Abstract
Pseudomonas aeruginosa produces a number of proteases that are associated with virulence and disease progression. A substrate able to detect P. aeruginosa-specific proteolytic activity could help to rapidly alert clinicians to the virulence potential of individual P. aeruginosa strains. For this purpose we designed a set of P. aeruginosa-specific fluorogenic substrates, comprising fluorescence resonance energy transfer (FRET)-labeled peptides, and evaluated their applicability to P. aeruginosa virulence in a range of clinical isolates. A FRET-peptide comprising three glycines (3xGly) was found to be specific for the detection of P. aeruginosa proteases. Further screening of 97 P. aeruginosa clinical isolates showed a wide variation in 3xGly cleavage activity. The absence of 3xGly degradation by a lasI knock out strain indicated that 3xGly cleavage by P. aeruginosa could be quorum sensing (QS)-related, a hypothesis strengthened by the observation of a strong correlation between 3xGly cleavage, LasA staphylolytic activity and pyocyanin production. Additionally, isolates able to cleave 3xGly were more susceptible to the QS inhibiting antibiotic azithromycin (AZM). In conclusion, we designed and evaluated a 3xGly substrate possibly useful as a simple tool to predict virulence and AZM susceptibility.
Introduction
Pseudomonas aeruginosa, is a gram-negative rod-shaped bacterium, which is an important cause of infection in individuals suffering from a wide range of underlying disease conditions, including individuals with a compromised host defence, burn patients and in patients suffering from the genetically inherited respiratory tract disease cystic fibrosis (CF). P. aeruginosa is also a frequent cause of hospital acquired pneumonia, wound infections and bacteraemia [1]. Early detection of a P. aeruginosa infection facilitates effective antimicrobial treatment, reduces inappropriate antibiotic prescription and could possibly contribute to preventing irreversible lung disease in CF patients. Additionally, this bacterial pathogen produces a number of protease enzymes that are associated with virulence and disease progression [2]. In this respect, a substrate able to detect P. aeruginosa-specific proteolytic activity could help to rapidly alert clinicians to the virulence potential of individual P. aeruginosa strains isolated from different patients. Specifically, detection of these protease virulence factors could potentially be used to monitor the severity of infection and to predict disease outcome, thereby providing useful information to support tailor-made patient monitoring and treatment, a first step towards introducing “personalized medicine”.
In theory, bacterial proteases may be suited as biomarkers for the rapid and sensitive identification of microorganisms in clinical samples. Further, the ability to utilize protease based detection methods to diagnose bacterial infections has been previously described [3,4]. In this respect, it is known that P. aeruginosa is equipped with a large arsenal of virulence factors that aid to successfully infect the host [2]. The majority of these virulence factors include proteases produced under control of the Las and Rhl quorum sensing (QS) systems of P. aeruginosa [5], examples of which include LasA and LasB elastases and alkaline protease [6]. These proteases play an important role in P. aeruginosa pathogenesis through the degradation of biologically active proteins present in human tissue [7]. In fact, the significant role of the QS-system in P. aeruginosa-related disease progression has resulted in much research into QS-inhibiting compounds that could potentially reduce the organism’s virulence potential [8–12]. Further, the therapeutic efficacy of this anti-QS, anti-virulence, strategy in the treatment of P. aeruginosa has already been demonstrated for the QS-inhibiting antibiotic azithromycin [12]. Though anti-virulence treatment based on QS-inhibition is only useful when an active, virulence factor secreting, QS-system is present in the P. aeruginosa strain to be targeted.
In this study we describe the design of a novel P. aeruginosa-specific Fluorescence Resonance Energy Transfer (FRET)-peptide substrate and examine its applicability for the detection of P. aeruginosa proteolytic activity in clinical specimens. In addition, we studied the link between substrate cleavage efficiency, virulence factor production and susceptibility towards QS-inhibiting antibiotics, such as azithromycin.
Materials and Methods
Bacteria
The P. aeruginosa strains used for substrate specificity testing are listed in Table 1. Clinical strains of P. aeruginosa were collected from a bacterial biobank present within the Department of Medical Microbiology and Infectious Diseases of the Erasmus Medical Center, Rotterdam, Netherlands, collected between the years 2008-2012. In total 97 clinical isolates were selected; 13 from blood, 56 from sputum (35 CF patients and 21 non-CF patients), and 28 strains were isolated from wounds. The VITEK 2 system (bioMérieux, Marcy L`Etoile, France) was used for identification and antibiotic susceptibility testing of the clinical isolates. For the preparation of culture supernatants, all bacteria were grown in 5 ml Brain Heart Infusion (BHI) medium (bioTrading, Mijdrecht, The Netherlands) at 37 °C. After 16 h of culture, the bacteria were pelleted by centrifugation for 10 min at 3000 x g, and the enzyme containing supernatant was filter sterilized through a 0.22 μm filter (Millipore, Amsterdam, The Netherlands).
