Summary
Translational regulation plays an essential role in Drosophila ovarian germline stem cell (GSC) biology. GSC self-renewal requires two translational repressors, Nanos (Nos) and Pumilio (Pum), which repress the expression of differentiation factors in the stem cells. The molecular mechanisms underlying this translational repression remain unknown. Here, we show that the CCR4 deadenylase is required for GSC self-renewal and that Nos and Pum act through its recruitment onto specific mRNAs. We identify mei-P26 mRNA as a direct and major target of Nos/Pum/CCR4 translational repression in the GSCs. mei-P26 encodes a protein of the Trim-NHL tumor suppressor family that has conserved functions in stem cell lineages. We show that fine-tuning Mei-P26 expression by CCR4 plays a key role in GSC self-renewal. These results identify the molecular mechanism of Nos/Pum function in GSC self-renewal and reveal the role of CCR4-NOT-mediated deadenylation in regulating the balance between GSC self-renewal and differentiation.
Highlights
-
•
The CCR4 deadenylase is required for female germline stem cell self-renewal
-
•
Nos/Pum recruit CCR4-NOT for translational repression in germline stem cells
-
•
mei-P26 mRNA is a major target of translational repression by Nos/Pum/CCR4
-
•
Fine-tuning of mei-P26 by CCR4 is required for germline stem cell self-renewal
Introduction
A major issue in stem cell biology concerns understanding the mechanisms controlling the balance between self-renewal and differentiation. Drosophila germline stem cells (GSCs) have proven to be an excellent model for studying adult stem cells in vivo (Fuller and Spradling, 2007). In the Drosophila ovary, two to three GSCs are localized at the anterior of the germarium, the anteriormost region of each ovariole, and give rise to the female germline. The GSCs divide asymmetrically to produce a GSC and a cell that differentiates as a cystoblast. The cystoblast then divides four times to produce a cyst of 16 germline cells.
Translational controls have a central role in the regulation of stem cell biology. The importance of translational regulations has been reported in mouse embryonic stem cells, which display a considerable increase in mRNA levels and translation during their differentiation (Sampath et al., 2008). In Drosophila GSCs, two major factors for stem cell self-renewal are the translational repressors Nanos (Nos) and Pumilio (Pum) (Gilboa and Lehmann, 2004; Wang and Lin, 2004). Females mutant for nos and pum have empty ovaries due to the loss of GSCs by differentiation. Nos and Pum are thus required in the stem cells to repress their differentiation, indicating that stem cell self-renewal corresponds in part to the repression of the differentiation program (Gilboa and Lehmann, 2004; Wang and Lin, 2004). The microRNA (miRNA) pathway also plays an essential role in GSC self-renewal. Mutations in Dicer-1, Argonaute1 (Ago1), and loquacious result in a phenotype of stem cell loss consistent with the potential role of the miRNA pathway in translational repression of differentiation factors in the GSCs (Jin and Xie, 2007; Park et al., 2007; Yang et al., 2007).
Bag of marbles (Bam) is the major factor of GSC differentiation (McKearin and Ohlstein, 1995; Ohlstein and McKearin, 1997). bam mutant females have tumorous ovaries full of stem cell-like germ cells, whereas overexpression of bam in stem cells leads to their differentiation. bam transcription in GSCs is repressed by the short-range bone morphogenetic protein (BMP) signaling that emanates from the niche, the microenvironment provided by somatic cells surrounding the GSCs (Song et al., 2004; Xie and Spradling, 1998). Upon division, the daughter cell still in contact with the niche continues to receive the BMP signal and thus remains a stem cell, whereas the daughter cell localized posteriorly expresses bam due to the lack of BMP signal and thereby differentiates into a cystoblast (Harris et al., 2011; Xia et al., 2012). Genetic data suggest that Bam promotes differentiation by relieving the Nos/Pum-dependent translational repression of differentiation factors (Chen and McKearin, 2005; Szakmary et al., 2005). Consistent with this, Bam downregulates Nos expression in cystoblasts through the regulation of nos mRNA (Li et al., 2009).
To date, a single mRNA target of Nos/Pum regulation has been identified: the brain tumor (brat) mRNA, which encodes a Trim-NHL domain-containing protein with a known function in stem cell biology (Harris et al., 2011). Brat is a key differentiation factor in neural stem cells (Betschinger et al., 2006; Lee et al., 2006) and was recently shown to be involved in female GSC differentiation by repressing the translation of self-renewal mRNAs, including Mad, which encodes a component of BMP signaling (Harris et al., 2011). Mei-P26 is another Trim-NHL protein with an essential function in the ovarian stem cell lineage. It promotes differentiation and restricts proliferation in cyst cells by inhibiting the miRNA pathway through its direct association with Ago1 (Neumüller et al., 2008). More recently, a distinct role for Mei-P26 in GSC self-renewal was described (Li et al., 2012). Mei-P26 was found to repress Brat expression in GSCs, thus allowing BMP signaling and Bam repression, which are required for GSC self-renewal. However, Mei-P26 overexpression in GSCs leads to GSC loss, highlighting the importance of the precise regulation of Mei-P26 expression levels for GSC biology (Neumüller et al., 2008).
The mechanisms of action of Nos and Pum in GSC self-renewal remain unknown. In the embryo, Nos and Pum act by two mechanisms: inhibition of translation initiation (Sonoda and Wharton, 1999) and recruitment of the CCR4-NOT deadenylation complex (Kadyrova et al., 2007). Direct interactions between Pum and the deadenylation complex are conserved from yeast to human (Goldstrohm et al., 2006; Kadyrova et al., 2007). Interactions between the CCR4-NOT deadenylation complex and Nanos2, a mouse homolog of Nos, have also been reported in mouse gonocytes where Nanos2 represses mRNAs involved in meiosis (Suzuki et al., 2010; Suzuki et al., 2012).
The Drosophila CCR4-NOT complex is composed of seven proteins: NOT1–NOT4, CAF40, and two potential deadenylases, CCR4 and CAF1 (Barckmann and Simonelig, 2013; Temme et al., 2004, 2010). Drosophila CCR4 deadenylase is encoded by the twin gene, which is required for early oogenesis. twin has a role in the control of germ cell divisions leading to 16-cell cysts, in germ cell survival, and in oocyte specification (Morris et al., 2005; Zaessinger et al., 2006). Here, we address the molecular mechanisms underlying Nos/Pum translational repression in the GSCs. We find that CCR4 is required for GSC self-renewal and interacts with Nos and Pum for this function. We identify mei-P26 mRNA as a direct target of the Nos/Pum/CCR4 complex. mei-P26 is a major target of this complex for GSC self-renewal, as GSC loss in twin mutants is rescued by lowering the gene dosage of mei-P26. In addition, we show that increased expression of Mei-P26 in twin mutant GSCs correlates with longer poly(A) tails of its mRNA. These data reveal that Nos and Pum translational repression in the GSCs depends on deadenylation by the CCR4-NOT complex. They also show that GSC fate requires the precise regulation of Mei-P26 levels and that this fine-tuning is achieved by CCR4-NOT-mediated repression.
