Abstract
Epidemiological, genetic, transcriptome, postmortem, peripheral biomarker, and therapeutic studies of schizophrenia all point to a dysregulation of both innate and adaptive immune systems in the disease, and it is likely that these immune changes actively contribute to disease symptoms. Gene expression disturbances in the brain of subjects with schizophrenia show complex, region-specific changes with consistently replicated and potentially interdependent induction of serpin peptidase inhibitor, clade A member 3 (SERPINA3) and interferon inducible transmembrane protein (IFITM) family transcripts in the prefrontal cortex. Recent data suggest that IFITM3 expression is a critical mediator of maternal immune activation. As the IFITM gene family is primarily expressed in the endothelial cells and meninges, and as the meninges play a critical role in interneuron development, we suggest that these two non-neuronal cell populations might play an important role in the disease pathophysiology. Finally, we propose that IFITM3 in particular might be a novel, appealing, knowledge-based drug target for treatment of schizophrenia.
Gene*environment interactions play a critical role in the emergence of schizophrenia pathophysiology. Epidemiological, genetic, transcriptome, postmortem, peripheral biomarker, and therapeutic studies of schizophrenia all point to a dysregulation of both innate and adaptive immune systems in the disease (1-3) and it is likely that these immune changes actively contribute to disease symptoms (1, 4, 5). Regardless of the abundance of data obtained to date, our understanding of the mechanism by which the immune system disturbances arise is limited: we do not have a good insight into the origin or sequence of events by which the immune dysregulation develops, and to date we have not taken full advantage of these changes as potential therapeutic targets.
Keywords: IFITM, IFITM3, SERPINA3, CHI3L1, CD14, postmortem, immune, schizophrenia, blood vessels, meninges, pia mater, blood-brain barrier
I. Origin of immune changes
What is the origin of the immune transcript disturbances? Are they coincidental, lifestyle-related, of genetic origin, “immune scars” related to early life events, or representations of lifelong immune system activation in the brain? While we do not have definite answers to these questions, knowledge integrated across several research disciplines provides us with important clues.
i) Genetic susceptibility studies
Recent genome-wide association studies identified immune-related susceptibility genes for schizophrenia in the major histocompatibility complex region of chromosome 6p21.3-22.1.2 (6-10). Using long-range phasing haplotypes tagging the major alleles at the HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1 and HLA-DQB1 loci, these studies revealed significant associations of SNPs in the 5-Mb region on 6p21.3-22.1 encompassing the classical human leukocyte antigen (HLA) alleles. Rs3131296 showed substantial correlation with the classical HLA alleles DRB1*03 and HLA-B*08. In the case of both DRB1*03 and HLA-B*08, the classical HLA allele was paired with the protective allele of rs3131296, making the results consistent with previous reports of under-transmission of DRB1*03 to schizophrenic offspring (11). However, while highly significant, the odds ratio was modest, and it is still somewhat debatable which (and if any) HLA/MHC genes directly increase risk for schizophrenia. Furthermore, these findings are clearly not sufficient to explain the immune system disturbances in the brains of subjects with schizophrenia: the dysregulation of immune system transcripts was observed in a much larger proportion of subjects (12-15) than one would expect on the bases of genetic studies.
ii) Biomarker studies
A recent meta-analysis by Miller and colleagues reported an increase in the inflammatory cytokines IL-1β, IL-6 and TGFβ during acute episodes of the illness (16). Furthermore, high hallucination and delusion scores appear to be correlated with the dysfunction of top canonical pathways related to interleukin signaling (17), and in recent-onset patients, up-regulation of IL 1β, IL-6 and TGFβ in peripheral blood mononuclear cells was observed (18). In addition, a study by Song et al, confirmed increased TNFα and IL-1β mRNA and protein levels in unmedicated first episode patients (19) and at least one study reported that elevated levels of pro-inflammatory cytokines is accompanied by reduced levels of GABAergic markers SST, PV and GAD67 in a subset of subjects with schizophrenia (15). Furthermore, recent results reveal that IgG, IgA, and IgM antibodies against NMDA-R were detected in 9.9% of patients with schizophrenia (20). These are perhaps the strongest arguments that the immune system changes are active and continuous contributors to the SCZ disease process, and are perhaps directly related to the symptoms of the disease.
iii) Anti-inflammatory treatment of schizophrenia
Aspirin is a non-steroidal anti-inflammatory drug (NSAID), and an irreversible inhibitor of both COX-1 and COX-2. Aspirin is known to reduce the plasma levels of inflammatory biomarkers such as CRP, IL-6 and TNF-α in patients with cardiovascular metabolic syndrome. A recent study showed that 1,000 mg of daily aspirin consumption reduced the core symptoms of schizophrenia over a course of 3-month treatment and resulted in Positive and Negative Symptoms Scale (PANSS) score improvements [for review, see (21) and (5) ]. Furthermore, in a separate double-blind, 6-week long placebo-controlled study observed a significantly better outcome in both positive and negative symptoms in the group treated with adjunct celecoxib (a COX-2 inhibitor) compared with the placebo group (22). However, it should be pointed out that the beneficial effect of NSAIDs has not been replicated in all clinical studies (5).
Interestingly, there is also a growing body of literature supporting the idea that antipsychotic medication reduces cytokine levels in SCZ patients and in various experimental models. In maternal immune activation (MIA), it appears that atypical antipsychotics suppress production of proinflammatory cytokines. Clozapine, olanzapine and risperidone are able to suppress tumor necrosis factor (TNF)-alpha and interleukin (IL)-6 induction in lipopolysaccharide-treated mice (23) suggesting that antipsychotic medication might act, at least partially, as a suppressor of the immune response. Furthermore, human data suggest measuring levels of inflammation-related proteins in blood may be useful in monitoring antipsychotic drug treatment responsiveness (24).
