Abstract
Crypts are the basic structural and functional units of colonic epithelium and can be isolated from the colon and cultured in vitro into multi-cell spheroids termed “colonoids”. Both crypts and colonoids are ideal building blocks for construction of an in vitro tissue model of the colon. Here we proposed and tested a microengineered platform for capture and in vitro 3D culture of colonic crypts and colonoids. An integrated platform was fabricated from polydimethylsiloxane which contained two fluidic layers separated by an array of cylindrical microwells (150-μm diameter, 150-μm depth) with perforated bottoms (30-μm opening, 10-μm depth) termed “microstrainers”. As fluid moved through the array, crypts or colonoids were retained in the microstrainers with a >90% array-filling efficiency. Matrigel as an extracellular matrix was then applied to the microstrainers to generate isolated Matrigel pockets encapsulating the crypts or colonoids. After supplying the essential growth factors, epidermal growth factor, Wnt-3A, R-spondin 2 and noggin, 63±13% of the crypts and 77±8% of the colonoids cultured in the microstrainers over a 48–72 h period formed viable 3D colonoids. Thus colonoid growth on the array was similar to that under standard culture conditions (78±5%). Additionally the colonoids displayed the same morphology and similar numbers of stem and progenitor cells as those under standard culture conditions. Immunofluorescence staining confirmed that the differentiated cell-types of the colon, goblet cells, enteroendocrine cells and absorptive enterocytes, formed on the array. To demonstrating the utility of the array in tracking the colonoid fate, quantitative fluorescence analysis was performed on the arrayed colonoids exposed to reagents such as Wnt-3A and the γ-secretase inhibitor LY-411575. The successful formation of viable, multi-cell type colonic tissue on the microengineered platform represents a first step in the building of a “colon-on-a-chip” with the goal of producing the physiologic structure and organ-level function of the colon for controlled experiments.
Introduction
Microengineered devices are unique tools for the culture and interrogation of cells and tissues in vitro by virtue of their ability to control the cellular microenvironment both temporally and spatially.1 Microdevices specifically designed to mimic in vivo organ microarchitecture and function, called “organ-on-chips”, are envisioned to expand the capabilities of cell culture models and provide better controlled experimental alternatives to animal studies.2–4 An excellent example of organ-on-chips is a physiologically functional “lung-on-a-chip” that reconstitutes the dynamic mechanical strain and alveolar-capillary interface of the human lung.5 Various other organ-on-chips have been reported including liver,6 heart,7 blood vessel,8 muscle,9 kidney,10 and gastrointestinal tract,11–13 by recapitulating a specific feature of the organ microenvironment (e.g. topography, tissue-tissue interface, mechanical movement, shear stress, biochemical gradient).
While these organ-on-chips have created novel in vitro models that permit the study of some aspects of human physiology, many of them still rely on the use of immortalized cell lines derived from tumors. For example, Caco-2 cells derived from a colon carcinoma were used in several “gut-on-chips” to mimic the intestinal epithelium.11–13 Although these tumor cell lines can form a contiguous monolayer, their cancer phenotype poorly reflects normal tissue physiology or microarchitecture found in vivo. This issue points to one of the major challenges of organ-on-chips which is the use of primary cells derived from normal tissue to form systems more representative of in vivo organ systems.3
The subunit of the living colon is the crypt which is a micron-scale tubular structure comprised of a single layer of columnar epithelium that invaginates into the underlying connective tissue of the lamina propria. The colonic epithelium is the most rapidly renewing tissue in the mammalian body with a renewal time of 3–5 days for mice.14 This tissue regeneration is driven by a pool of multipotent colonic epithelial stem cells at the base of the crypts.15 The stem cells give rise to transient amplifying progenitor cells that terminally differentiate into three major types of epithelial cells as they migrate from the base of the crypt towards the lumen: goblet cells (secreting mucus), absorptive colonocytes (absorbing water and electrolytes), and enteroendocrine (secreting hormones). The self-renewal property of crypts provides homeostasis to the colonic epithelium, while the different cell types enable a range of functions to be accomplished by the colon. Therefore, crypts are ideal building units for constructing an in vitro tissue model within a microdevice.
A previous effort to design a microdevice for capture and biological assay of colonic crypts used polymer crypt-surrogates and fixed crypts.16 A freestanding film microfabricated from epoxy photoresist containing an array of micron-scale capture sites, termed a micromesh (open holes), was used to capture fixed crypts with high efficiency in an ordered and properly oriented fashion.16 However, this micromesh structure was less effective at capturing and retaining live crypts likely because crypts are much softer and more deformable than crypt surrogates and fixed crypts. For example, when live crypts approached the holes via fluidic flow, they deformed and did not properly enter the holes. Additionally, the structures were readily dislodged when reagents were added to the device and the crypts were viable for only a few hours.
In vitro culture of live crypts and intestinal stem cells has been attempted for decades with little success until the pioneering work by the Clevers and colleagues in 2009 in which long-term culture of crypts and stem cells from the small and large intestines was achieved by virtue of the identification of a number of critical factors needed for cell maintenance and proliferation.17–21 One of the most substantial difficulties overcome was the blockage of rapid apoptosis following removal of crypts from the basement membrane by addition of a ROCK inhibitor to the initial culture media. A second accomplishment was identification of growth factors (Wnt-3A, R-spondin 1, noggin and epidermal growth factor [EGF]) that were required for support of the colonic epithelium. These factors enable long term survival of colonic epithelial cells when added exogenously to a 3D extracellular matrix (ECM). This 3D culture system supports the growth of colonoids (defined as colonic organoids without mesenchyme),22 which contain self-renewing stem cells as well as all of the differentiated cell types present in crypts. The development of this culture technology provides the opportunity to design microdevices to support a living colonic epithelium for in vitro studies in a user-controlled microenvironment.