Results
Function of CCR4 in GSC Self-Renewal
To address a potential function of CCR4 in GSCs, we first analyzed CCR4 expression in these cells. GSCs were identified by their anterior localization in the germarium and by the presence and anterior localization of a spherical organelle called the spectrosome, visualized using the Hu-li tai-shao marker (Hts/1B1 antibody) (Lin et al., 1994) (Figure 1A). CCR4 was present in the GSCs as well as in other cells in the germarium (Figure 1C) where it was mostly cytoplasmic and accumulated in discrete foci, as reported in other cell types in the ovary and embryo (Rouget et al., 2010; Temme et al., 2004; Zaessinger et al., 2006). CCR4 is encoded by the twin gene. CCR4 levels were strongly decreased in a strong hypomorphic allele twinDG24102 (see below) (Figure 1D).
Figure 1.
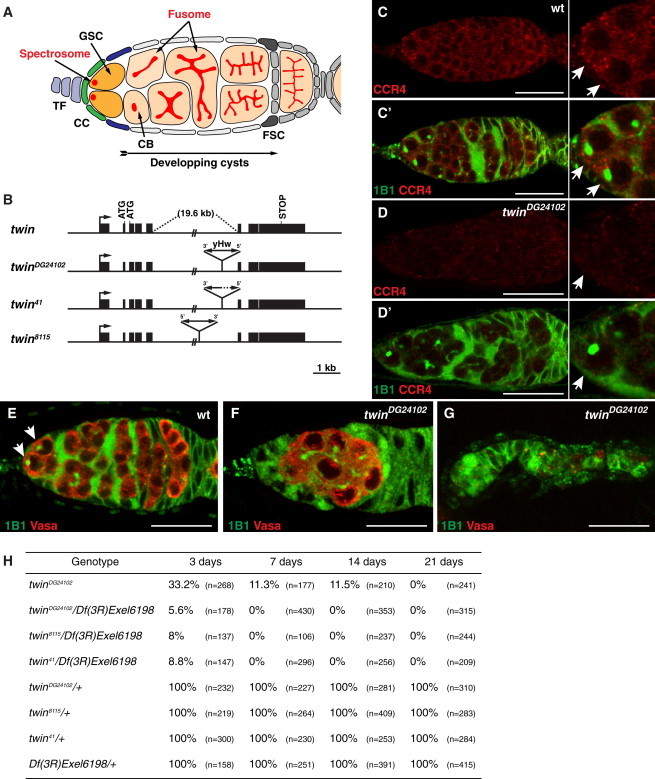
CCR4 Is Expressed in the GSCs and Is Required for Their Self-Renewal
(A) Schematic representation of a germarium. GSC, germline stem cell; CB, cystoblast; TF, terminal filament, CC, cap cell; and FSC, follicle stem cell.
(B) Schematic representation of the twin locus, twinDG24102, twin41, and twin8115 mutants. Black boxes indicate exons. The arrow indicates the transcription start site. The P-Hobo (yHw) transposable element (not drawn to scale) inserted in twinDG24102 is shown. The coordinates of the insertion sites are 20027036 for twinDG24102 and 20032277 for twin8115, according to the AE014297 sequence in NCBI.
(C–D′) Expression of CCR4 in GSCs. Wild-type (C and C′) and twinDG24102 mutant (D and D′) germaria labeled with anti-CCR4 antibody (red) and 1B1 (green), which marks the spectrosome and fusome. The merge is shown in C′ and D′. Right panels show higher magnifications of the anterior tips of germaria shown in the left panels. White arrows indicate GSCs.
(E–G) twin mutant phenotype of loss of GSCs. Wild-type (E) and twinDG24102 (F and G) mutant germaria labeled with 1B1 (green) and anti-Vasa antibody (red). Vasa is used as a germ cell marker. White arrows indicate GSCs. (F) Lack of GSCs and the presence of a differentiating cyst. (G) Lack of GSCs and germ cells. Scale bars represent 20 μm in (C–G).
(H) Quantification of germaria containing at least one GSC in different twin mutant and control genotypes in 3-, 7-, 14-, and 21-day-old females. n represents the number of germaria scored.
See also Figure S1.
We next analyzed GSC self-renewal in twin mutant ovaries using three alleles: twin8115, which we previously characterized as a strong hypomorphic allele (Zaessinger et al., 2006); twinDG24102 (FlyBase); and twin41, obtained after mobilization of the hybrid P-Hobo-element inserted into twinDG24102 (Figure 1B). twin mRNA levels quantified by RT-PCR and quantitative RT-PCR (qRT-PCR) were strongly reduced in the ovaries of twinDG24102 and twin41 mutants, indicating that they are strong hypomorphic alleles (Figure S1 available online). Ovaries from different allelic combinations were dissected from 3-, 7-, 14-, and 21-day-old females. All germ cells were rapidly lost in twin mutant ovaries (Figures 1E–1G). Quantification of germaria containing GSCs at the four time points showed that 100% of germaria were devoid of GSCs in 7-day-old females hemizygous for all three twin alleles over Df(3R)Exel6198 (Figure 1H). GSC loss was slightly slower in twinDG24102 homozygous females, consistent with the fact that this is not a null allele (Figure 1H). These results show that CCR4 is required for GSC self-renewal.
CCR4 Is Required in the GSCs for Their Self-Renewal
We used clonal analysis to determine if CCR4 was required intrinsically in the GSCs for their self-renewal. FLP-mediated FRT recombination was used to generate wild-type or twin mutant clonal GSCs that were analyzed at four time points after clone induction. Clonal GSCs were identified by the lack of GFP. In the wild-type, the percentage of germaria with at least one clonal GSC decreased from 31% 3 days after clone induction to 24% 21 days after clone induction, reflecting GSC turnover (Jin and Xie, 2007) (Figures 2A and 2B). In twin8115 and twinDG24102, the percentage of germaria with one clonal GSC was similar to wild-type 3 days after clone induction (34% and 31%) but strongly decreased at later time points, with only 5% and 3% of germaria containing a clonal GSC 21 days after clone induction, respectively (Figures 2A and 2B). This indicated that twin mutant GSCs were rapidly lost. This loss was more rapid in twinDG24102 mutant than in twin8115, indicating that twinDG24102 is a stronger allele.
Figure 2.
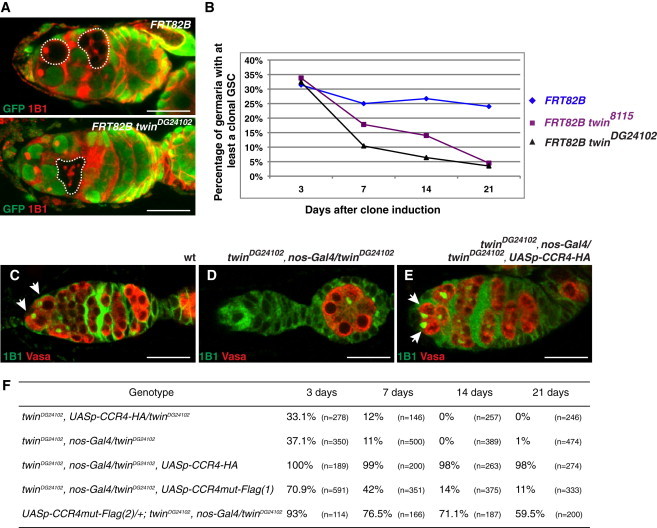
twin Is Cell-Autonomously Required in GSC Self-Renewal
(A) Clonal twin mutant GSCs do not self-renew. Control (top panel) and twinDG24102 (bottom panel) mosaic germaria labeled with GFP (green) and 1B1 (red) 7 days after clone induction. Clonal cells, marked by the lack of GFP, are outlined with white dotted lines.