iv) Maternal immune activation
Maternal immune activation (MIA) disrupts normal fetal brain development (25-27). It is estimated that >30% of schizophrenia cases would be prevented if infection could be averted in pregnant women (3). Most commonly, MIA occurs as a result of various bacterial, viral or parasitic infections, and presence of antibodies against influenza or toxoplasmosis in maternal serum during pregnancy is associated with increased risk of schizophrenia of the offspring (28). These pathogens trigger a maternal immune response, which consists of activation of various cytokine pathways, including IL-1, IL-6, TNFα and IFNγ (25). The effect of MIA on fetal brain does not require direct infection: even in the absence of the virus the cytokine induction with polyI:C (a synthetic dsRNA) is sufficient to produce long-lasting effects, suggesting that mother's response to the infection is critical for altering fetal brain development (29). Pathological findings in patients with schizophrenia encompass alterations in multiple domains and include increased GABAA receptor α2 immunoreactivity, dopamine hyperfunction, delayed hippocampal myelination, reduced NMDA receptor expression in hippocampus, reduced numbers of reelin- and parvalbumin-positive cells, reduced dopamine D1 and D2 receptors in prefrontal cortex and enhanced tyrosine hydroxlyase in striatal structures, and similar changes are present in the adult offspring of MIA-exposed mice (reviewed by (1, 2, 30)). Furthermore, in adulthood, MIA exposed offspring display behavioral deficits in social interaction, prepulse inhibition (PPI), and open field and novel object exploration (25), as well as heightened response to hallucinogen exposure (31). These cellular/molecular/behavioral deficits appear to be directly related to the deficits observed in schizophrenia, further emphasizing the potential role of MIA in the disease pathophysiology. In addition, stress, genetic predisposition, and MIA interact: combined exposure to prenatal immune challenge and peripubertal stress induces synergistic pathological effects on adult behavioral functions and neurochemistry (32) and DISC1 point mutations exacerbate the outcome of MIA (33). Importantly, it appears that induction of IL-6 is both a necessary and sufficient component of MIA, as a single injection of the cytokine IL-6 in pregnant mice can recapitulate most of the deficits observed across the various models of MIA (29).
In summary, these converging findings strongly argue that immune system disturbances are the integral part of schizophrenia pathophysiology. However, schizophrenia is a uniquely human brain disease. As a result, postmortem brain transcriptome studies are of particular importance, as they can identify disturbed genes and pathways arising from the synergistic effects of diverse genetic predispositions and various environmental insults (34-36). Thus, in the absence of true animal models of schizophrenia, postmortem studies of brains of subjects with schizophrenia are essential contributors to the overall understanding of disease pathophysiology.
II. Transcriptome changes in schizophrenia
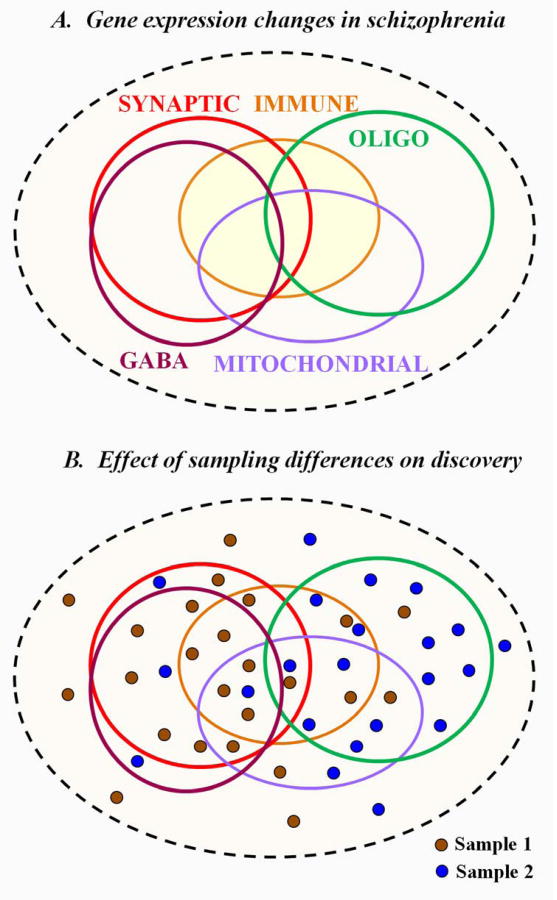
Schizophrenia is a syndrome, characterized by a considerable symptomatic diversity. It appears that the underlying molecular diversity of the disease is even more varied than the clinical manifestations (37). Transcriptome studies since the beginning of the millennium report that schizophrenic subjects show great, almost individual diversity in gene expression disturbances. Still, it appears that the overall data argue for a sub-stratification of disease-related mRNA profiles within the affected subjects, with molecular signatures commonly encompassing synaptic (38, 39), GABA-ergic (40, 41), mitochondrial (42-44), immune (12-15) and oligodendrocyte (45-47) disturbances (Figure 1A). These signatures overlap in a complex pattern, with many subjects showing altered expression across multiple biological domains, making it challenging to classify the individual subjects with schizophrenia into a single the molecular phenotype. Yet, the overall data suggest that the most distinct molecular signatures are the oligodendrocytic and GABA-ergic phenotypes and the ones that show the most overlap are the GABA-ergic and synaptic gene expression disturbances. Unfortunately, due to the limited postmortem brain availability and less than ideal clinical record access, we have no clear understanding of how the molecular disturbances might relate to the behavioral phenotypes or outcomes. As a result, different brain banking methods (e.g. recruitment from chronically hospitalized patients vs. obtaining samples for the coroner's office) might preferentially draw from different subpopulations of patients and the various “omics” analyses yield subpopulation-specific datasets that are not easily replicated (Figure 1B).
Figure 1. Transcriptome changes in the prefrontal cortex of subjects with schizophrenia.
A. Molecular deficits in schizophrenia are perhaps even more diverse than the symptomatology if the disease. Synaptic, mitochondrial, GABA-ergic, oligodendrocyte-specific, and immunological transcript disturbances have been all reported, although it appears that any single patient might show disturbances in multiple domains at the same time, making molecular classification of schizophrenia extremely challenging. However, it appears that synaptic and GABA-ergic disturbances are highly overlapping deficits, while GABA-ergic and oligodendrocyte-specific deficits are almost never found in the same cohort of subjects with schizophrenia, making them the two most distinct molecular phenotypes. B. Effect of sampling differences on the reported results. Recruitment strategies and specific characteristics of cohorts are likely to be correlated with the disease-underlying molecular phenotypes. Thus, sampling differences will significantly define the experimental outcome. In the case of sample 1, GABAergic and synaptic transcriptome changes are reported, while sample 2 will reveal mostly oligodendrocyte transcript deficits.
