, around 13 antibodies are required to hold the MB–cell equimolar complex together due to tension forces (25). High density of antibody coating is achieved by using high mole-percent of DSPE-PEGmaleimide for MB preparation.
Centrifugation speed is critical. At high speed, cells may detach from MBs and sediment. At 1000g, over 130 attachments are needed to hold the MB–cell equimolar complex together (25). Since the expected number of EpCAM molecules at the cross-section of MB–cell attachment (0.2–0.5 μm2) is less than 130 (27), the centrifugation force should be limited to sub-1,000g forces (25).
5. CONCLUSIONS
Buoyant MBs coated with anti-EpCAM antibody efficiently and quickly isolate various tumor cells spiked into biological media including blood and buffy coats. The isolated cells could be immobilized, immunostained and enumerated. Despite the fact that MBs and beads showed similar isolation efficiency, it is plausible to suggest that MBs can deliver cells in a more concentrated manner (Fig. 4A). Also, our previous experience suggests that MBs result in less leukocyte contamination than magnetic beads (25). MB method could be a useful tool in life sciences as an alternative to or in combination with immunomagnetic beads.
Acknowledgments
This project was funded by the NIH IMAT CA137721 and NIH CA16488 to D.S. and by the UCSD Cancer Center Specialized Support Grant P30 CA23100. W.C. was partially supported by the NIH P50CA128346 (ICMIC) grant. We wish to thank Drs. Michael Bouvet and Robert Hoffman (both from UCSD Department of Surgery) for providing us with fluorescencent tumor cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors maybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fehm T, Sagalowsky A, Clifford E, Beitsch P, Saboorian H, Euhus D, Meng S, Morrison L, Tucker T, Lane N, Ghadimi BM, Heselmeyer-Haddad K, Ried T, Rao C, Uhr J. Clin Cancer Res. 2002;8:2073–84. [PubMed] [Google Scholar]
- 2.Steeg PS. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Pantel K, Brakenhoff RH. Nat Rev Cancer. 2004;4:448–56. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 4.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr J Clin Oncol. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 5.Molnar B, Ladanyi A, Tanko L, Sreter L, Tulassay Z. Clin Cancer Res. 2001;7:4080–5. [PubMed] [Google Scholar]
- 6.Singletary SE, Larry L, Tucker SL, Spitzer G. J Surg Oncol. 1991;47:32–6. doi: 10.1002/jso.2930470108. [DOI] [PubMed] [Google Scholar]
- 7.Palmieri G, Ascierto PA, Perrone F, Satriano SM, Ottaiano A, Daponte A, Napolitano M, Caraco C, Mozzillo N, Melucci MT, Cossu A, Tanda F, Gallo C, Satriano RA, Castello G. J Clin Oncol. 2003;21:767–73. doi: 10.1200/JCO.2003.01.128. [DOI] [PubMed] [Google Scholar]
- 8.Moreno JG, O’Hara SM, Gross S, Doyle G, Fritsche H, Gomella LG, Terstappen LW. Urology. 2001;58:386–92. doi: 10.1016/s0090-4295(01)01191-8. [DOI] [PubMed] [Google Scholar]
- 9.Baker MK, Mikhitarian K, Osta W, Callahan K, Hoda R, Brescia F, Kneuper-Hall R, Mitas M, Cole DJ, Gillanders WE. Clin Cancer Res. 2003;9:4865–71. [PubMed] [Google Scholar]
- 10.Houghton RL, Dillon DC, Molesh DA, Zehentner BK, Xu J, Jiang J, Schmidt C, Frudakis A, Repasky E, Maltez Filho A, Nolasco M, Badaro R, Zhang X, Roche PC, Persing DH, Reed SG. Mol Diagn. 2001;6:79–91. doi: 10.1007/BF03262038. [DOI] [PubMed] [Google Scholar]
- 11.Portet D, Denizot B, Rump E, Lejeune JJ, Jallet P. J Colloid Interface Sci. 2001;238:37–42. doi: 10.1006/jcis.2001.7500. [DOI] [PubMed] [Google Scholar]
- 12.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 13.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Clin Cancer Res. 2007;13:920–8. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 14.Kraeft SK, Ladanyi A, Galiger K, Herlitz A, Sher AC, Bergsrud DE, Even G, Brunelle S, Harris L, Salgia R, Dahl T, Kesterson J, Chen LB. Clin Cancer Res. 2004;10:3020–8. doi: 10.1158/1078-0432.ccr-03-0361. [DOI] [PubMed] [Google Scholar]
- 15.Bauer KD, de la Torre-Bueno J, Diel IJ, Hawes D, Decker WJ, Priddy C, Bossy B, Ludmann S, Yamamoto K, Masih AS, Espinoza FP, Harrington DS. Clin Cancer Res. 2000;6:3552–9. [PubMed] [Google Scholar]
- 16.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. Proc Natl Acad Sci U S A. 2004;101:10501–4. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tveito S, Maelandsmo GM, Hoifodt HK, Rasmussen H, Fodstad O. Clin Exp Metastasis. 2007;24:317–27. doi: 10.1007/s10585-006-9052-8. [DOI] [PubMed] [Google Scholar]
- 18.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, Board R, Clack G, Hughes A, Blackhall F, Valle JW, Dive C. Br J Cancer. 2011 doi: 10.1038/bjc.2011.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. Journal of the American Chemical Society. 2008;130:8633–41. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu T, Lu B, Tai YC, Goldkorn A. Cancer Research. 2010;70:6420–26. doi: 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schutt EG, Klein DH, Mattrey RM, Riess JG. Angew Chem Int Ed Engl. 2003;42:3218–35. doi: 10.1002/anie.200200550. [DOI] [PubMed] [Google Scholar]
- 23.Von Bibra H, Voigt JU, Froman M, Bone D, Wranne B, Juhlin-Dannfeldt A. Echocardiography. 1999;16:733–41. doi: 10.1111/j.1540-8175.1999.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 24.Talu E, Hettiarachchi K, Powell RL, Lee AP, Dayton PA, Longo ML. Langmuir. 2008;24:1745–49. doi: 10.1021/la703065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi G, Cui W, Benchimol M, Liu Y-T, Mattrey R, Kesari S, Esener SC, Simberg D. PLoS One. 2013 doi: 10.1371/journal.pone.0058017. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AP, Hettiarachchi K, Talu E, Longo ML, Dayton PA. Lab on a Chip. 2007;7:463–68. doi: 10.1039/b701481n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Jr, Terstappen LW. Int J Oncol. 2005;27:49–57. [PubMed] [Google Scholar]