(B) Quantification of germaria containing at least one clonal GSC 3, 7, 14, and 21 days after clone induction (n = 111 to 497 germaria).
(C–E) Rescue of twin mutant phenotype of GSC loss with CCR4 expression in germ cells. Wild-type (C); twinDG24102, nos-Gal4/twinDG24102 (D); twinDG24102, nos-Gal4/twinDG24102, UASp-CCR4-HA (E) labeled with 1B1 (green) and anti-Vasa (red). White arrows indicate GSCs. Scale bars represent 20 μm in (A) and (C–E).
(F) Quantification of germaria containing at least one GSC in twin mutant and rescued contexts, in 3-, 7-, 14-, and 21-day-old females. n represents the number of germaria scored.
See also Figure S2.
We determined the division rate of twin8115 and twinDG24102 GSCs by counting the number of cysts produced by a clonal marked mutant GSC and dividing it by the number of cysts produced by a wild-type unmarked GSC in the same germarium (Jin and Xie, 2007). The division rate of wild-type GSCs (FRT82B chromosome) was close to 1 (0.93, n = 89), while those of twin8115 and twinDG24102 GSCs were 0.44 (n = 37) and 0.50 (n = 33), respectively, indicating that division in twin mutant GSCs is slower than in wild-type.
The loss of twin mutant GSCs could result either from apoptosis or differentiation. Clonal twin mutant GSCs were able to give rise to differentiated cysts identified by a branched fusome (Figure 2A), indicating that these mutant GSCs can differentiate. Apoptosis was recorded using activated caspase-3 antibody. The twin mutant clonal GSCs were not stained with this antibody (n = 35) (Figures S2A–S2C), thus ruling out their potential death by apoptosis.
As independent evidence in favor of the cell-autonomous requirement of CCR4 for GSC self-renewal, we rescued the twinDG24102 phenotype of GSC loss by expressing CCR4 in the GSCs. We used the UAS/Gal4 system to express CCR4-hemagglutinin (CCR4-HA) specifically in the germ cells with the nos-Gal4 driver. This led to a nearly complete rescue of the twinDG24102 GSC loss phenotype (Figures 2C–2F).
In the CCR4-NOT complex, both CCR4 and CAF1 are potential deadenylases, and CAF1 has been reported to be the major deadenylase in Drosophila S2 cells, with CCR4 acting as a structural subunit in the complex (Temme et al., 2010). To determine whether the deadenylase activity of CCR4 is required for GSC self-renewal, we generated the CCR4mut-Flag transgene, in which CCR4 has a double point mutation inactivating the deadenylase activity (Temme et al., 2010). When expressed with nos-Gal4, this deadenylase-dead CCR4 was able to partially rescue the twinDG24102 phenotype of GSC loss, with 59.5% of germaria containing GSCs in 21-day-old females (Figures 2F and S2D). This indicated that the deadenylase activity of CCR4 could be compensated to some extent for GSC self-renewal. This could result from CAF1 acting as a deadenylase in the GSCs, as in S2 cells, and/or from an additional role of the CCR4-NOT complex in translational repression, independently of deadenylation, as proposed in other systems (Chekulaeva et al., 2011; Cooke et al., 2010).
To address whether CCR4 acts in the GSCs as part of the CCR4-NOT complex, we knocked down NOT1, the key structural subunit of the complex, using RNAi in the germ cells. This led to a strong phenotype of lack of GSCs and germ cells (Figure S2E). In addition, CCR4 was found in complex with NOT1 and CAF1 in GSC-like cells, using coimmunoprecipitation (co-IP) experiments on bamΔ86 mutant ovaries, which contain only undifferentiated precystoblasts (Figure S2F).
Taken together, these results show that CCR4, most likely as part of the CCR4-NOT complex, is required intrinsically in the GSCs to control their division rate and for their self-renewal. They also show that twin mutant GSCs are not maintained because they differentiate.
CCR4 Functions Together with Pum and Nos
To genetically determine if twin could act together with nos and pum, we first tested whether the GSC loss phenotype in the hypomorphic twinDG24102 allele could be increased by reducing the gene dosage of pum or nos. GSC loss in twinDG24102 was accelerated in the presence of both heterozygous pumMSC or nos18 mutations (Figure 3A). This is consistent with a role for twin and pum, and twin and nos, together in the same pathway.
Figure 3.
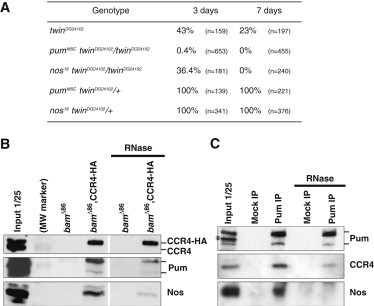
twin Genetically and Physically Interacts with nos and pum in GSCs
(A) Quantification of germaria containing at least one GSC in twin mutant 3- and 7-day-old females, in combination or not with pum or nos heterozygous mutants. n represents the number of germaria scored.
(B) Coimmunoprecipitations with CCR4-HA in GSC-like cells. Ovarian extracts from bamΔ86 and bamΔ86, nos-Gal4/bamΔ86, UASp-CCR4-HA flies were immunoprecipitated with anti-HA, either in the absence or the presence (right panel, RNase) of RNase A. Western blots were revealed with anti-CCR4, anti-Pum, and anti-Nos antibodies.
(C) Coimmunoprecipitations with Pum in GSC-like cells. Ovarian extracts from bamΔ86 flies were immunoprecipitated with anti-Pum (Pum IP) or mock immunoprecipitated (Mock IP), either in the absence or the presence (RNase) of RNase A. Western blots were revealed with anti-Pum, anti-CCR4, and anti-Nos antibodies. The asterisk marks a nonspecific band recognized by the anti-Pum antibody in the input. Input is the protein extract (1/25) prior to immunoprecipitation in (B) and (C).
See also Figure S3.
We analyzed intracellular colocalization in GSCs between Pum or Nos and CCR4 using CCR4-HA. Pum, Nos, and CCR4-HA were present diffusely in the cytoplasm and accumulated in cytoplasmic foci. Colocalization occurred mostly in diffusely distributed pools of proteins (Figures S3A–S3D), consistent with the hypothesis that deadenylation takes place outside foci (Barckmann and Simonelig, 2013).
We addressed whether CCR4, Pum, and Nos form a complex in GSC-like cells using co-IP experiments in bamΔ86 mutant ovaries. The CCR4-HA protein was able to coprecipitate both Pum and Nos in the absence of RNase (Figure 3B). Both co-IPs still occurred but were decreased in the presence of RNase. In the reverse experiment, Pum was able to coprecipitate Nos and CCR4. The presence of RNase decreased CCR4 co-IP and abolished that of Nos (Figures 3C and S3E). Nos and Pum are known to form a ternary complex with RNA, in which Nos stabilization depends on its interaction with both Pum and the RNA (Sonoda and Wharton, 1999). In light of these data, our results are consistent with the presence of CCR4, Pum, and Nos in the same complex and with the role of RNA in stabilizing Nos in the complex.
Taken together, these results indicate that CCR4, Pum, and Nos form a complex in the GSCs that is required for their self-renewal.
