Summary
Interactions among suprachiasmatic nucleus (SCN) neurons are required for robust circadian rhythms entrained to local time. To investigate these coupling mechanisms, we developed a novel functional coupling assay that uniquely captures the dynamic process by which SCN neurons interact. As a population, SCN neurons typically display synchronized rhythms with similar peak times, but will peak 6–12h apart after in vivo exposure to long days. Once removed from these conditions, SCN neurons resynchronize through a phase-dependent coupling process mediated by both vasoactive intestinal polypeptide (VIP) and GABAA signaling. Notably, GABAA signaling contributes to coupling when the SCN network in an anti-phase configuration, but opposes synchrony under steady-state conditions. Further, VIP acts together with GABAA signaling to couple the network in an anti-phase configuration, but promotes synchrony under steady-state conditions by counteracting the actions of GABAA signaling. Thus, SCN neurons interact through non-redundant coupling mechanisms influenced by the state of the network.
Keywords: circadian, suprachiasmatic nucleus, coupling, VIP, GABA, photoperiod, bioluminescence imaging
Introduction
Temporal coding within oscillating neuronal networks is an important organizational principle (Buzsaki, 2006; Schnitzler and Gross, 2005). The suprachiasmatic nucleus is a neuronal network that controls daily rhythms in mammalian behavior and physiology (Mohawk and Takahashi, 2011). Individual SCN neurons display self-sufficient rhythms in gene expression and electrical activity (Welsh et al., 2010) generated at the molecular level by interacting feedback loops involving the transcription and translation of clock genes (e.g., period2) (Takahashi et al., 2008). Network-level properties of the SCN sustain robust and coherent oscillations at the population level (Welsh et al., 2010), and intercellular interactions appear to be absent in most non-SCN tissues (Stratmann and Schibler, 2006). In particular, SCN neurons interact to maintain identical period lengths and specific phase relationships, and this coordination is lost following neuronal dissociation (Welsh et al., 2010). In this manner, intercellular interactions among SCN neurons (i.e., coupling) determine population-level properties that are required for the transmission of coherent output signals to downstream tissues and adjustment to changing environmental conditions (Meijer et al., 2012; Meijer et al., 2010).
While critical for pacemaker function, the process by which SCN neurons interact remains ill-defined. Candidates for SCN coupling factors have been identified (Aton and Herzog, 2005; Maywood et al., 2011), with vasoactive intestinal polypeptide (VIP) known to play an especially important role. Without competent VIP signaling, SCN neurons display desynchronized rhythms and a lower propensity for sustained cellular oscillations (Aton et al., 2005). However, SCN neurons also communicate through other signaling pathways that can compensate for the lack of VIP (Brown et al., 2005; Ciarleglio et al., 2009; Maywood et al., 2011; Maywood et al., 2006). GABA, the most abundant neurotransmitter within the SCN (Abrahamson and Moore, 2001), is also a putative coupling factor whose role remains unclear since GABAA signaling is sufficient (Liu and Reppert, 2000) but not required for synchrony (Aton et al., 2006). One obstacle in developing a mechanistic understanding of the role of different SCN coupling factors is the lack of analytical paradigms well suited for this purpose. Previous studies have relied largely on techniques that eliminate cellular interactions via physical, pharmacological, or genetic means to determine what forms of intercellular signaling are necessary or sufficient for period synchrony. While informative, this approach typically entails compromised neural function, which can complicate interpretation of the precise role played by the candidate coupling factor. Furthermore, this approach is unable to provide insight into how the intact, functional SCN network uses and integrates different coupling signals.
Here we develop a functional assay for SCN interactions that uses genetically intact animals with competent neuronal oscillatory and coupling mechanisms. Our research strategy was modeled on that previously employed to investigate coupling within an invertebrate pacemaker system (Roberts and Block, 1985), which involved shifting one of two coupled pacemakers and then tracking resynchronization between the pair over time in vitro. While it remains difficult to shift SCN subpopulations in vitro, the pacemaker network can be temporally reorganized in vivo by a variety of environmental lighting conditions (de la Iglesia et al., 2004; Inagaki et al., 2007; Meijer et al., 2010; Yan et al., 2005). Based on previous theoretical and experimental research (Inagaki et al., 2007; Pittendrigh and Daan, 1976b), we predicted that in vivo exposure to long day lengths would reorganize the SCN network into two subpopulations cycling out of phase, which we could use to investigate network resynchronization over time in vitro using bioluminescent reporters of clock protein expression in single SCN neurons. Using this novel assay of SCN coupling, we find that SCN neurons are coupled by both VIP and GABAA signaling, and that these SCN factors operate in a cooperative or antagonistic manner depending on the state of the network.
Results
Long Day Lengths Reorganize the SCN Network
Male PER2::LUC mice (Yoo et al., 2004) were bred and raised under a 24h light:dark cycle with 12h light and 12h darkness (LD12:12). At 7–9 wks of age, mice either remained under LD12:12 or were transferred to a long day length condition with 20h of light (LD20:4). As expected, LD20:4 produced a rapid decrease in the duration of the nocturnal active phase (Fig. 1A, Supplementary Fig. 1A–B). In addition, LD20:4 mice displayed a stable phase angle of entrainment and free-running rhythms that derived from the predicted phase (Supplementary Fig. 1A, 1C), both measures of true entrainment. Lastly, LD20:4 decreased free-running period by ~30 min (Supplementary Fig. 1D), similar to previous results using this species (Pittendrigh and Daan, 1976a). Collectively, these results indicate that PER2::LUC mice entrain to this long day length condition.
Figure 1. SCN reorganization under long day lengths.
A) Representative double-plotted actograms depicting wheel-running rhythms of individual PER2::LUC mice entrained to LD12:12 or LD20:4. Lighting conditions are represented by internal yellow shading and the light:dark bar above each actogram. Only the last two weeks of entrainment are illustrated for clarity. B) Still images of PER2::LUC expression from SCN slices collected from LD12:12 or LD20:4 mice (see also Supplementary Videos 1–2). Number in upper left corner indicates the number of hours in vitro, and the left lobe of each SCN is outlined in yellow to highlight the compartmental nature of PER2::LUC expression within the LD20:4 slice. C) Individual phase maps depicting peak time relative to the field rhythm of the whole slice on the first cycle in vitro. Cool and warm colors indicate early- and late-peaking regions, respectively. D) PER2::LUC time series from dorsal SCN (d, dSCN) and central SCN (c, cSCN) regions used to assess rhythms in the SCN shell and core, respectively. E) On the first cycle in vitro, LD20:4 altered the peak time of the cSCN region relative to that for the dSCN region (n=12/condition), such that the cSCN peaked earlier than the dSCN. * Student’s t-test, p < 0.0001. F) LD20:4 increased the level of PER2::LUC expression within the cSCN (n=12/condition). Still images of total PER2::LUC expression within representative SCN slices over the first 24h in vitro. PER2 expression level was quantified by summing brightness values for the cSCN region expressed relative to that for the dSCN region (n=12/condition). * Student’s t-test, p < 0.0001. See also Figure S1.