III. Immune transcriptome changes in the human brain
To understand the altered immune transcriptome in schizophrenia, first we have to ask a fundamental, yet elusive question - how do we define an “immune system gene”? Narrowly considered, any gene that directly participates in innate or acquired immune response can be considered an immune gene. However, this definition might be too conservative since a large number of genes do not directly participate in the immune response, yet they are potent regulators of various host responses. For example, responses that protect against excessive inflammation can be regulated through the cholinergic anti-inflammatory pathway: LPS-induced TNF-α release from macrophages is inhibited by selective CHRNA7 agonists (48). Furthermore, activation of GABA-ergic, cholinergic and adrenergic receptors suppresses microglial responses, whereas activation of ATP or adenosine receptors activates them (49). In addition, many of the cell adhesion molecules also serve as viral entry receptors (e.g. NCAM for Rabies, CD155 for Polio, nectin-1 for HSV-1 and CAR by adenoviruses) and alter neural cell behavior by promoting cytokine production, which in turn might lead to synaptic rearrangement and disrupted development (50). All these findings further blur the line between “immune” and “nonimmune” system genes. Thus, although many genes are best known for their primary role in neural transmission, cell communication, or synaptic development, a significant number of them also fit the broader definition of an immune system gene: one that encompasses any gene that alters the overall immune response and influences its outcome.
Using this definition, the majority of postmortem studies performed to date uncovered strong immune system transcript disturbances. These disturbances did not appear to be a result of treatment (14), and like most other gene expression changes in schizophrenia, showed a complex spatial pattern (51), arguing for a regional specificity of the changes. Yet, just like most of the other postmortem gene expression changes, the observed immune system disturbances are inconsistently replicated across the various cohorts of subjects.
IV. Expression changes in IFITM and SERPINA3
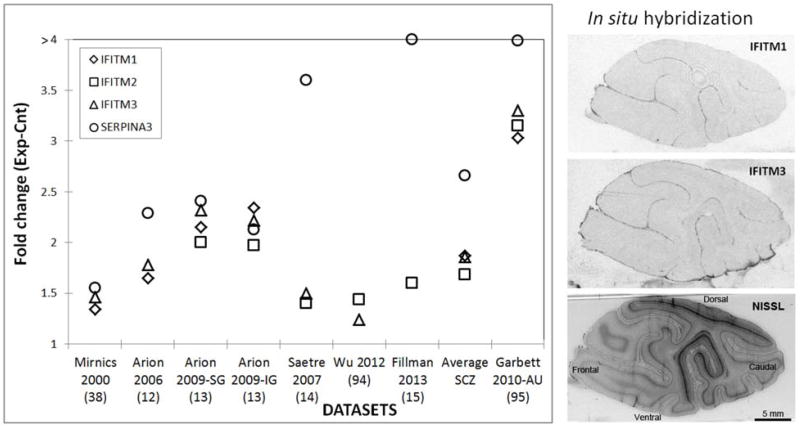
In this review, we will highlight a robust, replicated, and often overlooked immune signature in the prefrontal cortex of subjects with schizophrenia that consists of elevated SERPINA3 and interferon-induced transmembrane protein (IFITM) family mRNA expression levels (Figure 2A) (12-15, 38).
Figure 2.
A. IFITM and SERPINA3 changes across postmortem transcriptome profiling studies of schizophrenia (12-15, 38, 94) and autism (95). B. Autoradiography of 35S riboprobe in situ hybridization for IFITM1 and IFITM3 on parasagittal brain sections through the cortical mantle of a Cynomolgus monkey and a corresponding Nissl-stained section. Note that the highest expression of both transcripts is in the pia mater.
Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 (SERPINA3), also known as alpha-1-antichymotrypsin (AACT, α-1-ACT), is a serine protease inhibitor involved in a wide range of biological processes (52). The gene encoding SERPINA3 is clustered with 10 additional serpin genes on human chromosome 14q32.1. SERPINA3 expression is induced by various cytokines. It is expressed by hepatocytes, macrophages, endothelial and epithelial cells, as well as activated astrocytes. Importantly, in a recent study of sequencing hippocampal and cerebellar transcriptomes, SERPINA3 was found to be the most differentially expressed gene between the two tissues with a 208-fold enrichment in the hippocampus (53). The constitutive astrocyte-specific expression of SERPINA3 requires the activator protein-1 (AP-1) transcription factor and nuclear factor-1 (NFI-X) (54, 55). Its major target is the cathepsin family, a pro-inflammatory enzyme released at sites of inflammation contributing to activation of inflammatory cytokines, degradation of pathogens and remodeling of tissues (56). Therefore, by inhibiting cathepsin G, SERPINA3 should limit inflammation, coagulation, ECM remodeling and apoptosis (57). SERPINA3 has been implicated in the pathology of several devastating human diseases, including chronic obstructive pulmonary disease, Parkinson's disease, Alzheimer's disease, cystic fibrosis, stroke and cerebral hemorrhage (52). The gene product is considered an acute-phase inflammatory protein. SERPINA3 mRNA is also overexpressed in the placentas of preeclamptic pregnancies compared with controls, along with hypomethylation of the 5′ gene region (57, 58).
On the other hand, interferons (IFNs) defend against microbial infections and exhibit antiproliferative and differentiating activities (59). IFNα and IFNγ bind to interferon-stimulated response elements in the interferon inducible transmembrane (IFITM) family genes and induce their surface expression, which in turn modulate cell adhesion and influence cell differentiation (60). These are single pass type II membrane proteins and are expressed on the cell membrane, Golgi, endoplasmic reticulum and lysosomes (61). Family members include IFITM1, 2 and 3 and they are clustered together at the cytogenetic locus 11p15.5 in humans. The IFITM proteins are strongly induced by and mediate cellular resistance to influenza A, West Nile, Marburg, Ebola and dengue viruses by preventing cytosolic entry of viruses and restricting an early step in viral replication (62). Furthermore, IFNs prevent emergence of viral genomes from the endosomal pathway and that IFITM3 is both necessary and sufficient for this function. In addition, mice lacking the IFITM locus exhibited accelerated progression, greater mortality, and higher systemic viral burdens in response to influenza A virus infection (63). Importantly, IFITM3 can be strongly induced by interleukin 6 (IL-6) in rat forebrain cultures and IL-6 appears to be a critical modulator in the cascade of maternal immune activation (29). Most recently, preliminary results from the Volk laboratory demonstrate that schizophrenia subjects with higher IFITM mRNA levels in endothelial cells also have greater disturbances in cortical GABA neurons in the PFC (64).