The goal of the current work was to create a viable colonoids microarray with potential for a variety of uses including the study of colonic cell physiology and the stem-cell proliferation and differentiation. Microfabrication was combined with primary colonic tissue culture to create a viable colonoid microarray. A microengineered crypt-like architecture was developed on the arrays using biocompatible substrates. Crypts and colonoids captured on the array were embedded in Matrigel to form an array of extracellular matrix plugs encasing the living cells. Crypt survival and growth were quantitatively assessed in culture conditions that mimicked the native colonic crypt niche with provision of the essential growth factors of EGF, Wnt-3A, R-spondin and noggin. Finally, to explore the feasibility of using the device as an in vitro drug screening platform, the arrayed colonoids were subjected to small molecule inhibition of the Notch pathway, LY-411575.
Experimental Section
Materials
Fluorescein isothiocyanate–dextran (FITC-dextran, average molecular weight 2,000,000), Y-27632 dihydrochloride (ROCK inhibitor), N-acetyl-L-cysteine (NAC), and LY-411575 (γ-secretase inhibitor) were purchased from Sigma Aldrich. The 1002F epoxy photoresist was formulated according to a previous publication.23 Polydimethylsiloxane (PDMS) was prepared from the Sylgard 184 silicone elastomer kit (Dow Corning). Advanced DMEM/F-12 medium, EGF recombinant mouse protein, N-2 supplement, B-27 supplement, GlutaMAX supplement, penicillin-streptomycin, fetal bovine serum (FBS), HEPES (1M buffer solution), and G418 sulfate were obtained from Invitrogen. Mouse noggin recombinant protein was from eBioscience. Growth factor reduced Matrigel and collagen (type I from rat tail) were purchased from BD. Collagenase (type 4) was purchased from Worthington Biochemical. All other reagents including dithiothreitol (DTT), bovine serum albumin (BSA) and ethylenediaminetetraacetic acid (EDTA) were from Fisher Scientific.
Isolation of crypts from colons
Crypts were isolated from wild type and CAG-DsRed/Sox9-EGFP mice (6–9 week old) using previously described methods.24 The CAG-DsRed/Sox9-EGFP mouse used in this study is a cross between a bacterial-artificial-chromosome-transgenic mouse in which the enhanced green fluorescent protein (EGFP) is expressed as a function of the Sox9 regulatory region (Sox9-EGFP mouse),25, 26 and the CAG-DsRed mouse (CAG = CMV enhancer plus chicken actin promoter) that harbors a transgene for the DsRed protein.17, 26, 27 All experiments were performed in compliance with the relevant laws and institutional guidelines at the University of North Carolina. All experiments and animal usage was approved by the Institutional Animal Care and Use Committee at UNC.
Off-chip 3D culture of colon crypts
Isolated crypts were embedded in collagen for in vitro 3D culture according to previous publications with minor modification.21, 24 Briefly, isolated crypts were counted using a hemocytometer. A total of 5,000 crypts were suspended in 500 μL of type 1 rat tail collagen at 2 mg/ml and placed in a 12-well plate. After polymerization of the collagen at 37 °C for 30 min, 2 mL of crypt culture medium (CCM) was added to the well. CCM was prepared from a mixture of advanced DMEM/F12 medium, Wnt-3A-conditioned medium, and R-spondin 2-conditioned medium at a volumetric ratio of 2:1:1, and supplemented with noggin (100 ng/mL), EGF (50 ng/mL), N2 (1×), B27 (1×), Y27632 ROCK inhibitor (10 μM), NAC (1 mM), GlutaMAX (1×), HEPES (10 mM), penicillin (100 unit/mL), and streptomycin (100 μg/mL). The detailed steps to prepare Wnt3A and R-Spondin 2-conditioned media are described in the supplementary information. CCM was prepared in a bulk volume of 500 mL and split into 6-mL aliquots and stored at −80 °C until use. The medium was changed every 48 h. The crypts grew into colonoids in culture (Fig. 1). To harvest the colonoids, the collagen gel was digested in DMEM containing collagenase type 4 (500 U/mL) at 37 °C for 15 min. The released colonoids were washed with PBS containing 0.5% BSA, centrifuged at 300 g for 2.5 min, and suspended in DMEM for immediate use.
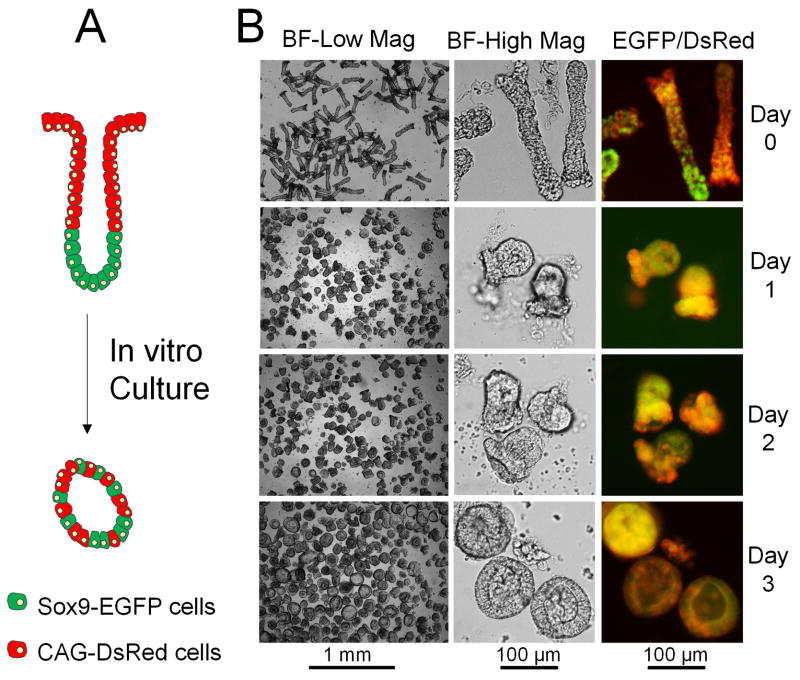
Fig. 1.