To investigate photoperiodic changes in pacemaker organization, coronal SCN slices were collected from PER2::LUC mice held under LD12:12 or LD20:4 (Fig. 1B). Real-time bioluminescence imaging of PER2::LUC expression was conducted in vitro and SCN spatiotemporal organization was mapped (see Methods). Consistent with previous work (Evans et al., 2011), SCN slices from LD12:12 mice showed regional PER2::LUC peak time differences ranging from 2–4 hours on the first cycle in vitro (Fig. 1C, Supplementary Fig. 1E, Supplementary Video 1). In contrast, LD20:4 slices displayed a much larger range of PER2::LUC peak times, with reorganization of two spatially distinct subpopulations (Fig. 1C, Supplementary Fig. 1E, Supplementary Video 2). In particular, LD20:4 slices were characterized by a central region that phase-led a surrounding semi-concentric region by ~6h on the first cycle in vitro (Fig. 1C–E, p < 0.0001), an organizational pattern resembling the functionally distinct SCN compartments oftentimes referred to as “core” and “shell” (Abrahamson and Moore, 2001; Antle et al., 2003). Indeed, the dense population of arginine vasopressin neurons that demarcates the SCN shell compartment was in spatial registry with the late-peaking shell-like region but not the early-peaking core-like region (Fig. 2). In addition to changing the spatiotemporal organization of the SCN network, LD20:4 increased the level of PER2::LUC expression within the central SCN on the first cycle in vitro (Fig. 1F, p < 0.0001). SCN slices from LD12:12 mice did not display these organizational or functional changes regardless of time of dissection (Supplementary Fig. 2A), indicating that the changes observed within LD20:4 slices are specifically associated with the long day length condition and not attributable to differences in the time of sample preparation. Importantly, LD20:4 produced region-specific changes in the timing and level of PER2 expression in vivo (Supplementary Fig. 2B–C), consistent with those observed in vitro. These results confirm our prediction that in vivo exposure to long days reorganizes the SCN network into two subpopulations that cycle out of phase, and demonstrate that the specific spatiotemporal pattern involves dissociation of SCN shell and core compartments.
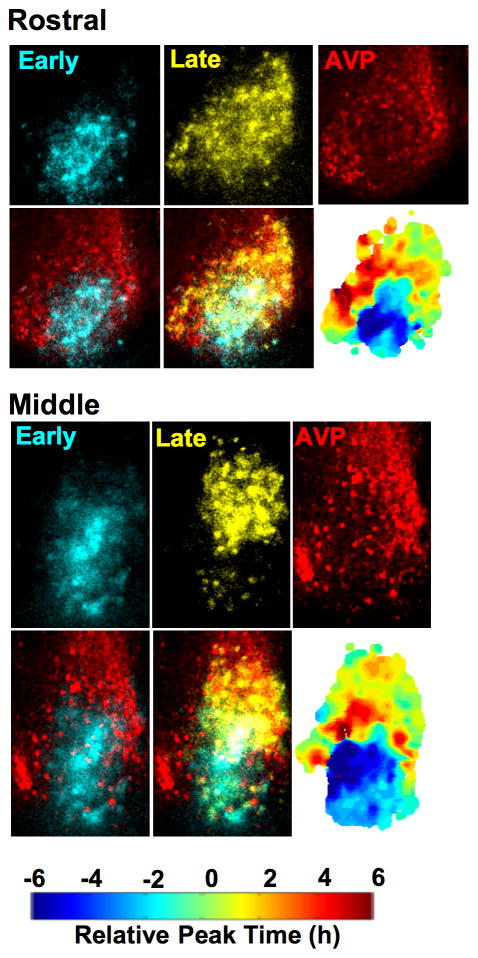
Figure 2. Photoperiodic reorganization of SCN shell and core regions.
Coronal SCN slices collected from LD20:4 mice were imaged for 2 days and then processed for arginine vasopressin (AVP, red)-immunoreactivity. Individual still images of PER2::LUC expression were selected to isolate the early- (blue pseudocolor) and late-peaking (yellow pseudocolor) regions on the first cycle in vitro before superimposition onto AVP-ir images. The results indicate that LD20:4 reorganizes the SCN into shell and core regions cycling in anti-phase, since the AVP-ir neurons that demarcate the SCN shell are in spatial registry with the late-peaking region but not with the early-peaking core-like region. Individual phase maps are indicated for each slice. See also Figure S2.
We further tested whether day length in vivo is proportional to the peak time difference between SCN shell and core regions in vitro by collecting SCN slices from PER2::LUC mice housed under a range of long day lengths (i.e., LD12:12, LD16:8, LD18:6, LD20:4, LD22:2). Average phase maps constructed for each photoperiod reveal that the magnitude of the shell-core peak time difference increased with day length (Fig. 3A). While SCN slices from LD12:12 mice lacked a clear distinction between shell and core compartments, the peak time difference between SCN shell and core regions increased in a manner proportional to day length (Fig. 3A-B, p < 0.0001, see also Supplementary Fig. 3). In contrast, the phase relationship between two spatially distinct shell regions (dorsal and lateral SCN) was not significantly influenced by day length (Supplementary Fig. 3, p = 0.12). Moreover, level of PER2::LUC expression within the SCN core increased with the magnitude of the shell-core peak time difference (Fig. 3C, R2 = 0.51, p < 0.0001, see also Supplementary Fig. 3). In contrast, long days did not systematically affect regional period length (Supplementary Fig. 3). Thus, while the SCN core is often described as a non-rhythmic or weakly rhythmic compartment in terms of intrinsic genetic or electrical expression (Antle et al., 2003), these data indicate that this region is capable of robust oscillations that are not evident under standard lighting conditions when the network is in a typical configuration. These results adds to a small body of research indicating that the SCN core is capable of robust gene expression rhythms depending on environmental conditions (Butler et al., 2012; Yan, 2009). While previous work could not distinguish between light-driven and intrinsically-rhythmic gene expression, here increased PER2::LUC expression within the SCN core was maintained in the absence of photic stimulation in vitro, which suggests that the basis of this plasticity derives from changes at the cellular and/or network level.
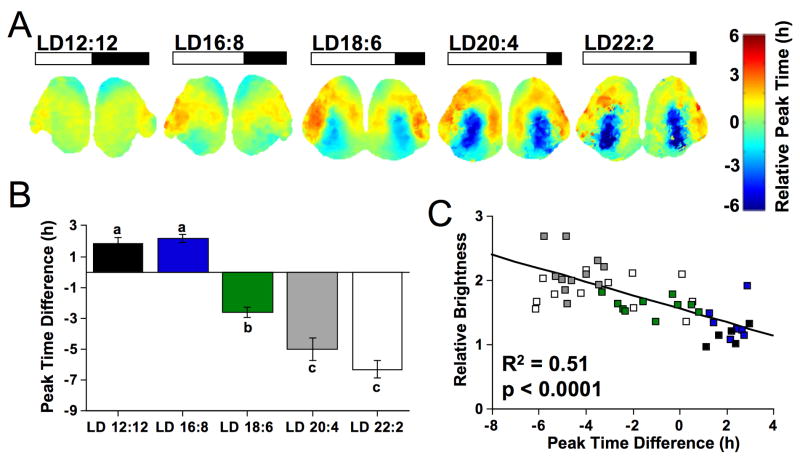
Figure 3. Photoperiodic changes in the organization and function of the SCN network.
A) Average phase maps illustrating SCN organization across a range of long day conditions (n =7–13/condition), with peak time relative to the field rhythm of the whole slice on the first cycle in vitro. B) The peak time difference between cSCN and dSCN regions increased with day length. Regions used for analyses as illustrated in Fig. 1D. Letters represent groups that differ significantly, ANOVA followed by Tukey’s honestly significant difference, p < 0.005. C) Total PER2::LUC expression within the cSCN (relative to the dSCN) over the first 24h in vitro correlates with the initial shell-core peak time difference. Symbol colors represent different day length conditions as indicated in B. See also Figure S3.