While previous microarray expression data argue that both the IFITM and SERPINA 3 signatures are strong in the prefrontal cortex of subjects with schizophrenia, they also suggest that they are characteristic of a subpopulation of diseased subjects and cannot be found in the unaffected individuals (12, 13). Furthermore, it is important to point out that IFITM family and SERPINA3 expression changes are tightly correlated across subjects with schizophrenia (r=0.79-0.86) (12). Furthermore, two other immune system related genes, Chitinase-3-like 1 (CHI3L1) and CD14 antigen, also show highly correlated changes with the IFITM family and SERPINA3 (Arion et al 2007), suggesting that these genes might act in concert during immune activation. Importantly, a multi-center case-control study and meta-analysis suggests that CHI3L1 confers a susceptibility to schizophrenia in Asian populations (65, 66) and the SNP in the CHI3L1 gene might be correlated with personality traits (67). In the human and non-human primate neocortex the IFITM genes are expressed at their highest levels in the pia mater (Figure 2B), which tightly adheres to the cortical surface, and show a modest expression level in the endothelial cells of small blood vessels. On the other hand, CD14 is a surface antigen that is preferentially expressed on monocytes/macrophages and outgrowth endothelial cells (68) and is known to mediate the innate immune response to bacterial lipopolysaccharide that is controlled through a p38(MAPK)-STAT3 molecular pathway (69), while CHI3L1 expression is prominent in macrophages, neutrophils, chondrocytes, and vascular smooth muscle cells (70) and has a chemotactic effect on endothelium and vascular smooth muscle cells (71). In contrast, SERPINA3 is primarily produced by activated astroglia (72). Thus, while IFITM, SERPINA3, CHI3L1 and CD14 all appeared to be related to immune activation, they are expressed in different cell types and the relationship between these changes remains to be further investigated.
V. Development and immune activation
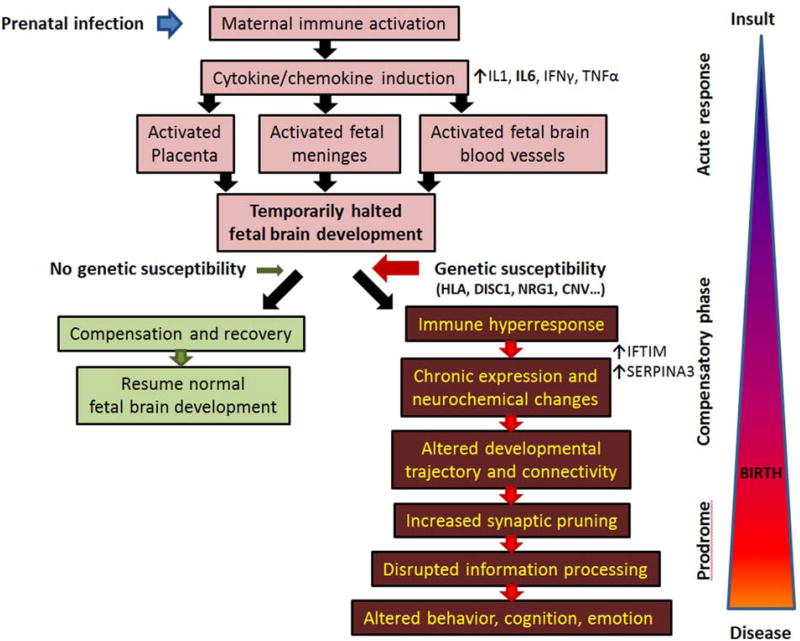
Based on the accumulated data across many fields of research, it is becoming apparent that immune system disturbances do not always directly target the developing brain cells (Figure 3). Maternal viral infection and subsequent immune activation have prominent effects on the developing brain, even though the virus itself is not detected in the fetal brain tissue (73). First, during maternal immune activation (e.g. viral infection) a strong induction of chemokines and cytokines occurs in the expecting mother. This maternal immune response consists, but is not limited to, a sharp rise in IL-1, IL-6, IFNγ and TNFα levels, which trigger a cascade of secondary events, targeting both the placenta and the developing fetus. Paracrine signaling between fetal and maternal cells is critical for establishing a state of immunosuppression (74). The placenta and fetal body acutely respond to the rise in IL-6 levels with a chemical imbalance and thousands of newly synthesized proteins floods the developing fetal brain (74). The fetal brain is vulnerable, the immature blood vessels are leaky (75), and the response of the fetal brain is acute and strong, encompassing gene expression changes in neuronal, glial, endothelial, and meningeal cells. In the brain cells there is a strong induction of crystallins (76), which are known to be also dysregulated in the postmortem brains of subjects with schizophrenia (38, 77, 78). We hypothesize that the cytokines/chemokines on the meninges and endothelial cells lead to secondary secretory changes, which diffuse into the developing brain. We propose that in the absence of a genetic risk to schizophrenia, these events might result in temporarily arrested/altered development. In such non-susceptible individuals this developmental arrest might relatively quickly normalize and development could resume with only a minimal disruption. However, we believe that in the presence of genetic susceptibility, the response in the fetus is likely to be much stronger and/or prolonged (33, 79). As a result, the developmental trajectory might permanently change, potentially resulting in slightly altered cell division, migration patterns, and axonal pathfinding. In our view, the developing brain tries to compensate with a complex pattern of gene expression changes, the acute inflammatory reaction subsides, becomes chronic, and leaves behind an “immune scar” or “immune memory” for a lifetime. Yet, this “immune scar” appears to be continuously active and might further interfere with postnatal brain development, especially with normal synaptic pruning mechanisms during adolescence. Ultimately, the capacity of the brain's compensatory mechanisms might be exceeded, altering brain connectivity and information processing, leading to the emergence of the disease symptoms.
Figure 3. Schematic representation of maternal immune activation and its proposed role in schizophrenia pathophysiology.