Table 1. Bacterial strains used in this study.
| Strain | |
|---|---|
| Pseudomonas aeruginosa | ATCC 15692, PAO1 |
| Pseudomonas aeruginosa | PA14 |
| Pseudomonas aeruginosa | PA14ΔlasI [31] |
| Pseudomonas fluorescens | Clinical isolate |
| Pseudomonas putida | S12 |
| Pseudomonas stutzeri | DSMZ 10701, JM300 |
| Staphylococcus aureus | ATCC 43300 |
| Staphylococcus simulans | ATCC 27851 |
| Staphylococcus capitis subsp. capitis | ATCC 35661 |
| Staphylococcus epidermidis | Clinical isolate |
| Streptococcus pneumoniae | ATCC 49619 |
| Streptococcus equi subsp. zooepidemicus | ATCC 43079 |
| Klebsiella pneumoniae | ATCC 43816 |
| Haemophilus influenzae | ATCC 49247 |
FRET- assay
The substrates used in this study were purchased at PepScan Presto B.V. (Lelystad, The Netherlands) with a purity > 90%. All substrates were C-terminally flanked with a fluorescent probe; FITC and N-terminally flanked with a lysine coupled quencher; Dabcyl. Identity of the substrates was confirmed by PepScan Presto B.V. using mass spectrometry. Assays were performed in black, clear bottom 96-well plates (Corning, Lowell, USA). Proteolytic activity was determined by incubating 16 μM substrate with 50 μl filtered bacterial culture supernatant or 50 µl lysostaphin (0.04 µg/µl diluted in BHI, Sigma, Zwijndrecht, The Netherlands) at 37 °C. Filtered BHI medium was used as a negative control. Plates were read for 60 min with 2 min intervals using a fluorescence microplate reader (FLUOstar Galaxy, BMG Laboratories, Offenburg, Germany) using an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Relative fluorescence (RF) values were obtained after correction against an un-inoculated BHI culture medium control. Protease activity was defined in RF per minute (RF/min).
Sensitivity testing of the 3xGly substrate in vitro
P. aeruginosa PAO1 was cultured overnight in BHI medium at 37 °C. Next day, 20 µl of the overnight culture was added to 20 ml fresh BHI medium. Subsequently, bacteria were grown for another 16 h and a sample was taken every hour. The number of bacteria in the sample was determined by colony counting by plating 10-fold serial dilutions on trypticase soy agar (TSA) plates (bioTrading, Mijdrecht, The Netherlands). Plates were incubated at 37 °C and bacteria were enumerated after overnight incubation. Proteolytic activity on the 3xGly substrate was examined using 50 µL samples in the FRET-assay as described above. Relative fluorescence (RF) values were obtained after correction against negative culture medium samples. The protease activity was defined in RF per minute (RF/min). Proteolytic activity with an RF/min > 5 was defined positive.
Quantification of pyocyanin production and LasA protease activity
Extracellular pyocyanin production was determined as previously described [13]. LasA protease activity was examined by measuring the ability of stationary phase P. aeruginosa culture supernatants (see FRET-assay section above) to lyse heat-killed Staphylococcus aureus cells as described by Kessler et al [14].
RNA isolation and cDNA synthesis
A selection of 12 P. aeruginosa isolates were chosen for RNA isolation. These isolates had been cultured from blood, wound and sputum clinical specimens, with two isolates that were 3xGly active (+, RF/min > 5) and two isolates that were 3xGly inactive (-, RF/min < 5), being chosen from each clinical specimen type .P. aeruginosa strain PAO1 (ATCC 15692) was used as a 3xGly positive reference control. The 12 P. aeruginosa isolates were initially grown overnight in BHI medium at 37°C,. Next day the overnight culture was diluted 1:10 in BHI medium and the bacteria again cultured at 37 °C until an OD600 of approximately 0.9 - 1.0 was reached. From these cultures total RNA was extracted using the FastRNA ProBlue kit (Promega, Leiden, The Netherlands) according to manufacturer’s instructions. To remove any contaminating DNA, 1 μg of extracted RNA was mixed with one unit DNase (Fermentas, Thermo Fisher, Landsmeer, The Netherlands) in 1x DNase reaction buffer. The mixture was then incubated at 37 °C for 30 min and the reaction stopped by the addition of 1 μl 50 mM EDTA and subsequent incubation for 10 min at 65 °C. For cDNA synthesis, DNase treated RNA (500 ng) was incubated for 60 min at 42 °C with 5 µM random hexamer primers, 10 units Ribolock, 1 mM dNTPs and 200 units RevertAid in 1x reaction buffer (Fermentas, Thermo Fisher, Landsmeer, The Netherlands). The reaction was stopped by incubation at 70 °C for 5 min.