Network resynchronization through dynamic changes in regional phase relationships
Next, we determined whether the reorganized SCN network could resynchronize when fully intact in vivo, a necessary prerequisite to studying resynchronization in vitro. PER2::LUC mice were entrained to LD12:12 or LD20:4 before release into constant darkness (DD) for 1, 4, 7, 14, or 21 days. As expected, SCN slices collected from mice held under LD20:4 displayed a large difference in the shell-core peak time, which was maintained on the first day after release into DD (Supplementary Fig. 4). After four days in DD, the shell-core peak time difference was still evident although diminished in magnitude relative to mice under LD20:4 (Supplementary Fig. 4). Finally, after one week in DD, the SCN network had returned to an organizational state like that observed under LD12:12 (Supplementary Fig. 4). Consistent with previous work (Evans et al., 2011), the spatiotemporal organization of LD12:12 slices was not markedly altered by DD (Supplementary Fig. 4). These data indicate that the network reorganization induced by LD20:4 is not permanent and that SCN neurons are able to resynchronize in vivo through a process that is complete within one week.
To test whether the reorganized SCN retains the ability to resynchronize in vitro, we tracked changes in network organization in LD20:4 and LD12:12 slices over time in culture (Fig. 4). While the spatiotemporal organization of LD12:12 slices changed little over time in vitro, LD20:4 slices displayed organizational changes and a decrease in the magnitude of peak time difference between shell and core regions (Fig. 4A). To further examine this process, we used regional analyses to quantify changes in the shell-core peak time difference over the first four cycles in vitro (Fig. 4B–D). In contrast to LD12:12 slices, LD20:4 slices displayed large changes in the shell-core phase relationship over time in vitro (Fig. 4B, p < 0.005), with the magnitude of change positively correlated with the initial peak time difference between SCN shell and core regions (Fig. 4C, R2 = 0.44, p < 0.001). When tracked on a cycle-by-cycle basis, half of the LD20:4 slices appeared to resynchronize with the SCN core shifting earlier (i.e., through phase advances, Fig. 4D), while the other half appeared to resynchronize with the SCN core shifting later (i.e., through phase delays, Fig. 4D). Directional differences in dynamic behavior over time in vitro depended on the magnitude of the initial peak time difference (post hoc t-test, p < 0.05), with the SCN core phase advancing or phase delaying depending on whether the initial shell-core phase difference was larger or smaller than 6h, respectively.
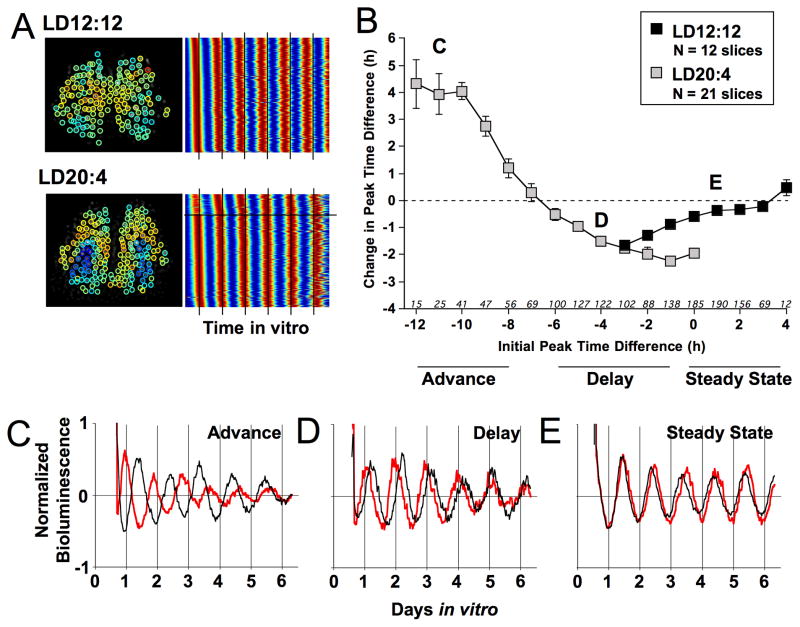
Figure 4. Changes in SCN network organization over time in vitro.
A) Average phase maps for LD12:12 and LD20:4 slices over time in vitro (first cycle in Fig 3A). Note that the magnitude of difference in peak time between shell and core regions deceases over the first four cycles in vitro. B) LD20:4 increased the change in peak time for the cSCN region. Absolute change in peak time difference between the cSCN and dSCN regions was calculated for the fourth cycle in vitro. C) In LD20:4 slices, change in peak time for the cSCN region was positively correlated with the initial shell-core peak time difference. D) Vertical plot depicting changes in the shell-core peak time difference over the first four cycles in vitro. On average, the cSCN region in LD12:12 slices (black squares) displayed a peak time similar to that for the dSCN (i.e., peak time difference is close to 0) and this relationship changed little over time. In LD20:4 slices (gray symbols), the cSCN region appears to gradually re-align into a configuration like that displayed by LD12:12 slices, with two distinct patterns of resetting evident. Note that LD12:12 data are double-plotted at −24 and 0 to visualize LD20:4 resetting in each direction. * Student’s t-test, p < 0.005. See also Figure S4.
To further investigate the phase-dependent nature of these resetting responses, we used cell-based computational analyses to track individual SCN neurons over time in vitro (Fig. 5). SCN neurons within LD12:12 slices showed stable phase relationships and similar period lengths over time in vitro, but SCN neurons within LD20:4 slices displayed larger differences in initial peak time and larger changes over time in vitro (Fig. 5A). Using all SCN core cells extracted from all slices, we next constructed a response curve to investigate whether the resetting responses of SCN core neurons were systemically related to the initial phase relationship with SCN shell neurons. The resulting “coupling” response curve is curvilinear in nature (Fig. 5B), with two main resetting regions: 1) a phase advance region when SCN core cells phase lead by 8–12h and 2) a phase delay region when SCN core cells phase lead by 1–6h (Fig. 5B–D). In addition, there is a steady-state region where the initial phase lag of 0–3h in LD12:12 slices is maintained over the recording period (Fig. 5B, 5E). As expected in a circadian response curve, the zero crossing at the phase relation of 4h indicates a continuity in responses (Fig. 5B) that is further evident when resetting responses are partitioned across consecutive cycles (Supplementary Fig. 5). Additionally, consistency in the phase-dependent nature of this resetting response was observed across consecutive cycles, across cells, and across most photoperiodic conditions (Supplementary Fig. 5). Since phase-dependence is a fundamental property of oscillator synchronization (Hansel et al., 1995), the curvilinear nature of this response curve, along with its consistency and continuity, strongly suggests that this dynamic behavior reflects coupling among SCN neurons. The coupling response curve generated here is analogous to a traditional phase response curve but is unique in that it characterizes the response of SCN neurons to a phase-shifting stimulus provided by the network itself, rather than an exogenous stimulus. Without knowledge of the precise signals SCN neurons use to influence one another, we view this formal analysis of SCN coupling mechanisms as a first step in understanding the functional roles of different signaling cues (Aton and Herzog, 2005; Maywood et al., 2011).
Figure 5. Phase-dependent changes in SCN organization over time in vitro.