GSCs Lacking Both twin and bam Can Differentiate
Extensive analyses of pum/bam relationships have shown that pum is not involved in the repression of Bam expression in the GSCs (Chen and McKearin, 2005) and that GSCs double mutant for pum and bam can differentiate (Chen and McKearin, 2005; Szakmary et al., 2005). These data led to the proposition that bam antagonizes pum function in the cystoblasts: Bam would relieve the translational repression of differentiation factors imposed by Pum/Nos in the GSCs, thus promoting differentiation.
We reasoned that if twin is involved together with pum in translational repression of mRNA targets in the GSCs, twin/bam relationships should be similar to pum/bam relationships. To test this, we analyzed Bam protein expression in twinDG24102 germaria and found that Bam was not overexpressed in twin mutant GSCs (0%, n = 35) (Figures 4A and 4B), as was the case in pum mutant GSCs (Chen and McKearin, 2005). We also verified the lack of Bam upregulation in pum and nos mutant GSCs (0%, n = 18 and n = 43, respectively) (Figure S4A).
Figure 4.
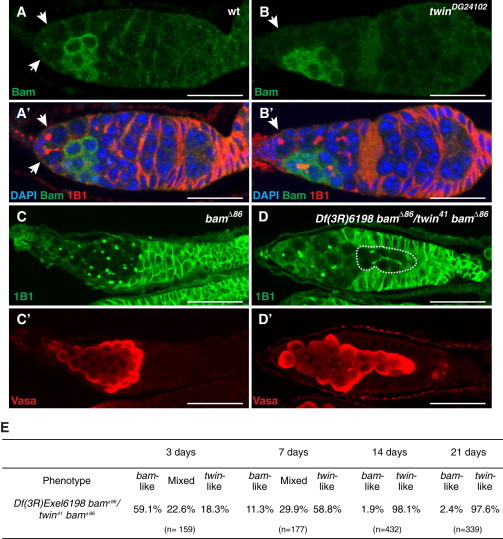
twin Function Is Epistatic to bam
(A and B) Bam levels are not upregulated in twin mutant GSCs. Wild-type (A) and twinDG24102 (B) germaria labeled with anti-Bam antibody (green). The merges between anti-Bam (green), DAPI (blue), and 1B1 (red) are shown in (A′) and (B′). White arrows indicate GSCs.
(C and D) twin bam double mutant GSCs can differentiate. bamΔ86 (C and C′) and Df(3R)Exel6198, bamΔ86/twin41, bamΔ86 (D and D′) germaria labeled with 1B1 (green) and anti-Vasa (red). A differentiated cyst marked by the presence of a branched fusome is outlined with a white dotted line. Scale bars represent 20 μm in (A–D).
(E) Quantification of germaria, labeled with 1B1 and anti-Vasa, presenting a bam-like phenotype (accumulation of GSC-like cells containing a spectrosome), a mixed phenotype (presence of GSC-like cells and of differentiating cysts, e.g., in D), or a twin-like phenotype (lack of GSCs or lack of germ cells) in Df(3R)Exel6198, bamΔ86/twin41, bamΔ86 double mutant germaria in 3-, 7-, 14-, and 21-day-old females. n represents the number of germaria scored.
See also Figure S4.
We then recorded the phenotype of twin bam double mutants. As reported for pum bam double mutants (Chen and McKearin, 2005), the twin bam phenotype was a mixture of both the bam phenotype (tumor of undifferentiated cells identified by the presence of a spectrosome; Figure 4C) and the twin phenotype (loss of GSCs). Similarly to pum bam, the twin bam phenotype evolved over time, with the percentage of bam phenotype decreasing and the percentage of twin phenotype increasing as the flies got older (Figures 4E and S4B). Importantly, twin bam GSCs were able to differentiate, as shown by the presence of cysts with branched fusomes and older cysts containing cells with polyploid nuclei or ring canals (Figures 4D, S4B, and S4C).
These results are consistent with twin and pum acting together in the repression of differentiation mRNAs in the GSCs for their self-renewal and with the role of bam in antagonizing twin/pum function for the differentiation in cystoblasts.
mei-P26 mRNA Is a Target of Nos/Pum/CCR4 Regulation in GSCs
Mei-P26 is a potent differentiation factor in the GSC lineage. Strong mei-P26 mutants develop ovarian tumors of cyst cells, and accordingly, Mei-P26 expression increases in -8-cell cysts and peaks in 16-cell cysts (Liu et al., 2009; Neumüller et al., 2008). However, Mei-P26 is already present in GSCs, although at lower levels, where it plays a role in self-renewal (Li et al., 2012). The tight regulation of Mei-P26 levels in the GSCs is essential, as overexpression of mei-P26 in GSCs leads to their loss (Neumüller et al., 2008) (Figure S5). We addressed whether mei-P26 regulation in the GSCs depended on the Nos/Pum/CCR4 complex. Because Pum is the specific RNA binding protein in the Nos/Pum complex, we used Pum immunoprecipitation (IP) experiments to investigate whether mei-P26 mRNA was in complex with Pum in GSC-like cells. Using RT-PCR and qRT-PCR, we found an enrichment of mei-P26 mRNA in Pum IP compared to in mock IP (Figures 5A and 5B). While CCR4 itself does not bind specifically to RNA, a specific interaction might be recorded between CCR4 and mRNAs resulting from the recruitment of CCR4 onto mRNAs by RNA binding proteins. We performed IP of CCR4-HA expressed with nos-Gal4 in bamΔ86 mutant ovaries and found an enrichment of mei-P26 mRNA in CCR4-HA IP compared to in mock IP (Figures 5A and 5B).
Figure 5.
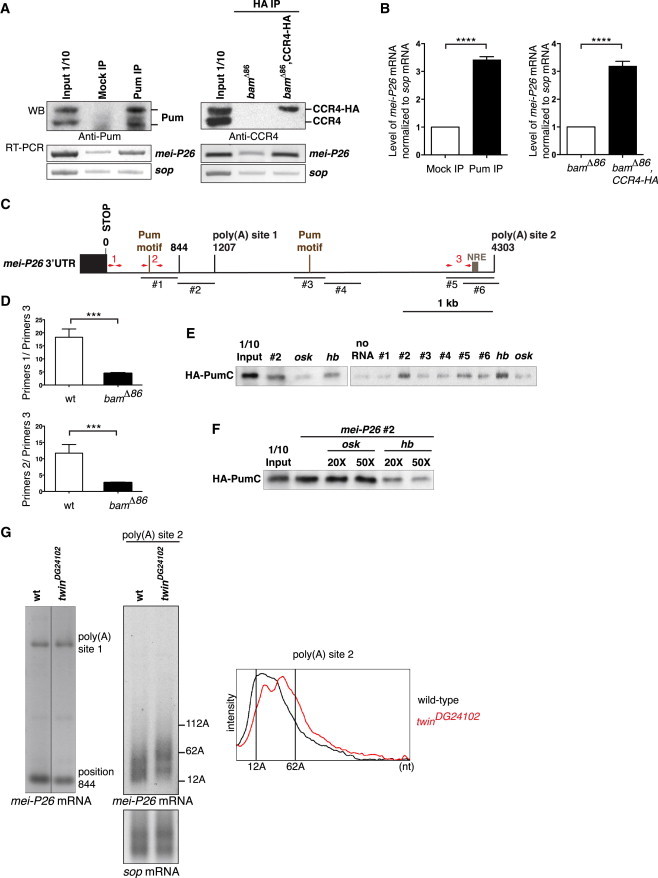
mei-P26 mRNA Is a Target of the Nos/Pum/CCR4 Complex in the GSCs
(A and B) RNA immunoprecipitations (IP) with the anti-Pum antibody in bamΔ86 ovarian extracts (left panels, Mock IP: preimmune serum) and with the anti-HA antibody in bamΔ86 and nos-Gal4, bamΔ86/UASp-CCR4-HA, bamΔ86 (bamΔ86, CCR4-HA) ovarian extracts (right panels). sop mRNA was used as a negative control. (A) Top panels show protein IP using western blots (WB). Bottom panels show the enrichment of mei-P26 mRNA in IP compared to in mock IP, visualized by RT-PCR. Inputs are protein or RNA extracts (1/10) prior to immunoprecipitation. Two (HA IP) and four (Pum IP) IPs were performed with similar results. (B) Quantification by qRT-PCR. Normalization was with sop mRNA. Quantifications were done in triplicate, and error bars represent SD. ∗∗∗∗p < 0.0001 using the t test.