Expression of quorum sensing genes
The detection of QS-gene expression was performed using real-time PCR amplification and specifically designed primers for the QS-related pathway genes lasI and rhlA, the non-QS related control gene trpD and the P. aeruginosa housekeeping gene rpsL [15–17]. Each PCR contained 1x FastStart SYBR Green mastermix (Roche, Woerden, The Netherlands), 4 mM MgCl2 and 0.5 pmol of each primer and 2 μl of 10x diluted cDNA. Amplification was performed using the LightCycler (Roche, Woerden, The Netherlands) The cycling parameters used were: 15 min at 95 °C, 40 amplification cycles of 95 °C for 20 s, 60 °C for 20 s and 72 °C for 30 s. Melting curve analysis, performed at the end of amplification, showed a single product peak, indicating that no non-specific products were amplified. Data were analyzed using LightCycler software (version 3.0) and the housekeeping gene rpsL was used as a reference (housekeeping) gene for normalizing gene expression. The 2-ΔΔCt method was used to calculate the expression of the genes of interest [18]. QS-gene expression was calculated as a relative percentage of the rpsL-normalized gene expression in the control P. aeruginosa PAO1 isolate.
Azithromycin susceptibility testing
The susceptibility of P. aeruginosa to the QS inhibiting antibiotic azithromycin (AZM) was evaluated for 35 P. aeruginosa strains isolated from the sputa of CF-patients using the disc diffusion method. For this purpose, a suspension of each bacterial isolate, overnight grown on tryptic soy agar (TSA) blood agar, was prepared in physiological saline at a turbidity of 0.5 McFarland units. Each bacterial suspension was streaked onto TSA plates with a sterile cotton swab, to obtain uniform bacterial growth, and a disc containing 15 µg AZM (Oxoid, Badhoevedorp, The Netherlands) was placed on the middle of the inoculated culture plate. Plates were then incubated at 37 °C, incubated overnight and the diameter of the zones of growth inhibition (mm) were measured.
Results
Design of P. aeruginosa- specific FRET-substrates
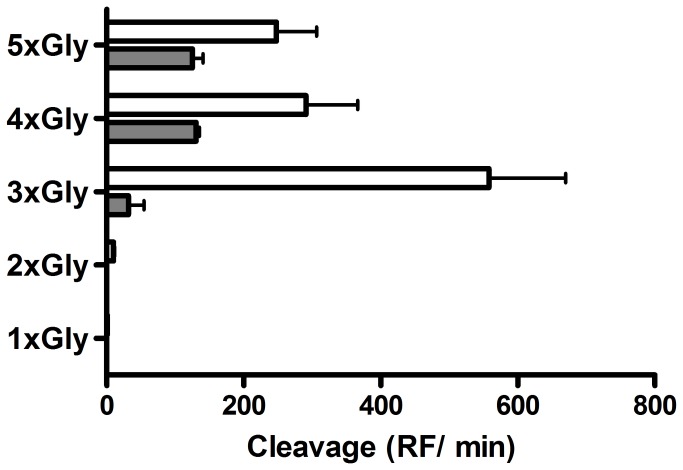
In a study by Vessillier et al. it was described that the P. aeruginosa specific protease LasA, recognizes and degrades glycine bonds [19]. Based on these cleavage characteristics we designed FRET-peptide substrates, comprising multiple glycine residues, and incubated these substrates with P. aeruginosa culture supernatant. These substrates included FITC-Gly-LysDbc (1xGly), FITC-(Gly)2-LysDbc (2xGly), FITC-(Gly)3-LysDbc (3xGly), FITC-(Gly)4-LysDbc (4xGly) and FITC-(Gly)5-LysDbc (5xGly). As a specificity control, we compared the activity of P. aeruginosa culture supernatant towards these substrates with cleavage activity with lysostaphin, an enzyme produced by Staphylococcus simulans, that recognizes and degrades penta-glycine bonds [20]. Experiments showed that the 5xGly substrate was cleaved by both lysostaphin and P. aeruginosa culture supernatant, and that a similar result was observed for the substrate containing four glycines (Figure 1). However, when the 3xGly substrate was tested, a significant decrease in proteolytic activity by lysostaphin was observed, whereas the proteolytic activity of P. aeruginosa actually significantly increased. Additional experiments showed minimal cleavage in case of the 2xGly substrate and no cleavage using the 1xGly substrate. These results indicated that the 3xGly substrate was the best candidate, amongst the substrates tested, to investigate as a potential substrate for the rapid detection of virulence in P. aeruginosa.
Figure 1. Cleavage activity of lysostaphin and P. aeruginosa culture supernatant on a range of glycine substrates.
Lysostaphin (2 µg, grey bar) and culture supernatant of P. aeruginosa PAO1 (white bar) were incubated with 16 µM FRET-substrate at 37 °C for 1 h. Cleavage of the substrates was defined in Relative Fluorescence per minute (RF/min). Results are expressed as mean ± SEM (n = 3).