A) Background-subtracted images of LD12:12 and LD20:4 slices illustrating cellular regions of interest (ROIs, with colored circle indicating cellular peak time relative to the field rhythm of the whole slice (color scale as in Fig. 1C), with heat-map style raster plots illustrating PER2::LUC rhythms from all cell-like ROIs extracted from each slices (high and low PER2::LUC expression indicated by red and blue, respectively). LD12:12 neurons display stable phase relationships over time in vitro, as evident by the stability of peak times relative to the line superimposed at 24h intervals. In contrast, LD20:4 neurons display larger initial differences in peak time, which diminish over time in vitro. Note that the horizontal line within the LD20:4 record separates data from core and shell neurons. B) Coupling response curve for SCN cells constructed with a 2h running average. Initial peak time difference was defined on the second cycle in vitro, with change quantified over the subsequent two cycles in vitro. According to convention, phase advances and delays are positive and negative values, respectively. Letters superimposed onto specific points of the coupling response curve correspond to representative cell data below. Italicized numbers along abscissa indicate number of core cell-like ROIs within each bin (LD20:4 sample size for initial phase difference of −3 to 0: 99, 70, 42, 12). C–E) PER2::LUC time series for representative dSCN (black) and cSCN cell-like ROIs (red) illustrating dynamic changes over time in vitro. After LD20:4, cSCN cell-like ROIs displayed either phase advances (C) or phase delays (D). Note that the SCN core cell moves toward a more typical phase relation with the SCN shell cell in both the advance and delay direction, with stable realignment occurring within the recording interval for the latter. After LD12:12, the peak time difference between dSCN and cSCN cell-like ROIs is small and relatively stable (E). See also Figure S5.
Dynamic changes in SCN network organization mediated by intercellular signaling mechanisms
SCN neurons influence one another through intercellular communication mediated by synaptic, electrical, and paracrine signaling (Aton and Herzog, 2005; Maywood et al., 2011). To directly test the hypothesis that dynamic changes in network organization in vitro reflect intercellular communication mediated by synaptic communication, we assessed whether dynamic changes in network organization would be abolished by tetrodotoxin (TTX). Since TTX attenuates bioluminescence rhythms of organotypic SCN slices (Buhr et al., 2010; Yamaguchi et al., 2003), but not acutely dissected SCN slices (Baba et al., 2008), we first tested the efficacy and side effects of TTX within the context of our preparation. SCN slices were collected from LD12:12 mice and immediately cultured with medium containing 2.5μM TTX. As expected, TTX increased the phase dispersion of SCN cells measured on the fifth cycle in vitro (Supplementary Fig. 6A), but did not alter the rhythmic properties of SCN core cells within LD12:12 slices (Supplementary Fig. 6D). Thus, TTX application within this preparation effectively suppressed cellular communication without compromising single cell oscillatory function.
SCN slices were collected from PER2::LUC mice entrained to either LD12:12 or LD20:4, then cultured with 2.5μM TTX. TTX did not markedly alter photoperiod-induced changes in SCN organization or function (Supplementary Fig. 6E, 6F), but greatly attenuated resetting responses over time in vitro (Fig. 6A, Supplementary Fig. 6F). Specifically, TTX reduced the area under the advance and delay portion of the coupling response curve by 82% and 55%, respectively (Fig. 6A, Fig. 7). Thus, dynamic changes in network organization were greatly attenuated by TTX, consistent with a primary mechanism dependent on Na+-dependent action potentials and conventional synaptic transmission. Further, these data indicate that dynamic changes in network organization over time in vitro is an active process mediated by neuronal coupling rather than a passive process mediated by regional period differences. Since TTX blocks period synchronization and enhances the ability to detect intrinsic period differences, TTX would be expected to increase the magnitude of phase changes due to regional period differences. Instead, TTX largely abolishes the coupling response curve. Small residual changes in the presence of TTX may reflect intrinsic regional differences in period length (Myung et al., 2012) or forms of intercellular communication that are less sensitive to TTX (Aton and Herzog, 2005; Maywood et al., 2011).
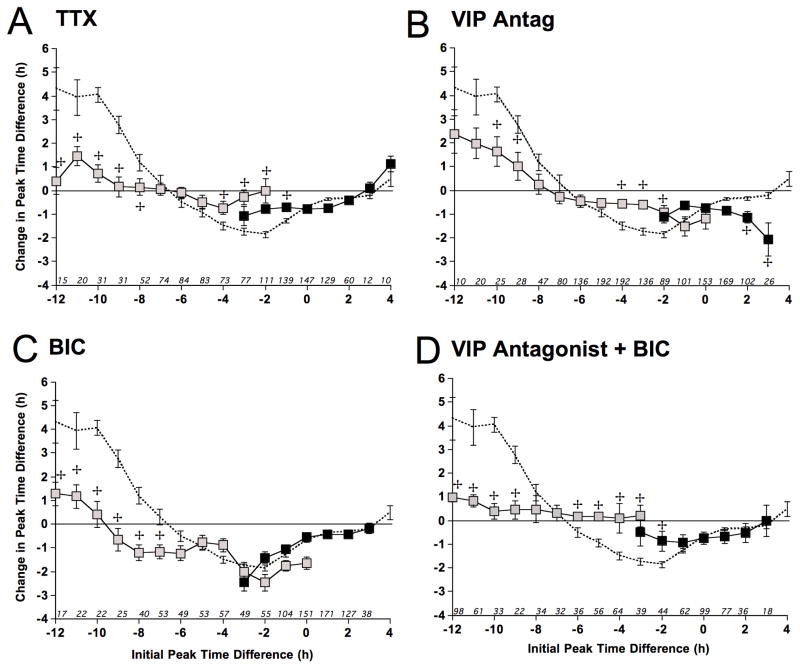
Figure 6. Intracellular signaling mechanisms mediating SCN resynchronization in vitro.
A) Coupling response curve for SCN cells cultured with 2.5μM TTX constructed with a 2h running average. Dashed curve illustrates the vehicle response curve from Figure 5. LD20:4 sample size for initial phase difference of −3 to −2: 57, 20. B) Coupling response curve for SCN cells cultured with 20μM [4Cl-D-Phe6, Leu17] VIP (VIP Antag) constructed with a 2h running average. LD20:4 sample size for initial phase difference of −2 to 0: 62, 23, 10. C) Coupling response curve for SCN cells cultured with 200μM BIC constructed with a 2h running average. LD20:4 sample size for initial phase difference of −3 to 0: 40, 38, 54, 35. D) Coupling response curve for SCN cells cultured with both 20μM [4Cl-D-Phe6, Leu17] VIP and 200μM BIC constructed with a 2h running average. LD20:4 sample size for initial phase difference of −3: 24. For A–D: LD20:4 and LD12:12 data are represented by gray and black symbols, respectively.
 Indicates significant differences relative to the same timepoint in the vehicle curve, Full Factorial ANOVA followed by Least Square Mean Contrasts, p < 0.003. See also Figure S6.
Indicates significant differences relative to the same timepoint in the vehicle curve, Full Factorial ANOVA followed by Least Square Mean Contrasts, p < 0.003. See also Figure S6.