Development of colonoids derived from intact crypts. (A) Simplified model of cell composition and tissue polarization for crypts (upper structure) and colonoids (lower structure). Stem/progenitor cells are depicted in green, and differentiated cells are red. (B) Microscopic images of in vitro growth of crypts over 3 days. The left panel shows a large number of crypts/colonoids in low magnification brightfield images. The middle panel shows 2–3 crypts/colonoids in high magnification brightfield images. The right panel shows the merged red/green fluorescence images of crypts/colonoids isolated from the murine model system expressing EGFP and DsRed.
Fabrication of a freestanding PDMS microstrainer array
A freestanding PDMS membrane containing an array of microstrainers was prepared by three microfabrication steps (Fig. S1), the details of which are described in the supplementary information. In the first step, a master mold composed of an array of microstrainers firmly adhered to the glass substrate was fabricated from 1002F photoresist by a two-layer photolithography process (Fig. S1A).16 A 10-μm thick 1002F film was fabricated first on the glass as the base. The structure was composed of a 10-μm thick grid within 30-μm square or circular openings. Then a 150-μm thick 1002F film was coated on the top of the base, and holes of 150 μm in diameter were fabricated in the top layer. In the second step, a PDMS mold was prepared by replicate molding of PDMS on the master mold (Fig. S1B). The PDMS mold contained an array of large posts (150 μm in height, 150 μm in diameter) with small posts (10 μm in height, 30 μm in diameter or length) at the top of each large post. In the third step, a PDMS microstrainer array was fabricated by replicate molding under pressure (Fig. S1C). The PDMS microstrainer had the same geometry as the epoxy microstrainer on the master mold.
Integration of the microstrainer array into a device
Soft lithography was used to fabricate two fluidic layers (a top layer and a bottom layer) that were integrated above and below a microstrainer array to create a 2-channel microfluidic platform in PDMS (Fig. 2D). The array and the top layer were plasma treated for 2 min, aligned and brought into conformal contact to form permanent bonding. Then the bottom layer was bonded to the other side of the array in the same manner. The assembled device was then baked at 95 °C overnight to enhance the bond strength.
Fig. 2.
The microstrainer array and integrated fluidic device. (A) A scheme to fabricate the PDMS microstrainer array by replicate molding under pressure. (B–C) Microscopic images of the array: (B) brightfield and (C) SEM. Scale bar = 200 μm. (D) The integrated platform was composed of two microfluidic layers and one array. They were in order: a PDMS top piece (green), a PDMS microstrainer array (purple), and a PDMS bottom piece (green). (E) Top view of the assembled device. The numbers label the different access ports between the reservoirs and the compartments. (F) A photograph of the device with an exterior dimension of 30 × 30 × 10 mm.
Capture of crypts, selective placement of Matrigel, and on-chip culture of crypts on the microstrainer array
The device was sterilized with 70% ethanol and rinsed with PBS buffer ×5. Trapped air bubbles inside the microstrainers were removed by degasing the device in a covered Petri dish. The microstrainer array was treated with 1% BSA for 1 h at room temperature, and rinsed with PBS buffer ×3 prior to loading crypts. To load crypts on the array, a suspension of crypts in DMEM was added to port 1 and 2 followed by addition of PBS buffer. Gravity drove a trans-array flow delivering the crypts to the wells in the microstrainer array. The ratio of crypts:wells was 2:1. Once the crypts were captured on the array, buffer was aspirated from the channels, which was then quickly filled with cold liquid Matrigel (50% dilution in CCM, 4 °C), and incubated at room temperature for 5 min. Aspiration of liquid Matrigel from the channels generated isolated Matrigel pockets embedding the crypts. After this step, the device was placed at 37 °C for 10 min to solidify the Matrigel. CCM (1 mL) was then added to the top and bottom fluidic layers for crypt culture. Medium was changed every 24 h. Colonoids were loaded and cultured on the device in the same manner as the isolated crypts.
Microscopy
The crypts and colonoids were imaged using a Nikon Eclipse TE300 inverted epifluorescence microscope equipped with DAPI/FITC/Texas Red filter sets. Wide-field imaging of the entire array was obtained using an Olympus MVX-10 research macro zoom fluorescence microscope equipped with FITC/Texas Red filter sets. 3D images of crypts embedded in solidified Matrigel pockets (mixed with 100 μg/mL FITC-dextran) on the array were obtained using an Olympus spinning disk confocal microscope equipped with FITC/Texas Red filter sets. The PDMS microstrainer array was inspected by SEM (FEI Quanta 200 ESEM, FEI Company).