Prenatal infection leads to strong maternal immune system activation, which induces a systemic release of cytokines, including, but not limited to IL1, IL6, TNFα and IFγ. These cytokines, and especially IL6, have multiple effects on the developing fetus involving activation of placenta, fetal blood vessel and fetal meninges. The developing brain, even in the absence of the virus, is flooded by cytokines and immune-response proteins. This leads to temporarily slowed/halted development, which in the absence of genetic susceptibility resumes normally in a relatively short period of time. Yet, in the presence of schizophrenia-predisposing genetic elements the fetal meninges and blood vessels are hyper responsive, the inflammatory response becomes chronic, and never quite disappears. This low-level, persistent blood vessel and meningeal immune activation results in continuous secretion of various chemokines and cytokines, changing postnatal developmental trajectory by interfering with proper neuronal connectivity and potentially altering normal synapse pruning. As a result, once the brain's intrinsic compensatory mechanisms are exhausted, information processing is disrupted, and the symptoms of the disease manifest themselves.
VI. Immune response: the role of meninges and blood vessels?
Meningeal activation in schizophrenia might be an important and overlooked aspect of pathophysiology. The pia mater is in intimate contact with the cortical surface and its function is critical for normal brain development. Recent studies clearly argue that the meningeal membranes are necessary and sufficient substrates for tangential migration of Cajal-Retzius cells (80), a reelin-expressing cell subpopulation that has been affected by schizophrenia (81). Furthermore, maternal immune activation by human influenza virus results in neuronal migration abnormalities in Cajal-Retzius cells via reduction in reelin production in neonatal brains (82). In addition, it appears that forebrain meninges are critical for the tangential migration of cortical interneurons along the cortical marginal zone and CXCL12 (a direct transcriptional target of Foxc1 in the meninges) is the factor controlling this process (83). Furthermore, CXCL12 expression appears to be under the direct control of cytokines such as IL-1beta and its expression is specifically altered in the blood-brain barrier in immune system disorders such as multiple sclerosis (84). Thus, we propose the following mechanism based on these findings. In genetically susceptible individuals, maternal immune activation results in increased IFN levels in the fetus, potentially targeting the meninges and inducing expression of IFITM genes as a preventive mechanism against the viral infection. The meninges would respond by secreting a host of soluble factors, including CXCL2, which would diffuse into the developing cortex. In genetically predisposed individuals, the acute response is likely to be prolonged, become chronic, and permanently alter the developmental trajectory of the brain. A similar, equally important process might also occur in the blood vessels and elegant studies of Volk and colleagues report higher IFITM mRNA levels were positively correlated with cortical GABA neuron disturbances in subjects with schizophrenia (64). This, together with the observed SERPINA3, CHI3L1 and CD14 might be directly related to impaired development or altered function of blood-brain barrier in a subset of patients with schizophrenia (85, 86).
In the light of these studies, increased expression of IFITM proteins, SERPINA3 CD14, and CHI3L1 in schizophrenia might be directly related to the pathophysiology of the disease. Of these genes, it appears that IFITM3 expression might be the most critical for schizophrenia pathophysiology, since IFITM3 expression and activation are necessary for the CNS effects of maternal immune activation: IFITM3 is strongly induced following neonatal immune challenge by polyI:C and the offspring of IFITM3-/- knockout mice did not show decreased MAP2 expression, reduced spine density, diminished dendrite complexity in the frontal cortex, or memory impairments which were all observed in control mice following MIA (87).
This view is also supported by the findings that the supragranular cortical layers, the closest to the putatively altered meningeal homeostasis, show pronounced abnormalities in the brain of subjects with schizophrenia. These encompass diverse findings including, but are not limited to reductions in average size of neuronal somata (88), reduced density of ankyrin-G-immunoreactive axon initial segments, reduced reelin expression, altered AMPA-trafficking (89), increased expression of stargazin, and increased methyltransferase expression in layer 1 interneurons (90), and multiple GABA-ergic disturbances (91-93).
Finally, although increased expression of IFITM and SERPINA3 occur in different cellular compartments, it is also possible that they are directly related to each other: the immune response activated endothelial and meningeal secretome might diffuse into the cortex and activate cortical perivascular astrocytes, which in genetically predisposed individuals respond by a sustained, chronically elevated SERPINA3 expression, explaining the high correlation between the IFITM and SERPINA3 transcript levels, and perhaps with CHIL3L1 and CD14 levels.
VII. Immune system based therapeutic approaches and challenges
Overall, the data from epidemiologic, genetic, gene expression, biomarker, pharmacological, and animal studies unequivocally suggest that immune system activation plays an important role in developing schizophrenia. Furthermore, the data strongly argue that the immune system activation is persistent throughout the disease, at least in a subpopulation of patients. However, exploiting this knowledge for developing specific therapeutic approaches will prove quite challenging. First, modifying the function of immune pathways, including IL-6 signaling or a JAK/STAT signaling pathway, are likely to have deleterious systemic side effects. Second, identification of patients who would benefit from such a future therapy will be challenging, as the peripheral or imaging biomarkers might not be sufficiently specific and sensitive to be diagnostic. Third, it is likely that the maximum benefit from an immune intervention would be in early life, during the initial immune system hyperactivation in the genetically susceptible individuals, long before the disease is diagnosed, and at a time point when a perceived risk of developing schizophrenia will hardly justify aggressive therapeutically interventions. As a result, combined genetics, biomarker and epidemiological studies might hold the key to identify subpopulations of patients (and potentially future patients) who might benefit the most from modifying the immune response. However, as the immune activation appears to be a chronic, sustained, and symptom-associated process in schizophrenia, we cannot exclude the possibility that modification of a chronic immune response will have a beneficial effect on the treatment of the disease even in adulthood. Thus, if the IFITM genes play a key role of dampening the detrimental effects of immune response on the brain (and are not only potential biomarkers of immune activation), they might turn out to be intriguing, knowledge-based drug targets. As they are membrane proteins, they would be much easier to target with novel, specific compounds than the various intracellular cascades. However, as our current understanding of IFITM function on immune activation and brain development is limited, further mechanistic experiments will be required before we can claim IFITM genes as legitimate drug targets for schizophrenia treatment or prevention.
Acknowledgments
The authors are grateful to Martin Schmidt for helpful suggestions and edits of the manuscript. KM's work is supported by NIMH grants R01MH067234 and R01 MH079299.