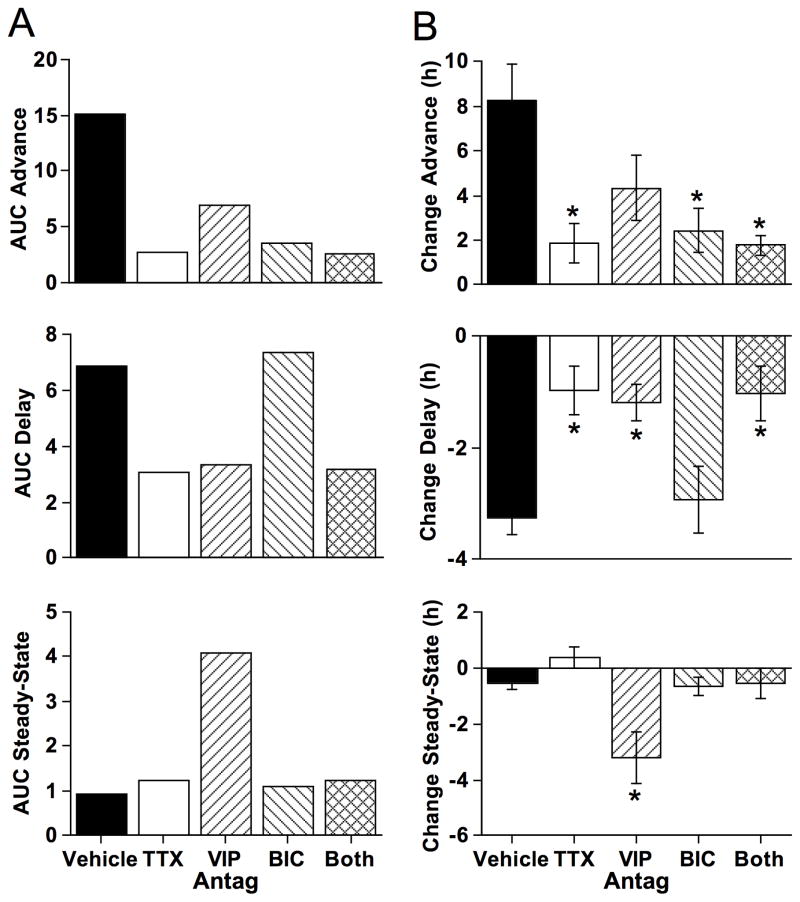
Figure 7. Pharmacological manipulation of SCN coupling responses.
A) Area under the curve (AUC) for the advance, delay, and steady-state regions (as defined below the abscissa in Fig. 5B). B) Differential effects of different pharmacological agents on resynchronization and maintenance of steady-state phase relationships. Effects of pharmacological manipulations were compared using cellular data within the advance portion of the curve (all cells with an initial peak time difference between −11 and −12) delay portion of the curve (all cells with an initial peak time difference between −4 and −3), and steady-state portion of the curve (all cells with an initial peak time difference between +2 and +3). Sample sizes for each group are indicated along the abscissa of Figure 5B and Figure 6A–D. * Full Factorial ANOVA followed by post hoc Student’s t test with Bonferroni correction, p < 0.01.
VIP signaling is important for both steady-state phase relations and network resynchronization
VIP meets many of the criteria for an important SCN coupling factor, including lack of synchrony among SCN neurons during pharmacological or genetic elimination of VIP-signaling (Aton and Herzog, 2005). Importantly, synchrony is re-established in VIP−/− SCN slices by in vitro application of a VIP receptor agonist (Aton et al., 2005), but can also be re-established by GRP or K+-induced depolarization (Brown et al., 2005; Maywood et al., 2006). Recent co-culture experiments with VIP−/− slices further highlight the import of VIP signaling and indicate that there is viable compensation through a variety of other signaling pathways (Maywood et al., 2011). That a subset of VIP knockout animals continue to display robust rhythms in behavior and SCN function further suggests that non-VIP signals can effectively couple the network (Brown et al., 2005; Ciarleglio et al., 2009). Since these studies using genetic knockout models provide strong evidence that VIP is an important SCN coupling factor, we next investigated its role in our functional coupling assay using a genetically intact SCN circuit.
Since the dynamic process of SCN coupling involves intercellular signaling over several days in vitro, we first determined the efficacy and side-effects of VIP receptor antagonism within the context of our preparation. LD12:12 slices were incubated with either vehicle (ddH20) or 20μM VIP receptor antagonist [4Cl-D-Phe6, Leu17] VIP, as in (Atkins et al., 2010). At the time of the fourth peak in vitro, either vehicle (ddH20) or 20μM VIP was added to the culture medium. VIP produced a large reduction in the amplitude of the PER2::LUC rhythm, consistent with (An et al., 2011), and this amplitude reduction was fully blocked by [4Cl-D-Phe6, Leu17] VIP (Supplementary Fig. 6B). Application of [4Cl-D-Phe6, Leu17] VIP did not alter the rhythmic properties of SCN cells or decrease the number of rhythmic cells within LD12:12 slices (Supplementary Fig. 6D). Based on this functional assay, we conclude that application of [4Cl-D-Phe6, Leu17] VIP within this preparation effectively suppresses VIP signaling for at least four days in vitro without the compromised single cell oscillatory function commonly observed in genetic models with deficient VIP signaling (Brown et al., 2005; Ciarleglio et al., 2009; Maywood et al., 2011; Maywood et al., 2006).
To test whether VIP signaling contributes to dynamic changes in network organization in vitro, SCN slices from LD12:12 and LD20:4 mice were cultured with 20μM [4Cl-D-Phe6, Leu17] VIP added to the medium at the start of the recording. VIP receptor antagonism did not eliminate photoperiod-induced changes in SCN organization or function (Supplementary Fig. 6E, 6F), but partially blocked network resynchronization over time in vitro (Fig. 6B, Supplementary Fig. 6F). In particular, [4Cl-D-Phe6, Leu17] VIP attenuated both the advance and delay portion of the coupling response curve, reducing the area under the curve by 56% and 44%, respectively (Fig. 6B, Fig. 7). Moreover, [4Cl-D-Phe6, Leu17] VIP destabilized the steady-state portion of the response curve such that LD12:12 slices did not maintain the typical network organization over time in vitro (Fig. 6B, Fig. 7, Supplementary Fig. 6F). These results reveal that VIP signaling not only contributes to the maintenance of steady-state phase relationships but also plays a role during network resynchronization after photoperiodic reorganization. Further, since TTX and VIP receptor antagonism had differential effects on the amplitude of phase advances (Fig. 7B), this suggests that other signals may contribute to resynchronization. Lastly, since VIP receptor antagonism, but not TTX, destabilized steady-state network organization (Fig. 7B), this suggests that network desynchrony is a response to another signaling mechanism that is typically inhibited by VIP signaling and blocked by TTX.
State-dependent role of GABAA signaling in regulating network organization
Previous research indicates that SCN neurons interact through multiple, seemingly redundant signaling mechanisms, but the specific roles of different coupling factors has been difficult to define (Aton and Herzog, 2005; Welsh et al., 2010). GABA is a putative SCN coupling factor that is expressed in nearly all SCN neurons (Abrahamson and Moore, 2001), which acts on the GABAA receptor to regulate the amplitude of SCN electrical rhythms in vitro (Aton et al., 2006), synchronize dispersed SCN neurons (Liu and Reppert, 2000), and facilitate communication between the ventral and dorsal SCN during propagation of photic input (Albus et al., 2005; Han et al., 2012). However, the most recent work on the role of GABAergic signaling found that it was not required for maintaining network synchrony within an intact organotypic SCN slice (Aton et al., 2006). Since GABA is the most abundant neurotransmitter within the SCN, clarifying its role in regulating network-level properties is essential for understanding SCN function.
As with [4Cl-D-Phe6, Leu17] VIP, we first determined the efficacy and side-effects of GABA antagonism within the context of our preparation. LD12:12 slices were cultured with either vehicle (ddH20) or 200μM of the GABAA receptor antagonist bicuculline (BIC), then provided with vehicle (ddH20) or 20μM GABA at the time of the fourth peak in vitro. GABA produced a phase delay in the PER2::LUC rhythm, consistent with (Liu and Reppert, 2000), and this phase delay was blocked by BIC (Supplementary Fig. 6C). Consistent with previous research (Aton et al., 2006), BIC did not alter the rhythmic properties of SCN core cells or decrease the number of rhythmic cells within LD12:12 slices (Supplementary Fig. 6D). Thus, BIC application effectively suppresses GABAA signaling over time in vitro without altering single cell oscillatory function.