(C) Schematic representation of mei-P26 3′ UTR showing position 844, the two poly(A) sites identified by RNA circularization and the three potential Pum binding motifs, including the NRE-type motif. RNA fragments tested in RNA pull-down assays (#1 to #6) and primer sets (1, 2, and 3) used to quantify the utilization of alternative poly(A) sites are also indicated. Coordinates are from the STOP codon, 1 being the first nucleotide of the 3′ UTR (see also Figure S6).
(D) Quantification by qRT-PCR of ratios of mRNA levels upstream of position 844 to just upstream of poly(A) site 2, in wild-type early ovarian stages and bamΔ86 mutant ovaries. Two primer sets (1 and 2) localized upstream of position 844 gave similar results. Means are from two independent RNA extracts quantified in triplicate, and error bars represent SD. ∗∗∗p < 0.0005 using the t test.
(E) RNA pull-down assays using the mei-P26 RNA fragments #1 to #6 shown in (C) and HA-PumC protein. Input is the in vitro synthesized protein (1/10) prior to RNA pull-down. osk and hb RNA fragments are negative and positive controls for Pum-C binding, respectively.
(F) RNA pull-down competition assays of mei-P26 RNA fragment #2 with increasing amounts of osk or hb unlabeled RNA fragments. Lane 2 is HA-PumC pull-down with fragment #2 in the absence of competitor. Input is as in (E).
(G) PAT assays of mei-P26 mRNA with specific primers allowing to measure potential poly(A) tails downstream of position 844 and poly(A) site 1 (left panel), and poly(A) site 2 (right panel), in wild-type and twinDG24102 early ovarian stages. sop was used as a control mRNA. PAT assay profiles of mei-P26 poly(A) site 2, using ImageJ, are shown.
A potential poly(A) site has been mapped in mei-P26 at position 844 after the stop codon in ovaries (Liu et al., 2009) (cDNA clone GH10646 from FlyBase). However, a more recent study identified another mei-P26 mRNA in ovaries with an extended 3′ UTR of about 4 kb (Smibert et al., 2012). We therefore used mRNA circularization to map mei-P26 poly(A) sites in early stages of oogenesis (germarium to stage 8 dissected from newly eclosed females). We identified two poly(A) sites at positions 1207 [poly(A) site 1] and 4303 [poly(A) site 2], respectively (Figures 5C and S6). This indicated the potential utilization of mei-P26 alternative poly(A) sites in ovaries. We sought to determine whether a specific poly(A) site was preferentially used in the GSCs. In particular, poly(A) test assays (PAT assays) used to measure poly(A) tail lengths at the abovementioned three poly(A) sites indicated that poly(A) sites at positions 844 and 1207 were poorly used in early ovaries (see below). We quantified by qRT-PCR the ratio of mRNA upstream of position 844 to mRNA cleaved at poly(A) site 2 and compared this ratio between early ovarian stages containing mostly differentiated cells (germarium to stage 8) and GSC-like cells from bamΔ86 mutant ovaries. Using two different sets of primers upstream of position 844 (primers 1 and 2) and one set just upstream of poly(A) site 2 (primers 3) (Figure 5C), we found that the ratios (primers 1 or 2/primers 3) were strongly reduced in GSC-like cells, indicating the increased utilization of poly(A) site 2 in these cells (Figure 5D).
To investigate potential direct interactions between the Nos/Pum/CCR4 complex and mei-P26 mRNA, we looked for Pum binding sites in mei-P26 3′ UTR. Two Pum binding sites have been defined: UGUAHAUA (Gerber et al., 2006) and the Nanos response element (NRE) GUUGN(3 to 45)AUUGUA, first identified in hunchback (hb) mRNA (Chen et al., 2008; Wharton and Struhl, 1991). Two UGUAHAUA Pum binding sites and one NRE are present in mei-P26 3′ UTR (Figures 5C and S6). We used RNA pull-down assays with the C-terminal moiety of Pum, which contains the RNA binding domain (HA-PumC) synthesized in vitro, to identify potential fragments of mei-P26 3′ UTR that directly interacted with PumC. A region of oskar (osk) coding sequence, not known to be bound by Pum, and the region of hb 3′ UTR containing the NRE were used as negative and positive control RNAs, respectively. Surprisingly, none of the fragments containing a Pum binding motif in mei-P26 3′ UTR were able to strongly pull down PumC, although fragments containing the NRE appeared to weakly do so (Figure 5E). The 5′-most Pum binding motif was also shown recently to be inactive in mei-P26 repression in male germ cell cysts (Insco et al., 2012). However, we identified another fragment of mei-P26 3′ UTR (#2) that was able to pull down PumC (Figures 5C and 5E). Competition assays were used to test the binding specificity of PumC to this fragment. Unlabeled osk or hb RNA fragments were added in excess (20× or 50×) to the binding reaction. The presence of osk RNA competitor had no effect on the binding of PumC to fragment #2. In contrast, increasing amounts of hb NRE fragment increasingly reduced the pull down of PumC by fragment #2, consistent with a specific binding of Pum to this region (Figure 5F). Although this fragment does not contain a canonical Pum binding site, it has a degenerated motif (UGUAACAA) that might be used to interact with Pum (Figure S6).
We then measured poly(A) tail length variations of mei-P26 mRNA in twin mutant early ovarian stages (germarium to stage 8) using PAT assays. Different primers were used to visualize potential poly(A) tails at position 844, poly(A) site 1, and poly(A) site 2. PAT assays in the region of position 844 and poly(A) site 1 did not produce the expected smear corresponding to different sizes of poly(A) tails, but did produce a discrete band (Figure 5G). This suggested a poor utilization of these poly(A) sites in early ovarian stages (although their usage was even weaker in GSC-like cells). In both cases, the discrete band would correspond to PCR amplification between the mei-P26 primer and the poly(A) stretch present downstream of position 844 and poly(A) site 1 (Figure S6), thus indicating an extended 3′ UTR. In contrast, PAT assays in the region of poly(A) site 2 produced the expected smear indicating the utilization of this poly(A) site (Figure 5G). The mei-P26 poly(A) tails were longer in twinDG24102 mutant than in wild-type, consistent with the repression of mei-P26 mRNA by CCR4-NOT-mediated deadenylation.