Characterization of 3xGly substrate cleavage
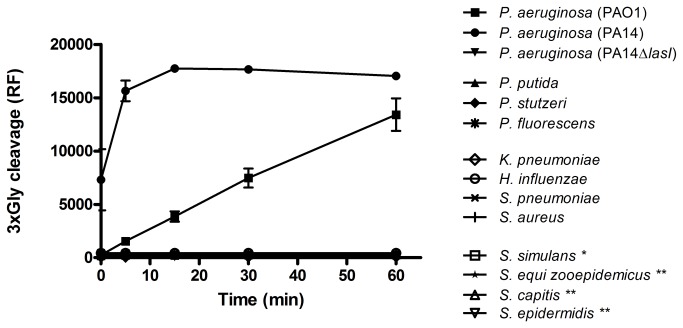
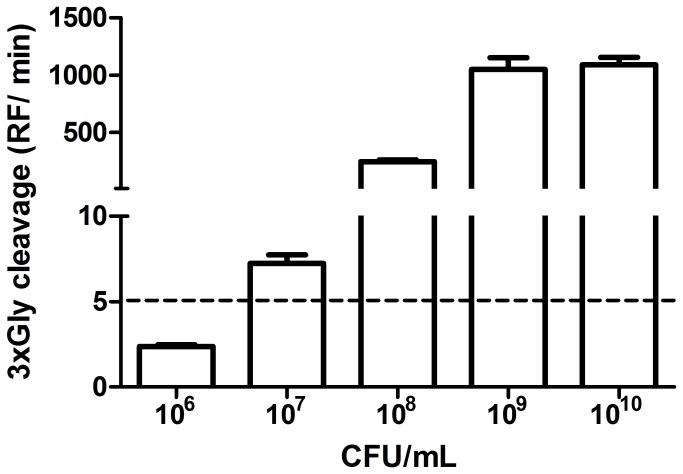
To characterize the specificity of the 3xGly substrate, we examined 3xGly cleavage using a range of culture supernatants (i) from a range of Pseudomonas non-aeruginosa species, and (ii) from a range of additional respiratory bacteria, and microorganisms known to produce lysostaphin (-like) proteases (Table 1). Results showed that the 3xGly substrate was exclusively cleaved by culture supernatants of P. aeruginosa (strains PA14 and PAO1), but not by P. putida, P. stutzeri or P. fluorescens. Further, no cleavage activity was observed for the range of additional respiratory bacteria, and microorganisms known to produce lysostaphin, that were tested. Interestingly, no cleavage activity was observed when the 3xGly substrate was incubated with culture supernatant of the P. aeruginosa strain PA14∆lasI which lacks the lasI QS-gene (Figure 2). Besides the above mentioned bacteria the 3xGly substrate was screened with an additional set of in total 17 bacterial supernatants. None of these bacteria was able to cleave the substrate (data not shown). To characterize 3xGly sensitivity, the substrate was incubated with a dilution series of P. aeruginosa PAO1. Using a cut-off of RF/min > 5, it was observed that the limit of detection for P. aeruginosa PAO1 using the 3xGly substrate was 107 CFU//ml (Figure 3).
Figure 2. Specificity testing of the 3xGly substrate.
Culture supernatants of Pseudomonas spp., respiratory microorganisms and bacteria producing lysostaphin (*) or lysostaphin-like (**) proteases were incubated with 16 µM 3xGly. Fluorescence was measured for 1 h at 37 °C. Results are expressed as mean ± SEM (n = 3).
Figure 3. Sensitivity testing of the 3xGly substrate.
Serial dilutions of P. aeruginosa PAO1 were incubated with 16 µM 3xGly at 37 °C. Cleavage activity was defined in Relative Fluorescence per minute (RF/min). The cut-off of the assay was estimated at an RF/min of 5. Results are expressed as mean ± SEM (n = 3).
In order to obtain a more comprehensive view of protease activity in an extended range of clinically relevant P. aeruginosa isolates, we tested a total of 97 randomly selected P. aeruginosa clinical isolates that had been cultured from wounds, blood and sputum. These strains were analyzed for their supernatant cleavage activity on the 3xGly substrate. Results revealed that a large percentage of the strains tested (60/97; 62%) were unable to cleave the 3xGly substrate. This percentage varied between clinical specimens from 50% in wound isolates to 73% in isolates from CF patients (Table 2).