To test whether GABAA signaling contributes to network resynchronization in vitro, LD12:12 and LD20:4 slices were cultured with 200μM BIC added to the medium. BIC did not eliminate photoperiod-induced changes in SCN organization or function (Supplementary Fig. 6E, 6F), but did inhibit network resynchronization over time in vitro (Fig. 6C, Supplementary Fig. 6F). In particular, BIC attenuated the phase advance portion of the coupling response curve by 71%, an effect similar to that produced by TTX and larger than that produced by VIP receptor antagonism (Fig. 6C, Fig. 7), which reveals that GABAA signaling contributes to network coupling when SCN core cells are close to anti-phase. In contrast, BIC did not attenuate phase delays like TTX or the VIP receptor antagonist and did not destabilize the steady-state portion of the coupling response curve like the VIP receptor antagonist (Fig. 6C, Fig. 7), indicating that non-GABAA signaling mechanisms facilitate synchrony when the network is in less polarized states. Lastly, the steady-state portion of the coupling response curve is stable when both BIC and the VIP receptor antagonist are applied (Fig. 6D, Fig. 7), indicating that the destabilization produced during VIP antagonism is a response caused by GABAA signaling. Collectively, this pattern of results suggests that GABAA signaling promotes network synchrony in an anti-phase state, but opposes network synchrony in a steady-state configuration. This state-dependent role for GABAA signaling may account for previous results indicating that GABA is sufficient to synchronize dissociated SCN neurons (Liu and Reppert, 2000), but its absence does not desynchronize the SCN network under steady-state conditions (Aton et al., 2006).
Discussion
Here we develop a functional assay of SCN coupling that uniquely captures the dynamic process by which SCN neurons interact. First, we confirm that long day lengths alter the spatiotemporal organization of the SCN network, finding that the specific pattern of reorganization involves temporal dissociation of SCN shell and core compartments. We then exploit this alternative operational state to investigate, for the first time, the process of network resynchronization with real-time bioluminescence imaging of single SCN neurons over time in vitro. Our results provide the first experimental evidence that SCN neurons interact in a dynamic manner through phase-dependent resetting responses, which is a staple of mathematical models of oscillator coupling but has been difficult to demonstrate experimentally, even in invertebrate pacemaker preparations. That phase-dependent responses likewise characterize SCN responses to environmental cues support the theory that this process is essential for all forms of synchronization (Hansel et al., 1995; Smeal et al., 2010). The coupling response curve constructed here is the first of its kind in circadian biology and its continued use has the potential to provide further insight into the mechanisms by which SCN neurons influence one another to regulate network-level properties.
By developing and employing this novel in vitro assay of SCN interactions, our work reveals that SCN neurons are coupled through non-redundant signaling mechanisms whose functional roles are influenced by the state of the network. Predating this work, there was strong evidence supporting a role for VIP in maintaining SCN network synchrony (Aton and Herzog, 2005). However, similar evidence for GABA was relatively modest, with no apparent role for GABAA signaling in maintaining network synchrony (Aton et al., 2006). Our results reveal that both VIP and GABAA signaling contribute to SCN coupling, but that the roles of these SCN coupling factors are functionally distinct. Notably, we find that GABAA signaling contributes to SCN coupling specifically when the network is in a polarized state, but opposes synchrony under steady-state conditions. Further, our results indicate that VIP acts together with GABAA signaling to promote resynchronization when the network is in an anti-phase configuration, but opposes the actions of GABAA signaling to promote network synchrony under steady-state conditions. That VIP and GABAA signaling exert opposing actions under steady-state conditions, with the latter destabilizing network synchrony, is consistent with a recent report investigating functional connections between SCN neurons in culture (Freeman et al., 2013). Our study complements and extends this work, demonstrating that GABAA signaling can either inhibit and promote network synchrony in a manner that depends on the state of the network. This state-dependent role of GABAA signaling may reflect phase-dependent resetting responses (i.e., GABA advances SCN cells near anti-phase, but delays those in-phase), as predicted by the phase response curve for GABA (Liu and Reppert, 2000). In addition, chronic light stimulation during long day entrainment may produce intrinsic changes in neuronal function, altered synaptic inputs to SCN cells, or other experience-dependent changes in network architecture (Koch et al., 2011; Marder and Goaillard, 2006). Potentially substantiating this latter possibility, we find that long day entrainment increases the level of PER2::LUC expression within the SCN core, which suggests that this condition induces changes in cellular and/or network signaling. Collectively, our study reveals new forms of SCN plasticity, and prompts further study of its mechanistic bases, biological relevance, and state-dependence.
After reorganization, SCN core and shell neurons resynchronize to re-establish a steady-state network organization, which indicates that these SCN compartments are coupled through bidirectional lines of communication. Since most studies to date have found anatomical connections traveling only from SCN core to shell neurons, this study provides the best evidence to date for the functional transmission of information in the opposite direction. First, we find that VIP signaling contributes to network synchronization in both steady-state and reorganized states, which confirms and extends previous work using genetic models deficient in VIP signaling. Since VIP is produced exclusively by neurons within the SCN core, VIP in this context likely acts as a cue transmitted from the SCN core to the SCN shell. Thus, this result indicates the presence of another coupling signal transmitted from the shell that directly resets SCN core neurons. We tested whether GABAA signaling may serve this role since GABA is synthesized and processed in nearly all SCN neurons (Abrahamson and Moore, 2001; Belenky et al., 2008). We find that GABAA signaling contributes to network resynchronization when the SCN network is in an anti-phase state but not in less polarized states. This further indicates that at least one other signal is transmitted from the shell to reset SCN core neurons and produce network resynchronization in less polarized states. Given the lack of compelling evidence for synaptic connections from shell to core neurons, this yet-to-be-identified signal may be paracrine in nature (Maywood et al., 2011; LeSauter and Silver, 1998).
Further use of this functional coupling assay has the potential to reveal additional aspects of SCN circuitry that would be difficult to detect with the exclusive use of loss-of-function genetic models. In addition to the common developmental confounds associated with germ line mutations, murine models lacking VIP or GABAA signaling display deficits in photic entrainment and resetting (Han et al., 2012; Dragich et al., 2010; Hughes et al., 2004), which can limit the utility of these models to investigate the specific role of these factors in intrinsic network coupling. The use of a genetically intact model within the present study circumvents these issues and allows us to exploit light-induced changes in network organization to investigate the functional roles of VIP and GABAA signaling in SCN coupling. This also provides an analytical tool to investigate in an unprecedented manner the mechanisms by which the network uses and integrates different SCN coupling factors. While our current approach necessitates the use of long-lasting, potent, and selective pharmacological agents that may incur non-specific effects (Teshima et al., 2003), previous work indicates that these non-specific effects are short-lasting and minimally affect long-term SCN recordings (Aton et al., 2006). Further, the pharmacological agents used here affected some, but not all, portions of the coupling response curve, which demonstrates the specific nature of these effects. Nevertheless, it would be of interest to take advantage of other techniques to manipulate SCN signaling pathways (Brancaccio et al., 2013; Miesenbock, 2011) within the context of this functional coupling assay.