We next analyzed the effect of mei-P26 mRNA deadenylation by measuring Mei-P26 protein levels in GSCs. As described above, Mei-P26 was present in GSCs and its amount increased in 16-cell cysts (Figures 6A and 6E). Mei-P26 levels were significantly increased in GSCs mutant for twin, pum, or nos, consistent with the translational repression of mei-P26 mRNA by the Nos/Pum/CCR4 complex in GSCs (Figures 6A–6E). Increased levels of Mei-P26 in twin mutant GSCs were confirmed using the clonal analysis (Figures 6F and 6G).
Figure 6.
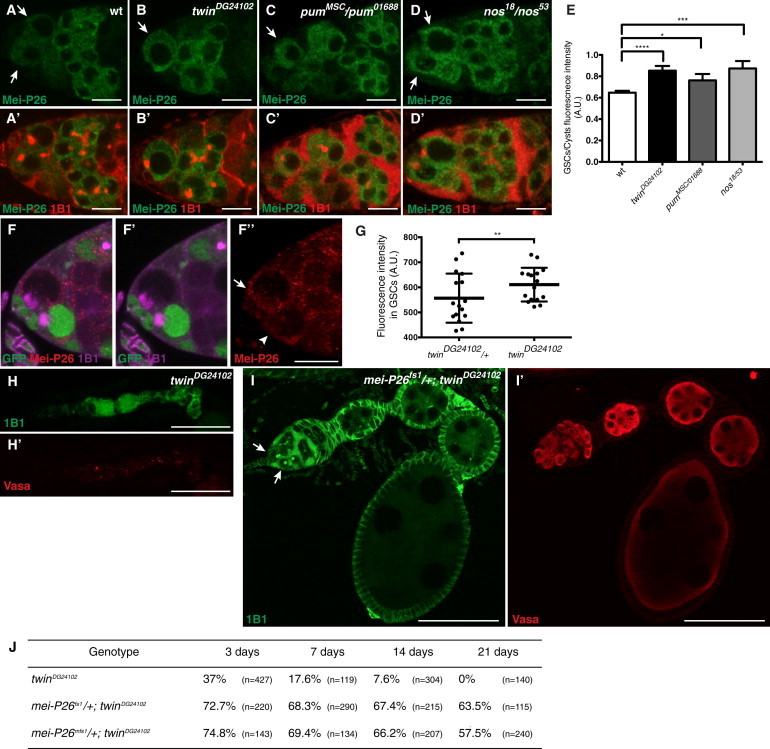
mei-P26 Is a Major Target of CCR4-Mediated Translational Repression for GSC Self-Renewal
(A–D) Upregulation of Mei-P26 protein levels in twin, pum, and nos mutant GSCs. Wild-type (A), twinDG24102 (B), pumMSC/pum01688 (C), and nos18/nos53 (D) germaria from 3-day-old females stained with anti-Mei-P26 antibody (green). The merges between anti-Mei-P26 (green) and 1B1 (red) are shown in (A′–D′). White arrows indicate GSCs. Scale bars represent 10 μm.
(E) Quantification of fluorescence intensity ratios between GSCs and cysts in the four genotypes shown in (A–D). The numbers of germaria used in the quantification were 44 wild-type, 22 twinDG24102, 24 pumMSC/pum01688, and 14 nos18/nos53. Error bars represent SD. ∗p = 0.01, ∗∗∗p = 0.0007, and ∗∗∗∗p < 0.0001 using the t test.
(F and G) Upregulation of Mei-P26 protein levels in twin mutant clonal GSCs. (F) Example of twinDG24102 mosaic germarium labeled with GFP (green), 1B1 (purple), and anti-Mei-P26 (red) 7 days after clone induction. The white arrow indicates the twinDG24102 clonal GSC (lack of GFP) and the white arrowhead indicates the twinDG24102/+ heterozygous GSC. Scale bars represent 10 μm. (G) Quantification of fluorescence intensity of Mei-P26 staining in twinDG24102/+ heterozygous and twinDG24102 clonal GSCs. n = 8 germaria. Bars represent means with SD. ∗∗p = 0.0088 using the t test.
(H and I) Rescue of twinDG24102 mutant phenotype of GSC loss by decreasing the gene dosage of mei-P26. twinDG24102 (H and H′) and mei-P26fs1/+; twinDG24102 (I and I′) ovaries labeled with 1B1 (green) and anti-Vasa (red) in 7-day-old females. (I) The white arrows indicate GSCs; oogenesis appears normal. Scale bars represent 20 μm in (H) and 60 μm in (I).
(J) Quantification of germaria containing at least one GSC in twinDG24102, mei-P26fs1/+; twinDG24102 and mei-P26mfs1/+; twinDG24102 mutant females of 3, 7, 14 and 21 days. n represents the number of germaria scored.
Together, these results show that mei-P26 mRNA is a direct target of the Nos/Pum/CCR4 complex through its interaction with Pum protein. Binding of the complex to mei-P26 mRNA leads to deadenylation and translational repression, resulting in the fine-tuning of Mei-P26 protein levels in the GSCs.
mei-P26 mRNA Is a Major Target of CCR4 for GSC Self-Renewal
We addressed whether this regulation of mei-P26 mRNA by Nos/Pum/CCR4 was functionally important by determining the effect of reducing mei-P26 gene dosage in twin mutant ovaries. Strikingly, reduction of one copy of mei-P26 gene strongly rescued the twinDG24102 phenotype of GSC loss and resulted in normal egg chamber development (Figures 6H–6J). About 60% of germaria contained GSCs in mei-P26−/+; twinDG24102 21-day-old-females compared to 0% in twinDG24102 females of the same age (Figure 6J). This shows that mei-P26 mRNA is an essential target of CCR4 for GSC self-renewal. Although mei-P26 is unlikely to be the only target of CCR4-mediated regulation in GSCs, its control by CCR4 has a major impact in GSC self-renewal, possibly because of its central role in GSC biology.
Discussion
Here, we have provided evidence that the twin gene that encodes the CCR4 deadenylase is essential for GSC self-renewal. GSCs are rapidly lost in twin mutants because they differentiate and cannot self-renew. Clonal analysis shows that twin is required cell autonomously in the GSCs for their self-renewal. Nos and Pum are major factors of GSC self-renewal and are translational repressors (Gilboa and Lehmann, 2004; Wang and Lin, 2004). Genetic and protein interactions among twin, nos, and pum indicate that CCR4 acts together with Nos and Pum to promote GSC self-renewal. This identifies the recruitment of the CCR4-NOT deadenylation complex as the molecular mechanism underlying Nos and Pum translational repression in the GSCs. Two mechanisms of action used by Nos/Pum have previously been described in the embryo. First, Nos/Pum represses hb mRNA translation by forming a complex with Brat, which in turn interacts with 4EHP and blocks initiation of translation (Cho et al., 2006; Sonoda and Wharton, 2001). Second, Nos/Pum represses cyclin B mRNA translation in the primordial germ cells by recruiting the CCR4-NOT complex through direct interactions between Pum and CAF1 and between Nos and NOT4 (Kadyrova et al., 2007). Brat is not expressed in GSCs (Harris et al., 2011), thus excluding the first mode of Nos/Pum translational repression in these cells. However, we did find Pum, Nos, and CCR4 present in a complex in GSC-like cells, consistent with the recruitment of the CCR4-NOT complex by Nos/Pum for GSC self-renewal.