Table 2. 3xGly cleavage activity among 97 randomly selected P. aeruginosa clinical isolates a .
|
Sputum
|
||||
|---|---|---|---|---|
| (n=56) |
||||
| Wound | Blood | CF | Non-CF | |
| (n=28) | (n=13) | (n=35) | (n=21) | |
| Cleavage | 14 (50) | 4 (31) | 9 (27) | 10 (48) |
| No cleavage | 14 (50 | 9 (69) | 26 (73) | 11 (52) |
Number in brackets denotes percentage (%)
Relationship between 3xGly cleavage and virulence
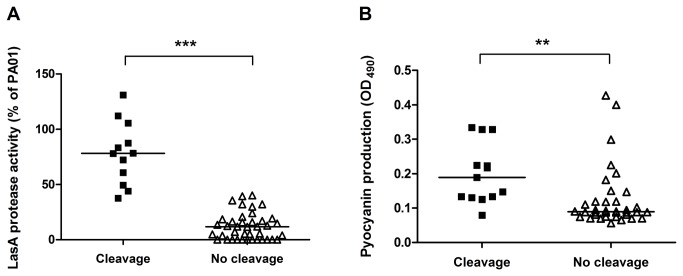
LasA is a member of the beta-lytic endopeptidase family and pyocyanin is a secondary metabolite which has the ability to oxidize and reduce other molecules [21,22]. The expression of both of these virulence factors by P. aeruginosa has been shown to be related to its QS system [2]. Results from both LasA and pyocyanin production indicated that both were significantly higher in 3xGly cleaving P. aeruginosa isolates, though some variation in individual isolates was observed (Figure 4A-B). The association between LasA activity and 3xGly cleavage was very significant (P < 0.0001, r = 0.542).
Figure 4. Link between 3xGly cleavage and secretion of QS-related proteins.
Culture supernatants of 56 P. aeruginosa strains isolated from sputum were analyzed for LasA protease activity (A) and pyocyanin production (B). QS-related LasA cleavage activity in 3xGly cleaving P. aeruginosa strains (RF/min > 5) was compared to LasA cleavage in 3xGly non-cleaving strains (RF/min < 5) using the unpaired, two-tailed students t-test (** P < 0.01, *** P < 0.0001). A horizontal line indicates the median LasA activity /pyocyanin production.
Relationship between 3xGly cleavage and the quorum sensing system
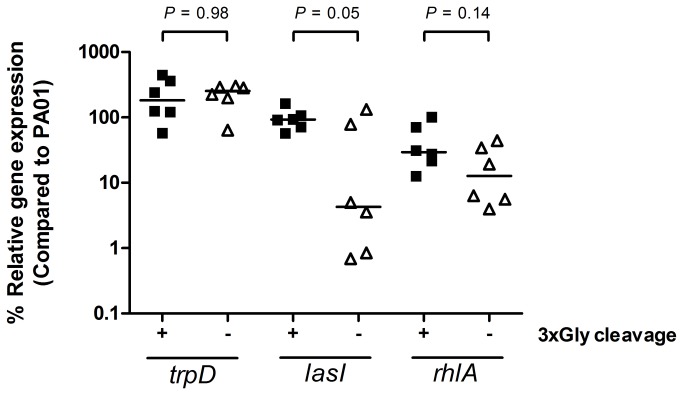
The observation of a lack of 3xGly activity using a P. aeruginosa lasI knock-out mutant strain (Figure 1) indicated that an absence of 3xGly cleavage activity might be related to an impaired QS-system expression. In order to verify this hypothesis we investigated the expression of the QS-genes lasI and rhlA using a selection of 3xGly cleaving and non-cleaving P. aeruginosa isolates. The strains which lacked 3xGly proteolytic activity showed a reduced expression of lasI and rhlA when compared to lasI and rhlA expression in 3xGly cleaving isolates. A significant reduction in normalized lasI, but not rhlA, was observed (P = 0.05). There was no difference observed in the normalized expression of the trpD control, a gene unrelated to the QS-system (Figure 5).
Figure 5. Link between 3xGly cleavage and expression of QS-genes.
QS-gene expression was determined in a subset P. aeruginosa strains as described in material and methods. Normalized expression of the QS-circuit gene lasI, QS-target gene rhlA and the QS-independent gene trpD measured in mid-log phase grown bacteria is shown as relative values (%) compared to the P. aeruginosa PAO1 control strain. A horizontal line indicates the median expression levels. P-values were calculated using unpaired, two-tailed students t-tests.