Footnotes
Financial Disclosures: Drs. Mirnics and Horvath reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Michel M, Schmidt MJ, Mirnics K. Immune system gene dysregulation in autism and schizophrenia. Dev Neurobiol. 2012;72:1277–1287. doi: 10.1002/dneu.22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2010;93:23–58. doi: 10.1016/j.pneurobio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown AS, Derkits EJ. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am J Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller N. COX-2 inhibitors as antidepressants and antipsychotics: clinical evidence. Curr Opin Investig Drugs. 2010;11:31–42. [PubMed] [Google Scholar]
- 5.Muller N, Myint AM, Schwarz MJ. Immunological treatment options for schizophrenia. Curr Pharm Biotechnol. 2012;13:1606–1613. doi: 10.2174/138920112800784826. [DOI] [PubMed] [Google Scholar]
- 6.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bamne M, Wood J, Chowdari K, Watson AM, Celik C, Mansour H, et al. Evaluation of HLA polymorphisms in relation to schizophrenia risk and infectious exposure. Schizophr Bull. 2012;38:1149–1154. doi: 10.1093/schbul/sbs087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agartz I, Brown AA, Rimol LM, Hartberg CB, Dale AM, Melle I, et al. Common sequence variants in the major histocompatibility complex region associate with cerebral ventricular size in schizophrenia. Biol Psychiatry. 2011;70:696–698. doi: 10.1016/j.biopsych.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 9.Consortium SPG-WAS. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia P, Wang L, Fanous AH, Chen X, Kendler KS, Zhao Z. A bias-reducing pathway enrichment analysis of genome-wide association data confirmed association of the MHC region with schizophrenia. J Med Genet. 2011;49:96–103. doi: 10.1136/jmedgenet-2011-100397. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Underhill J, Liu XH, Sham PC, Donaldson P, Murray RM, et al. Transmission disequilibrium analysis of HLA class II DRB1, DQA1, DQB1 and DPB1 polymorphisms in schizophrenia using family trios from a Han Chinese population. Schizophr Res. 2001;49:73–78. doi: 10.1016/s0920-9964(00)00111-0. [DOI] [PubMed] [Google Scholar]
- 12.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arion D, Horvath S, Lewis DA, Mirnics K. Infragranular gene expression disturbances in the prefrontal cortex in schizophrenia: signature of altered neural development? Neurobiol Dis. 2009;37:738–746. doi: 10.1016/j.nbd.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fillman SG, Cloonan N, Catts VS, Miller LC, Wong J, McCrossin T, et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2012;18:206–214. doi: 10.1038/mp.2012.110. [DOI] [PubMed] [Google Scholar]
- 16.Miller BJ, Buckley P, Seabolt W, Mellor A, Kirkpatrick B. Meta-analysis of cytokine alterations in schizophrenia: clinical status and antipsychotic effects. Biol Psychiatry. 2011;70:663–671. doi: 10.1016/j.biopsych.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurian SM, Le-Niculescu H, Patel SD, Bertram D, Davis J, Dike C, et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatry. 2009;16:37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- 18.Drexhage RC, Knijff EM, Padmos RC, Heul-Nieuwenhuijzen L, Beumer W, Versnel MA, et al. The mononuclear phagocyte system and its cytokine inflammatory networks in schizophrenia and bipolar disorder. Expert Rev Neurother. 2009;10:59–76. doi: 10.1586/ern.09.144. [DOI] [PubMed] [Google Scholar]
- 19.Song XQ, Lv LX, Li WQ, Hao YH, Zhao JP. The interaction of nuclear factor-kappa B and cytokines is associated with schizophrenia. Biol Psychiatry. 2009;65:481–488. doi: 10.1016/j.biopsych.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Steiner J, Walter M, Glanz W, Sarnyai Z, Bernstein HG, Vielhaber S, et al. Increased prevalence of diverse N-methyl-D-aspartate glutamate receptor antibodies in patients with an initial diagnosis of schizophrenia: specific relevance of IgG NR1a antibodies for distinction from N-methyl-D-aspartate glutamate receptor encephalitis. JAMA Psychiatry. 2013;70:271–278. doi: 10.1001/2013.jamapsychiatry.86. [DOI] [PubMed] [Google Scholar]
- 21.Berk M, Dean O, Drexhage H, McNeil JJ, Moylan S, Oneil A, et al. Aspirin: a review of its neurobiological properties and therapeutic potential for mental illness. BMC Med. 2013;11:74. doi: 10.1186/1741-7015-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller N, Krause D, Dehning S, Musil R, Schennach-Wolff R, Obermeier M, et al. Celecoxib treatment in an early stage of schizophrenia: results of a randomized, double-blind, placebo-controlled trial of celecoxib augmentation of amisulpride treatment. Schizophr Res. 2010;121:118–124. doi: 10.1016/j.schres.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Sugino H, Futamura T, Mitsumoto Y, Maeda K, Marunaka Y. Atypical antipsychotics suppress production of proinflammatory cytokines and up-regulate interleukin-10 in lipopolysaccharide-treated mice. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:303–307. doi: 10.1016/j.pnpbp.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Dean B. Understanding the role of inflammatory-related pathways in the pathophysiology and treatment of psychiatric disorders: evidence from human peripheral studies and CNS studies. Int J Neuropsychopharmacol. 2010;14:997–1012. doi: 10.1017/S1461145710001410. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Fatemi SH, Sidwell RW, Patterson PH. Maternal influenza infection causes marked behavioral and pharmacological changes in the offspring. J Neurosci. 2003;23:297–302. doi: 10.1523/JNEUROSCI.23-01-00297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nawa H, Takei N. Recent progress in animal modeling of immune inflammatory processes in schizophrenia: implication of specific cytokines. Neurosci Res. 2006;56:2–13. doi: 10.1016/j.neures.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Nawa H, Yamada K. Experimental schizophrenia models in rodents established with inflammatory agents and cytokines. Methods Mol Biol. 2012;829:445–451. doi: 10.1007/978-1-61779-458-2_28. [DOI] [PubMed] [Google Scholar]
- 28.Brown AS, Schaefer CA, Quesenberry CP, Jr, Liu L, Babulas VP, Susser ES. Maternal exposure to toxoplasmosis and risk of schizophrenia in adult offspring. Am J Psychiatry. 2005;162:767–773. doi: 10.1176/appi.ajp.162.4.767. [DOI] [PubMed] [Google Scholar]