Immunofluorescence
Crypts isolated from a wild-type mouse were used for immunofluorescence (IF). The freshly isolated crypts and in vitro cultured colonoids on the array were fixed with 4% paraformaldehyde for 20 min, followed by permeabilization with 0.5% Triton X-100 for 20 min. IF staining was performed using the following primary antibodies: rabbit anti-Muc2 (1:200, Santa Cruz, #SC-15334), rabbit anti-chromogranin A (1:1000, Bioss, #bs-0539R), mouse anti-carbonic anhydrase II (1:500, Santa Cruz, #SC-48351). The secondary antibodies were donkey anti-rabbit or mouse antibodies conjugated with NL594 and NL637 (1:200, Santa Cruz). DNA was stained with Hoechst 33342 (1 μg/mL, Sigma Aldrich, #B2261). The stained crypts and colonoids were imaged using the Nikon Eclipse TE300 microscope described above.
Time-lapse imaging of colonoids in response to the growth factor, Wnt-3A
To study the effect of Wnt-3A, crypts isolated from a CAG-DsRed/Sox9-EGFP mouse were captured and cultured on two arrays. One array was cultured in the absence of Wnt-3A, while the other one was cultured in CCM and served as the control. Images were collected with a cooled CCD camera (Photometrix Cool Snap HQ2; Roper Scientific, Tucson, AZ) using a Micro-Manager hardware control interface.28 Image analysis was performed using a custom script implemented in MATLAB (MathWorks; Natick, MA). Briefly, centers of the wells in the array were detected using an implementation of the Hough transform for detecting circles.29 Detected wells in overlapping the image edge were rejected from analysis and the remaining well centers were used to generate a segmentation mask for each field-of-view within the array. This mask was then used to integrate fluorescence intensities for each colonoid on the acquired images. For experiments in which dynamic properties were tracked, the first image in the sequence was selected for segmentation and the resulting mask applied to all images in the sequence.
Gamma-secretase inhibition
To study the effect of a γ-secretase inhibitor, crypts isolated from a CAG-DsRed/Sox9-EGFP mouse were captured on the two chips and encapsulated with Matrigel. One chip was cultured in CCM containing 1 μM LY-411575,30 while the other one without LY-411575 served as the control. The medium was changed every 24 h. At 48 h, the colonoids on the arrays were fixed and stained with Hoechst 33342. The arrays were imaged on an Olympus MVX-10 microscope to quantify Sox9 (EGFP) expression as well as DNA content (Hoechst 33342). Image analysis was performed using a custom script implemented in MATLAB based on user initialization of the capture array geometry. The fluorescence intensity was analyzed by two-sample student’s t-test. Statistical significance for comparisons was assigned at P < 0.05.
Results and Discussion
Use of a CAG-DsRed/Sox9-EGFP mouse model to facilitate quantification of colonoid growth and differentiation
Transgenic mouse models expressing multiple fluorescent proteins have enabled identification and tracking of cells within viable crypts.17, 26, 27 A CAG-DsRed/Sox9-EGFP mouse model was used in which DsRed was constitutively expressed while EGFP was expressed in intestinal epithelial stem and progenitor cells, but not in the differentiated colonic epithelium.25 This model allows monitoring and quantification of undifferentiated cells (Sox9EGFP+; red and green) and differentiated cells (red only) by fluorescence microscopy (top scheme in Fig. 1A).
Two types of colonic epithelial samples were prepared for experiments, freshly isolated crypts or colonoids cultured from previously isolated crypts. Production of colonoids enabled expansion of the primary cells and minimized the numbers of animals required for tissue procurement. To produce this expanded tissue sample, isolated crypts were embedded in a patty of collagen hydrogel and supplied with essential growth factors (EGF, Wnt-3A, R-spondin 2 and noggin) and an apoptosis inhibitor (Y27632).17, 21, 24 Of the isolated crypts, 78±5% (a total number of 60 crypts counted in 3 culture experiments) formed colonoids when cultured for 3 days (Fig. 1B). When cultured under these conditions, the cells in the luminal portion of the crypts rapidly died while the crypt base containing the stem cells persisted in culture and developed into colloid structures. Colonoids continued to grow and by day 3 possessed an enclosed central lumen (Fig. 1). Thus, the crypts underwent a dramatic change in morphology during in vitro culture from open and elongated (at day 0) to enclosed and spherical (at day 3). Freshly isolated crypts possessed a diameter at the luminal end of 100 ± 23 μm, a basal diameter of 50 ± 10 μm and a length of 241 ± 49 μm (n=20). By day 3, the spherical colonoids cultured displayed a diameter of 110 ± 43 μm (n=20). In addition to the change in shape, compartmentalization of the various cell types was lost during in vitro culture. Colonoids possessed a non-polarized structure with self-renewing stem/progenitor cells (EGFP) and differentiated epithelial cell types (DsRed) being randomly dispersed (Fig. 1B). These cultured colonoids could be harvested from the collagen gel by digestion with collagenase which also fragmented the larger structures to yield increased numbers of smaller colonoids. The tissue could then be utilized for assays or further expanded in culture.
The freshly isolated crypts and in vitro cultured colonoids demonstrated distinct advantages and disadvantages. Freshly isolated crypts resembled the in vivo state of the colonic epithelium in terms of morphology, cell segregation and tissue polarization, but their elongated shape made it difficult to control their orientation on microdevices. Colonoids were readily manipulated in microdevices due to their spherical morphology, but their shape and lack of a distinct stem-cell and differentiated-cell compartments did not mimic that of in vivo crypts. In this study, both freshly isolated crypts and in vitro cultured colonoids (48 h in off-chip culture) were prepared in suspension and used as a tissue sources for the microdevices.