Relationship between 3xGly cleavage and azithromycin susceptibility
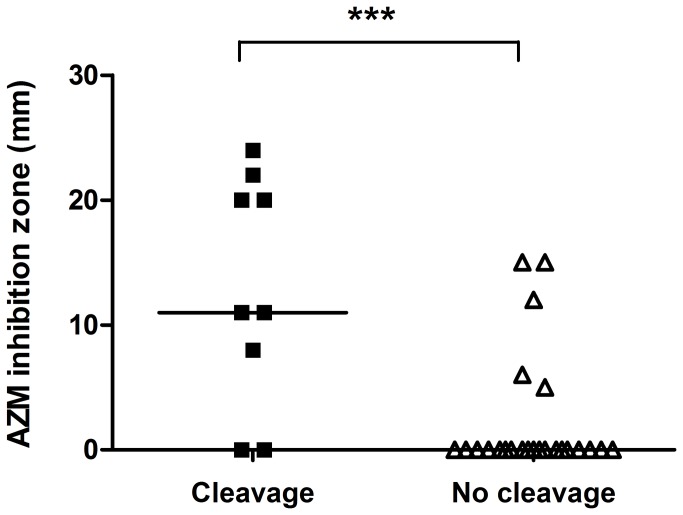
The apparent association between 3xGly cleavage and the QS-system led us to investigate a possible relationship between 3xGly cleavage and susceptibility to the QS-inhibiting antibiotic AZM. We examined the AZM susceptibility of P. aeruginosa clinical isolates using a disc diffusion assay, but found no relationship between 3xGly cleavage and AZM susceptibility for P. aeruginosa isolates cultured from blood and wounds, or from the sputum of non-CF patients. Interestingly however, a highly significant association was observed between AZM inhibition zone and 3xGly cleavage for P. aeruginosa isolates cultured from the sputa of CF patients (Figure 6). Finally, 3xGly cleavage in relation to a range of AZM unrelated antibiotic resistances was investigated, with no significant correlation being found between 3xGly cleavage and resistance to the other antibiotics tested. However, it was noted that among the group of P. aeruginosa isolates that lacked 3xGly cleavage more multi drug resistant (MDR) isolates were observed (Table S1).
Figure 6. Link between 3xGly cleavage and azithromycin susceptibility.
Azithromycin (AZM) susceptibility of P. aeruginosa isolates cultured from sputum was determined using the disc diffusion method. AZM inhibition zones for 3xGly cleaving (RF/min > 5) P. aeruginosa strains was compared to inhibition zones of 3xGly non-cleaving strains (RF/min < 5) using the unpaired, two-tailed students t-test (*** P < 0.0001). A horizontal line indicates the median AZM inhibition zone size.
Discussion
We designed and evaluated a fluorogenic substrate as a potential marker of virulence in the bacterial pathogen P. aeruginosa. Preliminary experiments indicated that this 3xGly substrate was specific for cleavage by P. aeruginosa, and that the sensitivity of the substrate was 107 CFU/ml within 1 h, though this limit of detection may vary among different P. aeruginosa strains (it was observed that PA14 cleaved the 3xGly substrate more efficient than PAO1). Only slight cross reactivity was observed with another bacterial protease, lysostaphin, which recognizes and degrades pentaglycin bonds [20]. Later experiments using a broader range of 97 clinical isolates showed that 3xGly cleavage activity differed between P. aeruginosa isolates, with a large percentage of the isolates being unable to cleave the 3xGly substrate. This is possibly related to difference in expression of the P. aeruginosa QS-system. We indeed observed a slight, but non-significant, decrease in expression of the QS-genes lasI and rhlI in P. aeruginosa strains which lacked 3xGly proteolytic activity. Because this experiment was performed with a subset of the P. aeruginosa strains future experiments are required in order to identify the exact genetic basis of the proteolysis / virulence correlation.
One of the most important proteases secreted under the direction of the P. aeruginosa QS system is the LasA protease. The LasA protease (staphylolysin) of P. aeruginosa can recognize and degrade glycine bonds, degrade elastin and is an important contributor to the pathogenesis of this organism. LasA (20 kDa) is a member of the beta-lytic endopeptidase family of extracellular bacterial proteases, and possesses high-level staphylolytic activity [7]. Further, Elston et al. showed that the LasA protease is capable of the degradation of peptides in which three glycines were present [23]. Therefore, based on the already established glycine-cleaving proteolytic activity of the P. aeruginosa LasA protease, its staphylolytic activity, its control via the P. aeruginosa QS system, and the results observed for 3xGly cleavage, it appears that the LasA protease is most likely the protease virulence factor measured in P. aeruginosa culture supernatant using our 3xGly substrate, but this needs to be confirmed experimentally.
Other factors, dependent on the expression of the QS-system of P. aeruginosa are pyocyanin production and antibiotic resistance. Pyocyanin is an important virulence factor and functions as an electron transfer facilitator [2]. In addition, P. aeruginosa isolates which lack production of QS-dependent virulence factors, have previously been shown to possess a higher resistance rate to antimicrobials, among which ciprofloxacin and tobramycin [24]. Although, we indeed observed a significant correlation between pyocyanin production and 3xGly cleavage, no significant correlation with resistance to the antimicrobials examined was found (Table 1). This discrepancy might be due to difference in culture conditions, as the secretion of QS-metabolites, such as the LasA protease, depends on the availability of nutrients in the environment [25].
Perhaps one of the most interesting findings of the study was the fact that the highest percentage of 3xGly non-cleaving isolates was observed in group of CF sputum isolates. This is possibly due to the fact that CF patients are often colonized with P. aeruginosa [26]. Kohler et al showed that during P. aeruginosa colonization of intubated patients the number of QS- mutants increases. These mutants lack the production of QS-dependent proteins, such as elastase and rhamnolipids, and take advantage of the “public goods” produced by wild-type isolates that reside within the total population of P. aeruginosa isolates that colonize the patient (“cheater” strains). This phenomenon may result in overgrowth of the wild-type P. aeruginosa isolate by the mutant isolate due to its fitness benefit [27].