- 29.Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007;27:10695–10702. doi: 10.1523/JNEUROSCI.2178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boksa P. Effects of prenatal infection on brain development and behavior: a review of findings from animal models. Brain Behav Immun. 2010;24:881–897. doi: 10.1016/j.bbi.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Moreno JL, Kurita M, Holloway T, Lopez J, Cadagan R, Martinez-Sobrido L, et al. Maternal influenza viral infection causes schizophrenia-like alterations of 5-HT(2)A and mGlu(2) receptors in the adult offspring. J Neurosci. 2010;31:1863–1872. doi: 10.1523/JNEUROSCI.4230-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giovanoli S, Engler H, Engler A, Richetto J, Voget M, Willi R, et al. Stress in puberty unmasks latent neuropathological consequences of prenatal immune activation in mice. Science. 2013;339:1095–1099. doi: 10.1126/science.1228261. [DOI] [PubMed] [Google Scholar]
- 33.Lipina TV, Zai C, Hlousek D, Roder JC, Wong AH. Maternal Immune Activation during Gestation Interacts with Disc1 Point Mutation to Exacerbate Schizophrenia-Related Behaviors in Mice. J Neurosci. 2013;33:7654–7666. doi: 10.1523/JNEUROSCI.0091-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath S, Janka Z, Mirnics K. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry. 2010;69:157–162. doi: 10.1016/j.biopsych.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Horvath S, Mirnics K. Breaking the gene barrier in schizophrenia. Nat Med. 2009;15:488–490. doi: 10.1038/nm0509-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirnics K, Middleton FA, Lewis DA, Levitt P. Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci. 2001;24:479–486. doi: 10.1016/s0166-2236(00)01862-2. [DOI] [PubMed] [Google Scholar]
- 38.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 39.Faludi G, Mirnics K. Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci. 2011;29:305–309. doi: 10.1016/j.ijdevneu.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, et al. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 45.Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE, et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- 46.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian V. A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen Psychiatry. 2012;69:1205–1213. doi: 10.1001/archgenpsychiatry.2012.704. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Mathieu SL, Harris R, Ji J, Anderson DJ, Malysz J, et al. Role of alpha7 nicotinic acetylcholine receptors in regulating tumor necrosis factor-alpha (TNF-alpha) as revealed by subtype selective agonists. J Neuroimmunol. 2011;239:37–43. doi: 10.1016/j.jneuroim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Lee M. Neurotransmitters and microglial-mediated neuroinflammation. Curr Protein Pept Sci. 2013;14:21–32. doi: 10.2174/1389203711314010005. [DOI] [PubMed] [Google Scholar]
- 50.McGavern DB, Kang SS. Illuminating viral infections in the nervous system. Nat Rev Immunol. 2011;11:318–329. doi: 10.1038/nri2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsel P, Davis KL, Gorman JM, Haroutunian V. Variations in differential gene expression patterns across multiple brain regions in schizophrenia. Schizophr Res. 2005;77:241–252. doi: 10.1016/j.schres.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 52.Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin) Front Biosci. 2007;12:2821–2835. doi: 10.2741/2275. [DOI] [PubMed] [Google Scholar]
- 53.Twine NA, Janitz C, Wilkins MR, Janitz M. Sequencing of hippocampal and cerebellar transcriptomes provides new insights into the complexity of gene regulation in the human brain. Neurosci Lett. 2013;541:263–268. doi: 10.1016/j.neulet.2013.02.034. [DOI] [PubMed] [Google Scholar]
- 54.Gopalan SM, Wilczynska KM, Konik BS, Bryan L, Kordula T. Nuclear factor-1-X regulates astrocyte-specific expression of the alpha1-antichymotrypsin and glial fibrillary acidic protein genes. J Biol Chem. 2006;281:13126–13133. doi: 10.1074/jbc.M601194200. [DOI] [PubMed] [Google Scholar]
- 55.Gopalan SM, Wilczynska KM, Konik BS, Bryan L, Kordula T. Astrocyte-specific expression of the alpha1-antichymotrypsin and glial fibrillary acidic protein genes requires activator protein-1. J Biol Chem. 2006;281:1956–1963. doi: 10.1074/jbc.M510935200. [DOI] [PubMed] [Google Scholar]
- 56.Horvath AJ, Irving JA, Rossjohn J, Law RH, Bottomley SP, Quinsey NS, et al. The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins. J Biol Chem. 2005;280:43168–43178. doi: 10.1074/jbc.M505598200. [DOI] [PubMed] [Google Scholar]
- 57.Chelbi ST, Wilson ML, Veillard AC, Ingles SA, Zhang J, Mondon F, et al. Genetic and epigenetic mechanisms collaborate to control SERPINA3 expression and its association with placental diseases. Hum Mol Genet. 2013;21:1968–1978. doi: 10.1093/hmg/dds006. [DOI] [PubMed] [Google Scholar]
- 58.Chelbi ST, Mondon F, Jammes H, Buffat C, Mignot TM, Tost J, et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension. 2007;49:76–83. doi: 10.1161/01.HYP.0000250831.52876.cb. [DOI] [PubMed] [Google Scholar]
- 59.Taylor KE, Mossman KL. Recent advances in understanding viral evasion of type I interferon. Immunology. 2013;138:190–197. doi: 10.1111/imm.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volk DW, Siegel BI, Sengupta EJ, Edelson JR, Lewis DA. Elevated Transcript Levels for Viral Restriction Factors in Cortical Endothelial Cells in Schizophrenia. Neuropsychopharmacology. 2012;38:T32. [Google Scholar]
- 65.Ohi K, Hashimoto R, Yasuda Y, Yoshida T, Takahashi H, Iike N, et al. The chitinase 3-like 1 gene and schizophrenia: evidence from a multi-center case-control study and meta-analysis. Schizophr Res. 2010;116:126–132. doi: 10.1016/j.schres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 66.Yang MS, Morris DW, Donohoe G, Kenny E, O'Dushalaine CT, Schwaiger S, et al. Chitinase-3-like 1 (CHI3L1) gene and schizophrenia: genetic association and a potential functional mechanism. Biol Psychiatry. 2008;64:98–103. doi: 10.1016/j.biopsych.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Yamamori H, Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Kasahara E, et al. A promoter variant in the chitinase 3-like 1 gene is associated with serum YKL-40 level and personality trait. Neurosci Lett. 2012;513:204–208. doi: 10.1016/j.neulet.2012.02.039. [DOI] [PubMed] [Google Scholar]