Fabrication of the microstrainer array and its integration into a device
Currently, the standard approach to in vitro culture of crypts uses conventional culture devices such as multi-well plates and Transwell permeable inserts.24, 26, 31 A microengineered device to efficiently array and maintain living crypt tissue would represent a highly efficient and cost effective platform and provide unprecedented user-controlled fluidic microenvironments. To achieve such a tool, a freestanding film containing a microstructure termed a microstrainer array was fabricated to capture and retain live crypts. The microstrainer array was composed of deep, cylindrical microwells (150-μm diameter, 150-μm depth) with a thin, grid-like bottom layer (30-μm opening, 10-μm depth) (Fig. 2A–C). The gridded bottom layer was designed to permit fluid to flow through the microwells while blocking crypt passage. The depth of the microwells was expected to enable crypt retention in the array sites during reagent manipulation. Arrays similar to that used here have been fabricated from epoxy photoresist by a two-layer photolithography process.16, 32 In this study, PDMS was selected as the material to fabricate the microstrainer array due to its low autofluorescence, biocompatibility, wide acceptance in the microfluidic community, and ease in bonding with other fluidic layers to form integrated devices.
A process composed of three microfabrication steps was used to fabricate the PDMS microstrainer array with the details described in the supplementary information. During the final fabrication step (Fig. 2A), a soft lithography-based replica molding method was employed similar to that used for fabricating a PDMS porous membrane.11 PDMS pre-polymer was sandwiched between a glass slide and a post-array mold under a uniform pressure generated by a 0.75 kg weight. Thermal cure and subsequent demolding yielded a high quality PDMS microstrainer array (Fig. 2B). The bottom layer was perforated with unobstructed holes (Fig. 2C). Each array was composed of 3×3 subgroups, and each subgroup contained 10×10 microstrainers, resulting in a total of 900 microstrainers per array. In order to track the growth of crypts, each microstrainer was provided an address designated by a combination of letters and numbers. For example, the address of the microstrainer in the upper/right corner of Figure 2B is “AA-5a” where “AA” denotes the subgroup, “5” and “a” are x and y coordinates of the microstrainer in this subgroup.
To form an integrated microfluidic device, the microstrainer array was sandwiched and sealed by plasma-activated bonding of two PDMS layers (Fig. 2D–F). The top layer contained a central chamber (8 mm in diameter) and four through holes (8-mm in diameter) that provided access points for fluid flow through the device. The bottom layer possessed three cylindrical chambers. The integrated device contained two fluidic layers separated by an array of microstrainers that divided the central chamber into luminal and basal compartments. Ports 1 and 2 connected the luminal compartment, while ports 3 and 4 connected the basal compartment (Fig. 2E). The fluidic flow in the platform was controlled by four ports: 1→2 for flow through the luminal compartment, 3→4 for flow through the basal compartment, and 1→4 or 2→3 for trans-array flow. Fig. 2F shows a photograph of the integrated platform.
Capture of crypts on the microstrainer array and selective placement of Matrigel
Crypts were loaded onto the array by pipette and moved through the channels by vacuum aspiration so that no pumps and valves were needed (Fig. 3A). A suspension of crypts was added to ports 1 and 2 followed by addition of PBS buffer to these reservoirs. At the same time PBS was removed from ports 3 and 4 driving a trans-array flow which transported the crypts to the microstrainer. As fluid moved through the porous base of the microstrainers the crypts were retained in the microstrainers (Fig. 3A-ii). This simple loading strategy yielded >90% capture efficiency (i.e. percentage of microstrainers on the array that was filled with crypts/colonoids) when a crypt:well ratio of 2:1 was employed (Fig. 3B,C). Since the crypts were on average 241 ± 49 μm in length, many crypts were deformed as they entered the microstrainer (diameter = 150 μm) (Fig. 3D). Some microstrainers captured more than one crypt which was deemed acceptable, since it was observed that two adjacent crypts within a microwell always fused into one colonoid during culture. Colonoids that were formed in culture prior to loading on the array were also captured on the microstrainer array with >90% capture efficiency (Fig. S2A) but without deformation due to their spherical shape and small size (Fig. 3E). If capture of single crypt per microstrainer is needed, e.g. in conducting clonal analysis of crypts in vitro, a low crypt:well ratio (e.g. 0.2:1) can be employed (Fig. S2B).
Fig. 3.
Capture of crypts/colonoids on the microstrainer array and selective placement of Matrigel. (A) Cross-sectional view of the device as the compartment contents were sequentially loaded. i) Device. ii) Capture of crypts. iii) Placement of liquid Matrigel. iv) Aspiration of liquid Matrigel from luminal and basal compartments and solidification of the Matrigel remaining within the microstrainer wells. v) Addition of medium to the luminal and basal compartments. (B–E) Capture of crypts/colonoids on the array. (B) Brightfield image of 64 capture sites filled with crypts. (C) Red fluorescence image of B. (D) A single crypt is shown captured in a microstrainer. (E) A single colonoid is captured in a microstrainer. (F) Confocal fluorescence image of Matrigel pockets formed in the microstrainer array. (G–H) Shown are nonconfocal fluorescence (G) and confocal fluorescence (H) images of a crypt encapsulated within a Matrigel pocket in the microstrainer. The panels display the merged red/green fluorescence images. The Matrigel was mixed with 100 μg/mL fluorescein-dextran in images F through G.