Currently, research is being performed to investigate the potential of QS-inhibiting compounds in the treatment of P. aeruginosa related infections [8–12]. The most thoroughly investigated QS-inhibiting antibiotic is AZM. AZM treatment is associated with improvement in disease outcome in P. aeruginosa infected CF-patients [28]. However, an anti-virulence agent such as AZM is only effective when an active, virulence factor secreting, QS-system is present. For this reason Kohler et al screened for rhamnolipid production to select patients for their clinical trial on AZM efficiency. Patients infected with P. aeruginosa strains which are unable to produce these QS-proteins were excluded from the protocol [12].
We observed a strong correlation between AZM susceptibility and 3xGly cleavage by P. aeruginosa isolates cultured from the sputum of CF patients, with growth inhibition zones in the 3xGly cleavage-positive isolates being significant larger compared to zones of 3xGly cleavage-negative isolates. Until recently it was stated that AZM is unable to eradicate P. aeruginosa by bacterial killing [29]. A recent publication by Buyck et al. however, revealed that growth inhibition of P. aeruginosa by AZM depends on the medium used [30]. P. aeruginosa strains grown in Mueller Hinton medium have a lower outer membrane permeability compared to strains cultured in for example RPMI. This results in an increase in susceptibility towards AZM and thus might explain the presence of AZM induced growth inhibition zones we observed on TSA agar plates.
In conclusion, we designed and evaluated a novel 3xGly FRET-peptide substrate for the assessment of virulence in P. aeruginosa. Cleavage of the 3xGly FRET-substrate was significantly associated with culture supernatant protease activity, staphylolytic activity, pyocyanin production and the expression of lasI in the P. aeruginosa QS system. This publication represents the first step in the development of a simple test to determine and monitor P. aeruginosa virulence in clinical samples, including the prediction of the effectiveness of QS-inhibiting antibiotic treatments. Preliminary evaluation experiments suggest that this methodology may achieve its greatest potential when monitoring CF patients colonized by, and undergoing treatment for, P. aeruginosa infections.
Supporting Information
Antibiotic susceptibility of the 97 P. aeruginosa strains used in this study.
(DOCX)
Acknowledgments
We would like to thank Dr. D. Hogan for providing the P. aeruginosa PA14 and the P. aeruginosa PA14∆lasI strain and Dr. T. Kohler for the sequences of the primers to determine the expression of the QS-genes.
Funding Statement
This research was funded by the European Community's Seventh Framework Programme FP7/2007-2013, TEMPOtest-QC, under grant agreement no. 241742 and an STW Valorisation Grant, TNO SBIR: Bacalyzer no. 16-001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bodey GP, Bolivar R, Fainstein V, Jadeja L (1983) Infections caused by Pseudomonas aeruginosa . Rev Infect Dis 5: 279-313. doi: 10.1093/clinids/5.2.279. PubMed: 6405475. [DOI] [PubMed] [Google Scholar]
- 2. Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH et al. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa . Microbiol Mol Biol Rev 76: 46-65. doi: 10.1128/MMBR.05007-11. PubMed: 22390972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wildeboer D, Hill KE, Jeganathan F, Williams DW, Riddell AD et al. (2012) Specific protease activity indicates the degree of Pseudomonas aeruginosa infection in chronic infected wounds. Eur J Clin Microbiol Infect Dis 31: 2183-2189. doi: 10.1007/s10096-012-1553-6. PubMed: 22278295. [DOI] [PubMed] [Google Scholar]
- 4. Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG et al. (2012) Highly specific protease-based approach for detection of Porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol 50: 104-112. doi: 10.1128/JCM.05313-11. PubMed: 22075590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Girard G, Bloemberg GV (2008) Central role of quorum sensing in regulating the production of pathogenicity factors in Pseudomonas aeruginosa . Future Microbiol 3: 97-106. doi: 10.2217/17460913.3.1.97. PubMed: 18230038. [DOI] [PubMed] [Google Scholar]
- 6. Brint JM, Ohman DE (1995) Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol 177: 7155-7163. PubMed: 8522523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrett AJ, Rawlings ND, Woessner JF (2004) Handbook of proteolytic enzymes. 387 p.