- 68.Medina RJ, O'Neill CL, Sweeney M, Guduric-Fuchs J, Gardiner TA, Simpson DA, et al. Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC Med Genomics. 2010;3:18. doi: 10.1186/1755-8794-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bode JG, Ehlting C, Haussinger D. The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal. 2012;24:1185–1194. doi: 10.1016/j.cellsig.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 70.Ringsholt M, Hogdall EV, Johansen JS, Price PA, Christensen LH. YKL-40 protein expression in normal adult human tissues--an immunohistochemical study. J Mol Histol. 2007;38:33–43. doi: 10.1007/s10735-006-9075-0. [DOI] [PubMed] [Google Scholar]
- 71.Junker N, Johansen JS, Hansen LT, Lund EL, Kristjansen PE. Regulation of YKL-40 expression during genotoxic or microenvironmental stress in human glioblastoma cells. Cancer Sci. 2005;96:183–190. doi: 10.1111/j.1349-7006.2005.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasternack JM, Abraham CR, Van Dyke BJ, Potter H, Younkin SG. Astrocytes in Alzheimer's disease gray matter express alpha 1-antichymotrypsin mRNA. Am J Pathol. 1989;135:827–834. [PMC free article] [PubMed] [Google Scholar]
- 73.Patterson PH. Immune involvement in schizophrenia and autism: etiology, pathology and animal models. Behav Brain Res. 2009;204:313–321. doi: 10.1016/j.bbr.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 74.Hsiao EY, Patterson PH. Placental regulation of maternal-fetal interactions and brain development. Dev Neurobiol. 2012;72:1317–1326. doi: 10.1002/dneu.22045. [DOI] [PubMed] [Google Scholar]
- 75.Daneman R. The blood-brain barrier in health and disease. Ann Neurol. 2013;72:648–672. doi: 10.1002/ana.23648. [DOI] [PubMed] [Google Scholar]
- 76.Garbett KA, Hsiao EY, Kalman S, Patterson PH, Mirnics K. Effects of maternal immune activation on gene expression patterns in the fetal brain. Transl Psychiatry. 2012;2:e98. doi: 10.1038/tp.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martins-de-Souza D, Maccarrone G, Wobrock T, Zerr I, Gormanns P, Reckow S, et al. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J Psychiatr Res. 2010;44:1176–1189. doi: 10.1016/j.jpsychires.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 78.Martins-de-Souza D, Schmitt A, Roder R, Lebar M, Schneider-Axmann T, Falkai P, et al. Sex-specific proteome differences in the anterior cingulate cortex of schizophrenia. J Psychiatr Res. 2010;44:989–991. doi: 10.1016/j.jpsychires.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 79.Abazyan B, Nomura J, Kannan G, Ishizuka K, Tamashiro KL, Nucifora F, et al. Prenatal interaction of mutant DISC1 and immune activation produces adult psychopathology. Biol Psychiatry. 2010;68:1172–1181. doi: 10.1016/j.biopsych.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borrell V, Marin O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat Neurosci. 2006;9:1284–1293. doi: 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- 81.Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci U S A. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fatemi SH, Emamian ES, Kist D, Sidwell RW, Nakajima K, Akhter P, et al. Defective corticogenesis and reduction in Reelin immunoreactivity in cortex and hippocampus of prenatally infected neonatal mice. Mol Psychiatry. 1999;4:145–154. doi: 10.1038/sj.mp.4000520. [DOI] [PubMed] [Google Scholar]
- 83.Zarbalis K, Choe Y, Siegenthaler JA, Orosco LA, Pleasure SJ. Meningeal defects alter the tangential migration of cortical interneurons in Foxc1hith/hith mice. Neural Dev. 2012;7:2. doi: 10.1186/1749-8104-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCandless EE, Budde M, Lees JR, Dorsey D, Lyng E, Klein RS. IL-1R signaling within the central nervous system regulates CXCL12 expression at the blood-brain barrier and disease severity during experimental autoimmune encephalomyelitis. J Immunol. 2009;183:613–620. doi: 10.4049/jimmunol.0802258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steiner J, Bogerts B, Sarnyai Z, Walter M, Gos T, Bernstein HG, et al. Bridging the gap between the immune and glutamate hypotheses of schizophrenia and major depression: Potential role of glial NMDA receptor modulators and impaired blood-brain barrier integrity. World J Biol Psychiatry. 2011;13:482–492. doi: 10.3109/15622975.2011.583941. [DOI] [PubMed] [Google Scholar]
- 86.Watanabe Y, Someya T, Nawa H. Cytokine hypothesis of schizophrenia pathogenesis: evidence from human studies and animal models. Psychiatry Clin Neurosci. 2010;64:217–230. doi: 10.1111/j.1440-1819.2010.02094.x. [DOI] [PubMed] [Google Scholar]
- 87.Ibi D, Nagai T, Nakajima A, Mizoguchi H, Kawase T, Tsuboi D, et al. Astroglial IFITM3 mediates neuronal impairments following neonatal immune challenge in mice. Glia. 2013;61:679–693. doi: 10.1002/glia.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry. 1998;55:215–224. doi: 10.1001/archpsyc.55.3.215. [DOI] [PubMed] [Google Scholar]
- 89.Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of AMPA receptor trafficking and signaling molecule transcripts in the prefrontal cortex in schizophrenia. Synapse. 2006;60:585–598. doi: 10.1002/syn.20329. [DOI] [PubMed] [Google Scholar]
- 90.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- 91.Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, Jr, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 92.Beasley CL, Zhang ZJ, Patten I, Reynolds GP. Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biol Psychiatry. 2002;52:708–715. doi: 10.1016/s0006-3223(02)01360-4. [DOI] [PubMed] [Google Scholar]
- 93.Glausier JR, Lewis DA. Selective pyramidal cell reduction of GABA(A) receptor alpha1 subunit messenger RNA expression in schizophrenia. Neuropsychopharmacology. 2011;36:2103–2110. doi: 10.1038/npp.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu JQ, Wang X, Beveridge NJ, Tooney PA, Scott RJ, Carr VJ, et al. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One. 7:e36351. doi: 10.1371/journal.pone.0036351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, Mirnics K, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]