Intestinal epithelial cells undergo rapid apoptosis termed anoikis when they are detached from basement membrane.33 To maintain the viability of crypts in the microstrainers, the crypts must be embedded in an ECM hydrogel as quickly as possible. Laminin is enriched in basement membranes surrounding the crypts and laminin-rich Matrigel has been shown to be effective in maintaining the viability of crypts and maintaining colonic stem cells.17 For this reason arrayed crypts were embedded in Matrigel pockets immediately after loading onto the array (Fig 3A). After crypts capture on the array (Fig. 3A-ii), buffer was aspirated from the channels which were then quickly filled with cold liquid Matrigel (Fig. 3A-iii). Liquid Matrigel was then aspirated from the channels leaving an isolated Matrigel plug in each microstrainer well. Incubation of the device at 37°C caused the Matrigel to gel and encase the crypts (Fig. 3A-iv). When the Matrigel was premixed with fluorescein-dextran, confocal, fluorescence microscopy confirmed that the crypts were embedded in isolated Matrigel pockets that had the same height and diameter as the microstrainers (Fig. 3F–H). Finally medium was added to the upper and lower compartments to supply the crypts with nutrients and growth factors (Fig. 3A-v). Although the diffusion of growth factors in Matrigel is slow, e.g. with a diffusion coefficient on the order of 106 μm2/hour for vascular endothelial growth factor (38.2 kDa),34 the micron-scale Matrigel pockets ensured the efficient delivery of growth factors to the crypts, and removal of metabolic wastes from the crypts. Addition and removal of medium demonstrated that the crypts or colonoids were held firmly in the Matrigel pockets as none were found to be dislodged from the array during medium exchange. After capture of crypts or colonoids, media was placed in the upper and lower compartments. After initial media placement, no fluid flow was present.
In vitro culture of crypts and colonoids in the microstrainer array
Within the microstrainers, crypts formed 3D colonoids over a 72-h period and the colonoids displayed a similar spherical morphology to present under standard growth conditions (Fig. 4A). Many colonoids filled the entire lumen of the microstrainers by 72 h. Cell debris was present on the surface of the microstrainer and likely originated from the expected apoptosis of cells in the luminal portion of the crypts. The efficiency of crypt growth into colonoids was assessed by comparing the DsRed fluorescence image after 1 and 72 h in culture for 60 wells in 3 independent culture experiments. Of the crypts loaded into the microstrainers, 63±13% grew into colonoids, a number similar to that under standard off-chip culture conditions (78±5%). All of the living crypt/colonoids expressed EGFP fluorescence and thus possessed Sox9-expressing cells indicating that stem/progenitor cells were present in the colonoids. These data suggested that the material (PDMS) and geometric constraints (microstrainer) did not exert a substantially negative effect on the growth of isolated crypts into colonoids.
Fig. 4.
Culture of crypts/colonoids in PDMS microstrainers. A crypt (A) or colonoid (B) was loaded into a microstrainer on the array. By 72 h, a colonoid formed filling the microstrainer. The top panels are brightfield images of the same microstrainer site while the lower panels are overlaid red/green fluorescence images of the array site. (C) Widefield images (7 × 7 mm) of the tissue array composed of viable, 3D colonoids. The array possessed 900 microstrainers. The upper panel is a brightfield image, and the lower panel is an overlaid red/green fluorescence image. (D) Immunofluorescence staining. Cell composition in freshly isolated crypts (left panel) and colonoids formed in the microstrainers (right panel). Immunofluorescence images are shown for samples stained for: (i) Sox9 (green, stem/progenitor cells), (ii) Muc2 (red, goblet cells), (iii) chromogranin A (red, enteroendocrine cells), and (iv) carbonic anhydrase II (red, enterocytes). Hoechst 33342 was used as a counter stain to mark the nuclei (blue) in all images. The lumen of crypts faces upward. White dashed circles in the bottom panel indicate the perimeter of the microstrainer well.
Colonoids cultured off-chip could also be captured and then cultured on the array (Fig. 4B) in a manner similar to that of the crypts. Sox9+ stem/progenitor cells were preserved in the colonoids during culture on the device as evidenced by the continued expression of EGFP (Fig 4B). Of the colonoids loaded onto the array, 77±8% (n=60 in 3 independent experiments) continued to expand in size on the array at 48 h which was similar to the survival rate of crypts loaded onto the array. By 48 h, the living colonoids filled the entire lumen of the microstrainers (Fig. 4B). At 72 h, over 70% of the 900-capture sites were filled with viable, 3D colonoids possessing stem/progenitor cells as indicated by the presence of EGFP in 100% of the colonoids (Fig. 4C). These experiments demonstrate that either freshly isolated crypts or in vitro cultured colonoids could be used as the tissue source for the microstrainer array.
To assess whether the colonoids on the microstrainer array possessed the full repertoire of differentiated lineages, immunostaining for lineage-specific markers was conducted on arrayed colonoids derived from a wild type mouse. At 72 h, the colonoids were fixed and stained for Muc2 (goblet cells), chromogranin A (enteroendocrine cells), and carbonic anhydrase II (enterocytes).21 In freshly isolated crypts, stem/progenitor cells (Sox9+) are localized at the base of the crypts (Fig. 4D-i, left,). In contrast, stem/progenitor cells in colonoids were the predominant cell type and were randomly dispersed (Fig. 4D-i, right). The Wnt-3A protein in the culture medium favored the expansion of stem/progenitor cells since Wnt acts to support the undifferentiated progenitor cell type.19 Mature, differentiated goblet cells (Muc2+), which secrete mucus to protect and lubricate the colon, are found at the luminal portion of the crypts but were dispersed throughout the colonoids (Fig. 4D-ii). Enteroendocrine cells (CGA+), which release hormones or peptides to control important physiological functions of the colon, are present in in low numbers in freshly isolated crypts, and were also rarely observed in the colonoids (Fig. 4D-iii). Colonic enterocytes (CAII+), which uptake water and ions from the solid waste in the colon, are located along the luminal portion of the crypts. These cells were present in the colonoids but dispersed throughout the colonoid at random locations (Fig. 4D-iv). The differentiated cells wre found in the majority of imaged wells (Fig. S3). The immunofluorescence staining of colonoids on the array was independent on the source of tissue (fresh crypts or off-chip cultured colonoids) (Fig. S4). These findings are consistent with the numerical distribution of stem/progenitor and differentiated cells in colonoids formed under standard culture conditions.19 Compared to freshly isolated crypts, colonoids lacked clear proliferative and differentiated regions.