- 8. Imperi F, Massai F, Ramachandran PC, Longo F, Zennaro E et al. (2013) New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob Agents Chemother 57: 996-1005. doi: 10.1128/AAC.01952-12. PubMed: 23254430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hodgkinson JT, Galloway WR, Wright M, Mati IK, Nicholson RL et al. (2012) Design, synthesis and biological evaluation of non-natural modulators of quorum sensing in Pseudomonas aeruginosa . Org Biomol Chem 10: 6032-6044. doi: 10.1039/c2ob25198a. PubMed: 22499353. [DOI] [PubMed] [Google Scholar]
- 10. Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD et al. (2012) Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother 56: 2314-2325. doi: 10.1128/AAC.05919-11. PubMed: 22314537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christensen LD, van Gennip M, Jakobsen TH, Alhede M, Hougen HP et al. (2012) Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J Antimicrob Chemother 67: 1198-1206. doi: 10.1093/jac/dks002. PubMed: 22302561. [DOI] [PubMed] [Google Scholar]
- 12. van Delden C, Köhler T, Brunner-Ferber F, François B, Carlet J et al. (2012) Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: a randomized controlled trial. Intensive Care Med 38: 1118-1125. doi: 10.1007/s00134-012-2559-3. PubMed: 22527075. [DOI] [PubMed] [Google Scholar]
- 13. Essar DW, Eberly L, Hadero A, Crawford IP (1990) Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172: 884-900. PubMed: 2153661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kessler E, Safrin M, Olson JC, Ohman DE (1993) Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268: 7503-7508. PubMed: 8463280. [PubMed] [Google Scholar]
- 15. Köhler T, Perron GG, Buckling A, van Delden C (2010) Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa . PLoS Pathog 6: e1000883 PubMed: 20463812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611-622. doi: 10.1373/clinchem.2008.112797. PubMed: 19246619. [DOI] [PubMed] [Google Scholar]
- 17. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 PubMed: 12184808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402-408. doi: 10.1006/meth.2001.1262. PubMed: 11846609. [DOI] [PubMed] [Google Scholar]
- 19. Vessillier S, Delolme F, Bernillon J, Saulnier J, Wallach J (2001) Hydrolysis of glycine-containing elastin pentapeptides by LasA, a metalloelastase from Pseudomonas aeruginosa . Eur J Biochem 268: 1049-1057. doi: 10.1046/j.1432-1327.2001.01967.x. PubMed: 11179971. [DOI] [PubMed] [Google Scholar]
- 20. Xu N, Huang ZH, de Jonge BL, Gage DA (1997) Structural characterization of peptidoglycan muropeptides by matrix-assisted laser desorption ionization mass spectrometry and postsource decay analysis. Anal Biochem 248: 7-14. doi: 10.1006/abio.1997.2073. PubMed: 9177719. [DOI] [PubMed] [Google Scholar]
- 21. Kessler E (1995) Beta-lytic endopeptidases. Methods Enzymol 248:740-56.: 740-756 [DOI] [PubMed] [Google Scholar]
- 22. Rada B, Leto TL (2009) Redox warfare between airway epithelial cells and Pseudomonas: dual oxidase versus pyocyanin. Immunol Res 43: 198-209. doi: 10.1007/s12026-008-8071-8. PubMed: 18979077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elston C, Wallach J, Saulnier J (2007) New continuous and specific fluorometric assays for Pseudomonas aeruginosa elastase and LasA protease. Anal Biochem 368: 87-94. doi: 10.1016/j.ab.2007.04.041. PubMed: 17553454. [DOI] [PubMed] [Google Scholar]
- 24. Karatuna O, Yagci A (2010) Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect 16: 1770-1775. doi: 10.1111/j.1469-0691.2010.03177.x. PubMed: 20132256. [DOI] [PubMed] [Google Scholar]
- 25. Duan K, Surette MG (2007) Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol 189: 4827-4836. doi: 10.1128/JB.00043-07. PubMed: 17449617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O et al. (2012) Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10: 841-851. doi: 10.1038/nrmicro2907. PubMed: 23147702. [DOI] [PubMed] [Google Scholar]
- 27. Köhler T, Buckling A, van Delden C (2009) Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci U S A 106: 6339-6344. doi: 10.1073/pnas.0811741106. PubMed: 19332772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saiman L, Marshall BC, Mayer-Hamblett N, Burns JL, Quittner AL et al. (2003) Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 290: 1749-1756. doi: 10.1001/jama.290.13.1749. PubMed: 14519709. [DOI] [PubMed] [Google Scholar]
- 29. Imamura Y, Higashiyama Y, Tomono K, Izumikawa K, Yanagihara K et al. (2005) Azithromycin exhibits bactericidal effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob Agents Chemother 49: 1377-1380. doi: 10.1128/AAC.49.4.1377-1380.2005. PubMed: 15793115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buyck JM, Plésiat P, Traore H, Vanderbist F, Tulkens PM et al. (2012) Increased susceptibility of Pseudomonas aeruginosa to macrolides and ketolides in eukaryotic cell culture media and biological fluids due to decreased expression of oprM and increased outer-membrane permeability. Clin Infect Dis 55: 534-542. doi: 10.1093/cid/cis473. PubMed: 22573850. [DOI] [PubMed] [Google Scholar]
- 31. Cugini C, Morales DK, Hogan DA (2010) Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156: 3096-3107. doi: 10.1099/mic.0.037911-0. PubMed: 20656785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibiotic susceptibility of the 97 P. aeruginosa strains used in this study.
(DOCX)