Interestingly, a mutant form of CCR4 that is inactive for deadenylation is able to partially rescue the lack of CCR4 in GSCs. This is consistent with CCR4 not being the only deadenylase in the complex (Temme et al., 2010). However, CCR4 does participate in the deadenylation activity of the complex, probably via a structural role. Furthermore, the CCR4-NOT complex has been shown recently to be involved in direct translational repression, in addition to its role in deadenylation (Chekulaeva et al., 2011; Cooke et al., 2010). This dual mode of action of CCR4-NOT might also be relevant to GSCs.
The miRNA pathway also plays a crucial role in GSC self-renewal (Jin and Xie, 2007; Park et al., 2007; Yang et al., 2007). A large body of evidence has shown that an important mechanism of silencing by miRNAs involves deadenylation resulting from the recruitment of CCR4-NOT by GW182 bound to Ago1 (for review, see Braun et al., 2012). Therefore, the CCR4-NOT complex is also likely to contribute to miRNA-mediated translational repression in the GSCs, thus making this complex a central effector of translational repression in the GSCs.
An important result from this study is that mei-P26 mRNA is a major target of Nos/Pum/CCR4 regulation for GSC self-renewal (Figure 7). Nos and Pum are known to be essential players in GSC self-renewal, and many mRNAs are expected to be regulated by this complex. However, to date only one mRNA target of this complex, brat, has been reported. Here, we have identified another target, mei-P26 mRNA, and have shown that its repression by the Nos/Pum/CCR4 complex has a key role in GSC self-renewal, because the loss of GSCs in the twin mutant is strongly rescued by decreasing mei-P26 gene dosage.
Figure 7.
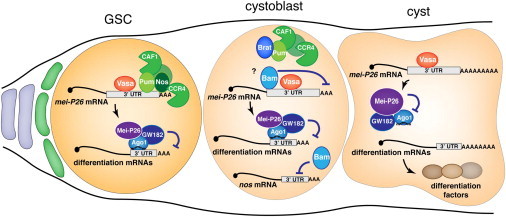
Model for the Role of CCR4 in GSC Self-Renewal
In the GSCs, the CCR4-NOT deadenylation complex interacts with Nos and Pum and is recruited to mei-P26 mRNA to repress its translation. Translational repression is counterbalanced by Vasa-mediated translational activation, leading to low levels of Mei-P26 protein, which cooperate with the miRNA pathway for silencing of mRNAs encoding differentiation factors. The CCR4-NOT complex is also likely to participate in miRNA silencing through its recruitment by GW182, as it is the case in other cell types. In cystoblasts, Bam represses nos mRNA translation (possibly together with Bgcn, Sex-Lethal, and Mei-P26; Li et al., 2013), resulting in the new association of Pum with Brat to target a different set of mRNAs (Harris et al., 2011). Mei-P26 levels do not increase, potentially due to a different mechanism of translational repression that might involve Bam. In eight-cell and 16-cell cysts, translational repression of mei-P26 mRNA is relieved, leading to an increase of Mei-P26 levels that antagonize miRNA-dependent silencing of mRNAs encoding differentiation factors.
Both Brat and Mei-P26 belong to the Trim-NHL family of proteins, which have conserved functions in stem cell lineages from C. elegans to mouse (for review, see Wulczyn et al., 2010). Proteins within this family are potential E3 ubiquitin ligases and can act by either activating or antagonizing the miRNA pathway, through their association with Ago1 and GW182. In particular, Mei-P26 function switches from activation of the miRNA pathway in the GSCs (Li et al., 2012) to inhibition of the pathway in differentiating cysts where Mei-P26 levels are higher (Neumüller et al., 2008). As such, Mei-P26 plays a central role in the control of cell fate in the GSC lineage. The rescue of the twin mutant phenotype of GSC loss by decreasing mei-P26 gene dosage suggests that the levels of Mei-P26 themselves might be important for this switch of its function. This might provide an explanation as to why such a precise regulation of its level is crucial for GSC self-renewal and differentiation.
Which molecular mechanisms underlie the fine-tuning of Mei-P26 in the GSC lineage? The translational repression of mei-P26 mRNA is not complete in GSCs. This differs from the complete repression by Nos/Pum of cyclin B mRNA in the primordial germ cells, or brat mRNA in the GSCs, and may result from the concomitant activation of mei-P26 by Vasa (Liu et al., 2009). Vasa does activate mei-P26 translation, leading to a peak of expression in -8- and 16-cell cysts. However, Vasa is expressed in all germ cells, suggesting that it is not the key regulator governing the timing of Mei-P26 peak of expression. We propose that translational activation of Mei-P26 by Vasa would be active already in GSCs but counterbalanced by translational repression by Nos/Pum and the CCR4-NOT complex (Figure 7). In cystoblasts, the presence of Bam overcomes Nos/Pum translational repression by decreasing Nos levels (Li et al., 2009), which would thus switch the balance to translational activation by Vasa. This does not lead to a peak of Mei-P26 expression in cystoblasts, but rather to a progressive increase of Mei-P26 levels in proliferating cysts. This progressive accumulation of Mei-P26 could depend on the necessity to build up Vasa-mediated translational activation. However, another possibility could be that a different factor still partially represses mei-P26 translation in cystoblasts and early cysts. A potential candidate is Bam, which has been defined as a translational repressor (Li et al., 2009; Shen et al., 2009) and has recently been reported to directly repress mei-P26 mRNA translation in the male GSC lineage (Insco et al., 2012). The Bam expression profile in female germ cells is consistent with this potential role in mei-P26 translational repression, because Bam protein is present from cystoblasts to 8-cell cysts but absent in 16-cell cysts, where Mei-P26 levels are the highest (Li et al., 2009).
Recent advances have established the generality of a central role for translational regulations in adult stem cell lineages (Crist et al., 2012; Harris et al., 2011; Insco et al., 2012). Translational repression is required to prevent the synthesis of differentiation factors whose mRNAs are already present in stem cells. In the Drosophila female GSC lineage, recent work has demonstrated that changes in cell fate are driven by different translational regulation programs; associations between translational repressors evolve to trigger stage-specific regulation of mRNA targets. For example, while Nos/Pum maintain female GSCs by repressing a specific set of mRNAs, Pum associates with Brat in cystoblasts to repress a different set (Harris et al., 2011). The Trim-NHL proteins appear to be of particular importance in the translational regulations essential for stem cell fate as exemplified by Mei-P26 (Li et al., 2012, 2013; Neumüller et al., 2008). Here, we add that the fine-tuning of Mei-P26 protein levels by translational repression is essential for GSC self-renewal and implicate CCR4 in this regulation.
The functions of Trim-NHL proteins are conserved in many adult stem cell lineages in different organisms, and mutations in the corresponding genes lead to highly proliferative tumors. Elucidating the molecular mechanisms behind their translational control is key to deciphering how these proteins regulate adult stem cell fates.
Experimental Procedures
Drosophila stocks are described in the Supplemental Experimental Procedures.
DNA Constructs
The DNA encoding the deadenylase-dead form of CCR4 (Temme et al., 2010) was cloned into the pUASP vector digested with KpnI and XbaI. In this mutant form of CCR4, amino acids 412 (aspartic acid) and 414 (asparagine) were substituted by alanine. Transgenic stocks were generated by BestGene. The HA-PumC construct was obtained by inserting the 2,236 pb SmaI to XbaI fragment from the pGEX-PumC (Zamore et al., 1997) (animo acids 849–1,533) into the StuI and XbaI sites of the pCSH3 (pCS2+ backbone vector with two HA tags).