Here we demonstrate that an ecologically relevant environmental manipulation (i.e., day length) markedly alters the operational state within the SCN network. While the precise mechanisms by which light alters SCN network organization and function remain unclear, it is possible that chronic long day exposure alters network signaling and cellular function. Changes in circadian function such as those demonstrated here may occur during exposure to the long day lengths that are common in industrialized countries. The significance of day length is underscored by photoperiodic changes in many behavioral and physiological systems of model animal species, including reproductive function (Goldman, 1999), immune function (Nelson, 2004), metabolic function (Bartness and Wade, 1985), cognitive function (Pyter et al., 2006), and affective behavior (Pyter and Nelson, 2006; Trainor et al., 2008). Humans and other primates possess the biological machinery for responding to photoperiod and display seasonal rhythms in a variety of physiological functions, including conception and susceptibility to illness (Foster and Roenneberg, 2008). Importantly, the changes in mood that manifest during Seasonal Affective Disorder are thought to be produced by light-induced alterations of circadian function (Wehr et al., 2001). Whether the changes in SCN organization and function contribute to such photoperiodic fluctuations demands further investigation, since this may be pertinent for understanding human health problems associated with nighttime exposure to artificial light and seasonal changes in daily light exposure. Lastly, SCN reorganization occurs under conditions that simulate jetlag and shift work (Albus et al., 2005; Sellix et al., 2012), and thus strategic manipulation of SCN coupling pathways may facilitate recovery under these conditions.
Progress in understanding network-level function requires rigorous mapping of circuit organization under various operating states and a mechanistic understanding of the processes that regulate network synchrony and plasticity (Buzsaki, 2006). Temporal coding within oscillating neuronal networks is an organizational principle important for many behavioral and physiological processes, including sensory processing, motor performance, intermodal integration, and higher-order cognitive function (Buzsaki, 2006; Schnitzler and Gross, 2005). Plasticity in the temporal organization of neural circuits is likewise proposed to be critical for the context-specific regulation of behavior and physiology (Buzsaki, 2006). Further, dysfunction in the process of neuronal synchronization is implicated in epilepsy and cognitive impairment (Schnitzler and Gross, 2005). Since the principles of neural synchronization apply to systems operating on a range of timescales (Buzsaki and Draguhn, 2004; Hansel et al., 1995), the present studies highlight organizational principles likely relevant for other oscillatory networks (Buzsaki, 2006).
Experimental Procedures
Animals and Husbandry Conditions
All procedures were approved by the Institutional Animals Care and Use Committee of the Morehouse School of Medicine in accordance with the guidelines of the US National Institutes of Health. Homozygous PERIOD2::luciferase (PER2::LUC) knock-in mice (Yoo et al., 2004), backcrossed to a C57Bl/6J background, were bred and raised under a 24h light:dark cycle with 12h light and 12h darkness (LD12:12, lights-on: 0600 EST). Ambient room temperature was maintained at 22 ± 2° C, and animals had ab libitum access to water and food (Purina Rodent Chow #5001, St. Louis, MO). For all experiments, adult male PER2::LUC mice (7–9 weeks of age) were transferred to individual wheel-running cages contained within light-tight secondary enclosures. Long day photoperiods were achieved by an abrupt and symmetrical reduction of the scotophase. Mice were entrained to either LD12:12, LD16:8, LD18:6, LD20:4, or LD22:2 for 12 weeks. LD20:4 entrainment for less than 12 weeks produced SCN reorganization, but with individual differences in whether the pattern was evident (data not shown). Wheel-running rhythms were monitored and analyzed with the Clocklab data collection and analysis system (Actimetrics, Evanston, IL).
Bioluminescence Imaging
Coronal SCN slices (150 μm) were collected and imaged as described previously (Evans et al., 2011). Unless otherwise stated, mice were sacrificed 2–4 hours before lights-off since dissections during late subjective day do not reset the phase of the SCN (Davidson et al., 2009). Each SCN slice was cultured on a membrane (Millicell-CM, Millipore) with 1.2mL of air-buffered medium containing 0.1mM beetle luciferin (Molecular Imaging Products) and imaged for 5–7 days using a Stanford Photonics XR Mega 10Z cooled intensified charge-coupled device camera. For drug treatments, tetrodotoxin citrate (TTX, 2.5μM, Tocris, Cat#1069), the VIP receptor antagonist [4Cl-D-Phe6, Leu17] VIP (20μM, Tocris, Cat#3054), or bicuculline methchloride (BIC, 200μM, Sigma, Cat#B7686) was added to the culture medium and remained for the duration of the recording. For each pharmacological agent, drug dose was selected from published literature (Atkins et al., 2010; Aton et al., 2006; Yamaguchi et al., 2003), and we independently validated dose efficacy in our preparation (Fig. S6A–C).
Computational Analyses
Rhythmic parameters of PER2::LUC expression were calculated for each slice and for cell-like regions of interest (ROIs) within each slice using ImageJ and Matlab-based computational analyses, as described previously (Evans et al., 2011). Briefly, individual phase maps were constructed by generating a time series for each 12-pixel diameter region of the image meeting criteria for circadian rhythmicity: autocorrelation coefficient with 24h lag significant at α = 0.05, local maximum in the autocorrelation between 18h and 30h, and signal-to-noise ratio ≥ 1. For composite phase maps, a representative sample was selected to which all other samples were aligned, and PER2::LUC peak time was averaged across samples. To locate and extract data from cell-like ROIs, an iterative process was employed after background and local noise subtraction (Evans et al., 2011). To avoid edge effects during wavelet fitting (Leise and Harrington, 2011), cell-like ROI data were analyzed starting on the second cycle in vitro. Analyses of change over time in vitro focused on cycles 2–4 due to avoid a slight drift in the Z axis plane that becomes noticeable after the 4th cycle in vitro. Statistical analyses were performed with JMP software (SAS Institute, City). Values in figures and text are mean ± SEM.
Immunohistochemistry
To determine the neuropeptide phenotype of regions affected by long day lengths, SCN slices were imaged for 2 days, treated with colchicine (25μg/ml) for 24h at 37°C, fixed with 4% paraformaldehyde for 24h before sucrose cryoprotection, as in (Evans et al., 2011). To assess PER2 expression in vivo, brains were removed at four timepoints spanning the circadian cycle (n = 2–3/timepoint/condition) and fixed in 4% paraformaldehyde for 24h before sucrose cyroprotection and sectioning. Free-floating slices (40μm) were incubated for 48h with primary antibodies for PER2 (Millipore, 1:500) and/or AVP (Bachem, 1:1K), followed by 2h incubation with secondary antibodies (Jackson ImmunoResearch, Dylight 488, Dylight 594; 1:200). Images were obtained with a Zeiss LMS 700 confocal laser scanning microscope.
Supplementary Material
Acknowledgments
This research was supported by NIH grants U54NS060659, F32NS071935, and S21MD000101, the Georgia Research Alliance, and the NSF Center for Behavioral Neuroscience. We wish to thank Stanford Photonics and the Morehouse School of Medicine animal husbandry staff. We are also grateful to Matt Ellis for research assistance, Dr Morris Benveniste for reagents, and Drs. Elliott Albers, Jason DeBruyne, Robert Meller, and David Welsh for discussions and advice.