In vitro response of colonoids to Wnt-3A and γ-secretase inhibitor LY-411575
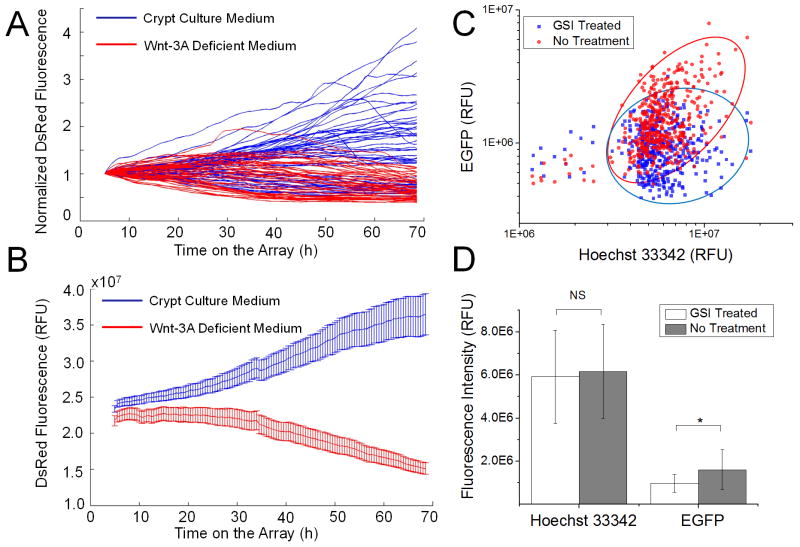
To demonstrate the utility of the microstrainer array platform, the arrays of CAG-DsRed/Sox9-EGFP crypts were exposed to two reagents, Wnt-3A and LY-411575. Wnt signaling is a major driving force behind intestinal cell renewal and is essential for in vitro growth of crypts into colonoids.17, 35 To confirm the role of Wnt-3A, crypts were cultured on the microstrainer arrays and images of DsRed fluorescence intensity were acquired every 30 min (Fig. 5A, B). In the presence of Wnt-3A, the DsRed fluorescence intensity of crypts increased steadily over time up to 68 h, indicating expansion of the crypts into colonoids. In the absence of Wnt-3A, however, DsRed fluorescence decreased over the time in culture as cell numbers declined. This result demonstrates the critical role of Wnt-3A in culturing colonic crypts and the utility of the arrays in tracking the of growth factors at the individual colonoid level.
Fig. 5.
Response of colonoids to Wnt-3A (A, B) and γ-secretase inhibitor LY-411575 (C, D). (A) DsRed fluorescence intensity normalized to the first time-point vs. time for 55 colonoids in the presence of Wnt-3A, and 47 colonoids in the absence of Wnt-3A. (B) Average fluorescence intensity vs. time for the colonoids. (C) Scatter plot showing EGFP and Hoechst 33342 fluorescence levels of individual colonoids after a 48-h exposure to 1 μM (blue) and 0 μM (red) LY-411575. (D) Fluorescence intensity of colonoids. Student’s t test: * (P < 0.05), NS (not significant, P > 0.05). The whiskers in the plot show, respectively, the upper and lower inner fence value, defined as the 25th percentile minus and 75th percentile plus 1.5 times the interquartile range.
LY-411575 is a γ-secretase inhibitor known to block Notch signaling.36 Inhibition of Notch signaling acts to increase differentiation signals along the secretory lineage and decrease stem cell numbers.37, 38 To study the effect of LY-411575 on crypts, crypts were cultured on the microstrainer arrays in the presence of 0 and 1 μM LY-411575 (Fig. 5C, D). After exposure of LY-411575 for 48 h, the colonoids were fixed and their nuclei were stained with Hoechst 33342. The location of each colonoid on the microstrainer array was determined by having a user initialize the positions of the array’s corners and interpolating the position of each well on the array. Hoechst and EGFP fluorescence were integrated over the area of each well after background subtraction. EGFP expression was reduced by 39.4% reduction compared to that of the control (P < 0.05, student’s t test) (Fig 5D). The results demonstrate that Sox9EGFP expression (Fig. 5C) was suppressed by administration of 1 μM of LY-411575 suggesting a decrease in functional stem/progenitor cells. No significant change in colonoid DNA content was observed (P = 0.087 >0.05, Fig. 5C) between the control and drug-exposed arrays indicating that total cell numbers were similar on both arrays and the drug did not act by killing cells. LY-411575 did not inhibit the survival of colonoids on the arrays at 48 h. Our data is consistent with a previous report that LY-411575 induces differentiation of intestinal stem cells and reduces expression of the stem cell marker Lgr5.21 While a significant difference in the mean level of EGFP fluorescence was observed, tracking individual colonoids revealed significant heterogeneity within each population.