Antibodies and Immunostaining
The rabbit and guinea pig anti-Pum antibodies were raised by Agro-Bio against amino acids 408–883 of the Pum protein (Zamore et al., 1997). Antibody dilutions for immunostaining and fluorescence quantification are described in the Supplemental Experimental Procedures.
Immunoprecipitations and RNA Pull-Down Assays
The procedures for immunoprecipitations and RNA pull-down assays are described in the Supplemental Experimental Procedures.
RNA Circularization
RNA circularization was performed as previously described (Chambeyron et al., 2002), with RT-PCR reactions followed by nested PCR. The primers used are listed in Table S1.
PAT Assays, RT-PCR, and qRT-PCR
PAT assays, RT-PCR, and qRT-PCR were performed as described previously using two to four independent RNA preparations (Juge et al., 2002; Zaessinger et al., 2006), and the ePAT method of PAT assays was also used (Jänicke et al., 2012). The primers used are listed in Table S1. Quantitative PCR experiments were performed with the LightCycler System (Roche Molecular Biochemical).
Acknowledgments
We are very grateful to D. Chen, T. Kai, J. Knoblich, D. McKearin, P. Lasko, R. Lehmann, H. Lipshitz, A. Nakamura, and E. Wahle for their gifts of DNA clones, antibodies, or Drosophila stocks and to F. Besse for her help with RNA pull-downs. We thank S. Pierson for her bioinformatic survey of Pum binding sites in mei-P26 3′ UTR. This work was supported by the CNRS UPR1142, ANR Blanche (ANR-06-BLAN-0343, ANR-2010-BLAN-1201 01), FRM (“Equipe FRM 2007,” “Equipe FRM 2013 DEQ20130326534,” and “Projets Innovants ING20101221078”), and ARC Libre 2009 (N°3192). W.J. held a salary from FRM and ANR Blanche. P.R.R. held a salary from the Labex EpiGenMed and Université Montpellier 1.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Barckmann B., Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. Biochim. Biophys. Acta. 2013;1829:714–724. doi: 10.1016/j.bbagrm.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Braun J.E., Huntzinger E., Izaurralde E. A molecular link between miRISCs and deadenylases provides new insight into the mechanism of gene silencing by microRNAs. Cold Spring Harb. Perspect. Biol. 2012;4:4. doi: 10.1101/cshperspect.a012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S., Brun C., Robin S., Bucheton A., Busseau I. Chimeric RNA transposition intermediates of the I factor produce precise retrotransposed copies. Nucleic Acids Res. 2002;30:3387–3394. doi: 10.1093/nar/gkf456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., McKearin D. Gene circuitry controlling a stem cell niche. Curr. Biol. 2005;15:179–184. doi: 10.1016/j.cub.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Chen G., Li W., Zhang Q.S., Regulski M., Sinha N., Barditch J., Tully T., Krainer A.R., Zhang M.Q., Dubnau J. Identification of synaptic targets of Drosophila pumilio. PLoS Comput. Biol. 2008;4:e1000026. doi: 10.1371/journal.pcbi.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho P.F., Gamberi C., Cho-Park Y.A., Cho-Park I.B., Lasko P., Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A., Prigge A., Wickens M. Translational repression by deadenylases. J. Biol. Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist C.G., Montarras D., Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Fuller M.T., Spradling A.C. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Gerber A.P., Luschnig S., Krasnow M.A., Brown P.O., Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L., Lehmann R. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 2004;14:981–986. doi: 10.1016/j.cub.2004.05.049. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Harris R.E., Pargett M., Sutcliffe C., Umulis D., Ashe H.L. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell. 2011;20:72–83. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco M.L., Bailey A.S., Kim J., Olivares G.H., Wapinski O.L., Tam C.H., Fuller M.T. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell. 2012;11:689–700. doi: 10.1016/j.stem.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänicke A., Vancuylenberg J., Boag P.R., Traven A., Beilharz T.H. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA. 2012;18:1289–1295. doi: 10.1261/rna.031898.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Juge F., Zaessinger S., Temme C., Wahle E., Simonelig M. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J. 2002;21:6603–6613. doi: 10.1093/emboj/cdf633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L.Y., Habara Y., Lee T.H., Wharton R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Wilkinson B.D., Siegrist S.E., Wharton R.P., Doe C.Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Li Y., Minor N.T., Park J.K., McKearin D.M., Maines J.Z. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. USA. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Maines J.Z., Tastan O.Y., McKearin D.M., Buszczak M. Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development. 2012;139:1547–1556. doi: 10.1242/dev.077412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Carreira-Rosario A., Maines J.Z., McKearin D.M., Buszczak M. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS ONE. 2013;8:e58301. doi: 10.1371/journal.pone.0058301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Yue L., Spradling A.C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Liu N., Han H., Lasko P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev. 2009;23:2742–2752. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D., Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121:2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- Morris J.Z., Hong A., Lilly M.A., Lehmann R. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development. 2005;132:1165–1174. doi: 10.1242/dev.01672. [DOI] [PubMed] [Google Scholar]
- Neumüller R.A., Betschinger J., Fischer A., Bushati N., Poernbacher I., Mechtler K., Cohen S.M., Knoblich J.A. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., McKearin D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 1997;124:3651–3662. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- Park J.K., Liu X., Strauss T.J., McKearin D.M., Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Rouget C., Papin C., Boureux A., Meunier A.C., Franco B., Robine N., Lai E.C., Pelisson A., Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P., Pritchard D.K., Pabon L., Reinecke H., Schwartz S.M., Morris D.R., Murry C.E. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Shen R., Weng C., Yu J., Xie T. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl. Acad. Sci. USA. 2009;106:11623–11628. doi: 10.1073/pnas.0903325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Miura P., Westholm J.O., Shenker S., May G., Duff M.O., Zhang D., Eads B.D., Carlson J., Brown J.B. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Wong M.D., Kawase E., Xi R., Ding B.C., McCarthy J.J., Xie T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- Sonoda J., Wharton R.P. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., Wharton R.P. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Igarashi K., Aisaki K., Kanno J., Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc. Natl. Acad. Sci. USA. 2010;107:3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Saba R., Miyoshi K., Morita Y., Saga Y. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS ONE. 2012;7:e33558. doi: 10.1371/journal.pone.0033558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A., Cox D.N., Wang Z., Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C., Zhang L., Kremmer E., Ihling C., Chartier A., Sinz A., Simonelig M., Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wulczyn F.G., Cuevas E., Franzoni E., Rybak A. MiRNA need a TRIM regulation of miRNA activity by Trim-NHL proteins. Adv. Exp. Med. Biol. 2010;700:85–105. [PubMed] [Google Scholar]
- Xia L., Zheng X., Zheng W., Zhang G., Wang H., Tao Y., Chen D. The niche-dependent feedback loop generates a BMP activity gradient to determine the germline stem cell fate. Curr. Biol. 2012;22:515–521. doi: 10.1016/j.cub.2012.01.056. [DOI] [PubMed] [Google Scholar]
- Xie T., Spradling A.C. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Yang L., Chen D., Duan R., Xia L., Wang J., Qurashi A., Jin P., Chen D. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- Zaessinger S., Busseau I., Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Williamson J.R., Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