Footnotes
Supplementary Information is available online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Current Biology. 2005;15:886–893. doi: 10.1016/j.cub.2005.03.051. [DOI] [PubMed] [Google Scholar]
- An S, Irwin RP, Allen CN, Tsai CA, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011 doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: A network model of the brain clock. J Biol Rhythms. 2003;18:339–350. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins N, Jr, Mitchell JW, Romanova EV, Morgan DJ, Cominski TP, Ecker JL, Pintar JE, Sweedler JV, Gillette MU. Circadian integration of glutamatergic signals by little SAAS in novel suprachiasmatic circuits. PLoS One. 2010;5:e12612. doi: 10.1371/journal.pone.0012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Herzog ED. Come together, right…now: Synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci U S A. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Ono D, Honma S, Honma K. A TTX-sensitive local circuit is involved in the expression of PK2 and BDNF circadian rhythms in the mouse suprachiasmatic nucleus. Eur J Neurosci. 2008;27:909–916. doi: 10.1111/j.1460-9568.2008.06053.x. [DOI] [PubMed] [Google Scholar]
- Bartness TJ, Wade GN. Photoperiodic control of seasonal body weight cycles in hamsters. Neurosci Biobehav Rev. 1985;9:599–612. doi: 10.1016/0149-7634(85)90006-5. [DOI] [PubMed] [Google Scholar]
- Belenky MA, Yarom Y, Pickard GE. Heterogeneous expression of gamma-aminobutyric acid and gamma-aminobutyric acid-associated receptors and transporters in the rat suprachiasmatic nucleus. J Comp Neurol. 2008;506:708–732. doi: 10.1002/cne.21553. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78:714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J Neurosci. 2005;25:11155–11164. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MP, Rainbow MN, Rodriguez E, Lyon SM, Silver R. Twelve-hour days in the brain and behavior of split hamsters. Eur J Neurosci. 2012;36:2556–2566. doi: 10.1111/j.1460-9568.2012.08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G. Rhythms of the Brain. Oxford: Oxford University Press; 2006. [Google Scholar]
- Buzsaki G, Draguhn A. Neuronal oscillations in cortical networks. Science. 2004;304:1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- Ciarleglio CM, Gamble KL, Axley JC, Strauss BR, Cohen JY, Colwell CS, McMahon DG. Population encoding by circadian clock neurons organizes circadian behavior. J Neurosci. 2009;29:1670–1676. doi: 10.1523/JNEUROSCI.3801-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur J Neurosci. 2009;29:171–180. doi: 10.1111/j.1460-9568.2008.06534.x. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One. 2011;6:e15869. doi: 10.1371/journal.pone.0015869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster RG, Roenneberg T. Human responses to the geophysical daily, annual and lunar cycles. Curr Biol. 2008;18:R784–R794. doi: 10.1016/j.cub.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Freeman GM, Jr, Krock RM, Aton SJ, Thaben P, Herzog ED. GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron. 2013;78:799–806. doi: 10.1016/j.neuron.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman BD. The circadian timing system and reproduction in mammals. Steroids. 1999;64:679–685. doi: 10.1016/s0039-128x(99)00052-5. [DOI] [PubMed] [Google Scholar]
- Han S, Yu FH, Schwartz MD, Linton JD, Bosma MM, Hurley JB, Catterall WA, de la Iglesia HO. Na(V)1.1 channels are critical for intercellular communication in the suprachiasmatic nucleus and for normal circadian rhythms. Proc Natl Acad Sci U S A. 2012;109:E368–377. doi: 10.1073/pnas.1115729109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel D, Mato G, Meunier C. Synchrony in excitatory neural networks. Neural Comput. 1995;7:307–337. doi: 10.1162/neco.1995.7.2.307. [DOI] [PubMed] [Google Scholar]
- Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Garcia AJ, 3rd, Ramirez JM. Network reconfiguration and neuronal plasticity in rhythm-generating networks. Integr Comp Biol. 2011;51:856–868. doi: 10.1093/icb/icr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leise TL, Harrington ME. Wavelet-based time series analysis of circadian rhythms. J Biol Rhythms. 2011;26:454–463. doi: 10.1177/0748730411416330. [DOI] [PubMed] [Google Scholar]
- Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–128. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A. 2011;108:14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Colwell CS, Rohling JH, Houben T, Michel S. Dynamic neuronal network organization of the circadian clock and possible deterioration in disease. Prog Brain Res. 2012;199:143–162. doi: 10.1016/B978-0-444-59427-3.00009-5. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Michel S, Vanderleest HT, Rohling JH. Daily and seasonal adaptation of the circadian clock requires plasticity of the SCN neuronal network. Eur J Neurosci. 2010;32:2143–2151. doi: 10.1111/j.1460-9568.2010.07522.x. [DOI] [PubMed] [Google Scholar]
- Miesenbock G. Optogenetic control of cells and circuits. Annu Rev Cell Dev Biol. 2011;27:731–758. doi: 10.1146/annurev-cellbio-100109-104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci. 2011;34:349–358. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J, Hong S, Hatanaka F, Nakajima Y, De Schutter E, Takumi T. Period coding of Bmal1 oscillators in the suprachiasmatic nucleus. J Neurosci. 2012;32:8900–8918. doi: 10.1523/JNEUROSCI.5586-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ. Seasonal immune function and sickness responses. Trends Immunol. 2004;25:187–192. doi: 10.1016/j.it.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents: I. The stability of spontaneous frequency. J Comp Physiol [A] 1976a;106:223–252. [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents: V. Pacemaker Structure: A Clock for All Seasons. J Comp Physiol [A] 1976b;106:333–355. [Google Scholar]
- Pyter LM, Nelson RJ. Enduring effects of photoperiod on affective behaviors in Siberian hamsters (Phodopus sungorus) Behav Neurosci. 2006;120:125–134. doi: 10.1037/0735-7044.120.1.125. [DOI] [PubMed] [Google Scholar]
- Pyter LM, Trainor BC, Nelson RJ. Testosterone and photoperiod interact to affect spatial learning and memory in adult male white-footed mice (Peromyscus leucopus) Eur J Neurosci. 2006;23:3056–3062. doi: 10.1111/j.1460-9568.2006.04821.x. [DOI] [PubMed] [Google Scholar]
- Roberts MH, Block GD. Analysis of mutual circadian pacemaker coupling between the two eyes of Bulla. J Biol Rhythms. 1985;1:55–75. doi: 10.1177/074873048600100107. [DOI] [PubMed] [Google Scholar]
- Schnitzler A, Gross J. Normal and pathological oscillatory communication in the brain. Nat Rev Neurosci. 2005;6:285–296. doi: 10.1038/nrn1650. [DOI] [PubMed] [Google Scholar]
- Sellix MT, Evans JA, Leise TL, Castanon-Cervantes O, Hill DD, DeLisser P, Block GD, Menaker M, Davidson AJ. Aging differentially affects the re-entrainment response of central and peripheral circadian oscillators. J Neurosci. 2012;32:16193–16202. doi: 10.1523/JNEUROSCI.3559-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeal RM, Ermentrout GB, White JA. Phase-response curves and synchronized neural networks. Philos Trans R Soc Lond B Biol Sci. 2010;365:2407–2422. doi: 10.1098/rstb.2009.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and dissorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima K, Kim SH, Allen CN. Characterization of an apamin-sensitive potassium current in suprachiasmatic nucleus neurons. Neuroscience. 2003;120:65–73. doi: 10.1016/s0306-4522(03)00270-7. [DOI] [PubMed] [Google Scholar]
- Trainor BC, Finy MS, Nelson RJ. Rapid effects of estradiol on male aggression depend on photoperiod in reproductively non-responsive mice. Horm Behav. 2008;53:192–199. doi: 10.1016/j.yhbeh.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr TA, Duncan WC, Jr, Sher L, Aeschbach D, Schwartz PJ, Turner EH, Postolache TT, Rosenthal NE. A circadian signal of change of season in patients with seasonal affective disorder. Arch Gen Psychiatry. 2001;58:1108–1114. doi: 10.1001/archpsyc.58.12.1108. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yan L. Expression of clock genes in the suprachiasmatic nucleus: Effect of environmental lighting conditions. Rev Endocr Metab Disord. 2009;10:301–310. doi: 10.1007/s11154-009-9121-9. [DOI] [PubMed] [Google Scholar]
- Yan L, Foley NC, Bobula JM, Kriegsfeld LJ, Silver R. Two antiphase oscillations occur in each suprachiasmatic nucleus of behaviorally split hamsters. J Neurosci. 2005;25:9017–9026. doi: 10.1523/JNEUROSCI.2538-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.