Conclusions
An integrated platform possessing microfluidic channels and a microstrainer array was fabricated and used to capture and culture colonic crypts that developed into colonoids. Captured colonic crypts or organoids were retained on the microstrainer array with >90% filling efficiency. A simple operation (filling and then aspiration) generated micron-scale Matrigel pockets that encapsulated the crypts and colonoids within the microwells on the array. The crypts and colonoids grew within the microstrainers to form or maintain viable 3D colonoids that possessed the entire cell lineages found in intact crypts. Large numbers of colonoids were rapidly screened on the microstrainer arrays without a need for computationally expensive image segmentation as the position of each microstrainer within the array was predefined. By combining the arrays with wide-field imaging, >400 colonoids could be captured per image frame. The arrays enabled rapid in vitro analysis of primary colonic tissues during drug exposure and may find usage as a screening tool to identify potential adverse gastrointestinal effects of orally administered drugs. This study demonstrated for the first time that extremely fragile specimens, such as crypts, can be captured and cultured in a microengineered device to form viable structures.
Supplementary Material
Acknowledgments
This research was supported by the NIH (EB012549, EB013803 and DK091427) and the University Cancer Research Fund (UCRF). We thank Adam Gracz for helpful discussions. We acknowledge the Chapel Hill Analytical and Nanofabrication Laboratory for access to the SEM and confocal microscope facilities.
References
- 1.Kovarik ML, Gach PC, Ornoff DM, Wang YL, Balowski J, Farrag L, Allbritton NL. Analytical Chemistry. 2012;84:516–540. doi: 10.1021/ac202611x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huh D, Hamilton GA, Ingber DE. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung JH, Esch MB, Prot JM, Long CJ, Smith A, Hickman JJ, Shuler ML. Lab Chip. 2013;13:1201–1212. doi: 10.1039/c3lc41017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 5.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S-A, No DY, Kang E, Ju J, Kim D-S, Lee S. Lab Chip. 2013 doi: 10.1039/c3lc50197c. [DOI] [PubMed] [Google Scholar]
- 7.Grosberg A, Alford PW, McCain ML, Parker KK. Lab Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Chen JM, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson K, Das M, Wahl KJ, Colton RJ, Hickman J. Plos One. 2010:5. doi: 10.1371/journal.pone.0011042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang KJ, Suh KY. Lab Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 11.Kim HJ, Huh D, Hamilton G, Ingber DE. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 12.Ramadan Q, Jafarpoorchekab H, Huang CB, Silacci P, Carrara S, Koklu G, Ghaye J, Ramsden J, Ruffert C, Vergeres G, Gijs MAM. Lab Chip. 2013;13:196–203. doi: 10.1039/c2lc40845g. [DOI] [PubMed] [Google Scholar]
- 13.Sung JH, Yu JJ, Luo D, Shuler ML, March JC. Lab Chip. 2011;11:389–392. doi: 10.1039/c0lc00273a. [DOI] [PubMed] [Google Scholar]
- 14.Barker N, van de Wetering M, Clevers H. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs E, Chen T. Embo Reports. 2013;14:39–48. doi: 10.1038/embor.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YL, Dhopeshwarkar R, Najdi R, Waterman ML, Sims CE, Allbritton N. Lab Chip. 2010;10:1596–1603. doi: 10.1039/b927316f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Nature. 2009;459:262–U147. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Nature. 2010 doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 20.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 21.Yui SR, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, Clevers H, Watanabe M. Nat Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 22.Stelzner M, Helmrath M, Dunn JCY, Henning SJ, Houchen CW, Kuo C, Lynch J, Li LH, Magness ST, Martin MG, Wong MH, Yu J N. I. H. I. S. C. Consortiu, . Am J Physiol-Gastroint Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pai JH, Wang Y, Salazar GT, Sims CE, Bachman M, Li GP, Allbritton NL. Anal Chem. 2007;79:8774–8780. doi: 10.1021/ac071528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T, Stange DE, Ferrante M, Vries RGJ, van Es JH, van den Brink S, van Houdt WJ, Pronk A, van Gorp J, Siersema PD, Clevers H. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 25.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Am J Physiol-Gastroint Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gracz AD, Ramalingam S, Magness ST. Am J Physiol-Gastroint Liver Physiol. 2010;298:G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 28.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer Control of Microscopes Using μManager. John Wiley & Sons, Inc; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballard DH. Pattern Recognit. 1981;13:111–122. [Google Scholar]
- 30.Ninov N, Borius M, Stainier DYR. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ootani A, Li XN, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Nat Med. 2009;15:1–U140. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ornoff DM, Wang Y, Allbritton NL. J Micromech Microeng. 2013:23. doi: 10.1088/0960-1317/23/2/025009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dufour G, Demers MJ, Gagne D, Dydensborg AB, Teller IC, Bouchard V, Degongre I, Beaulieu JF, Cheng JQ, Fujita N, Tsuruo T, Vallee K, Vachon PH. J Biol Chem. 2004;279:44113–44122. doi: 10.1074/jbc.M405323200. [DOI] [PubMed] [Google Scholar]
- 34.Miura T, Tanaka R. Math Model Nat Phenom. 2009;4:118–130. [Google Scholar]
- 35.Schuijers J, Clevers H. Embo J. 2012;31:2685–2696. doi: 10.1038/emboj.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong GT, Manfra D, Poulet FM, Zhang Q, Josien H, Bara T, Engstrom L, Pinzon-Ortiz M, Fine JS, Lee HJJ, Zhang LL, Higgins GA, Parker EM. J Biol Chem. 2004;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto R, Tsuchiya K, Nemoto Y, Akiyama J, Nakamura T, Kanai T, Watanabe M. Am J Physiol-Gastroint Liver Physiol. 2009;296:G23–G35. doi: 10.1152/ajpgi.90225.2008. [DOI] [PubMed] [Google Scholar]
- 38.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud A, Grosse AS, Gumucio DL, Ernst SA, Tsai YH, Dempsey PJ, Samuelson LC. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.