Abstract
Transformative impact in regenerative medicine requires more than the reprogramming of individual cells: advances in repair strategies for birth defects or injuries, tumor normalization, and the construction of bioengineered organs and tissues all require the ability to control large-scale anatomical shape. Much recent work has focused on the transcriptional and biochemical regulation of cell behaviour and morphogenesis. However, exciting new data reveal that bioelectrical properties of cells and their microenvironment exert a profound influence on cell differentiation, proliferation, and migration. Ion channels and pumps expressed in all cells, not just excitable nerve and muscle, establish resting potentials that vary across tissues and change with significant developmental events. Most importantly, the spatio-temporal gradients of these endogenous transmembrane voltage potentials (Vmem) serve as instructive patterning cues for large-scale anatomy, providing organ identity, positional information, and prepattern template cues for morphogenesis. New genetic and pharmacological techniques for molecular modulation of bioelectric gradients in vivo have revealed the ability to initiate complex organogenesis, change tissue identity, and trigger regeneration of whole vertebrate appendages. A large segment of the spatial information processing that orchestrates individual cells’ programs towards the anatomical needs of the host organism is electrical; this blurs the line between memory and decision-making in neural networks and morphogenesis in non-neural tissues. Advances in cracking this bioelectric code will enable the rational reprogramming of shape in whole tissues and organs, revolutionizing regenerative medicine, developmental biology, and synthetic bioengineering.
Cellular reprogramming – a wider context
Most problems in biomedicine boil down to the control of complex biological shape1: its self-assembly during embryogenesis, restoration in the face of traumatic injury or degeneration, and maintenance throughout life to resist cancer and aging. Even cancer is a kind of disease of geometry, where cells stop obeying the normal patterning cues of the body in favour of tumorigenesis2–4. If all the problems of stem cell differentiation were solved and any single cell type could be produced at will, we would still be faced with the intractable problem of having to directly micromanage the assembly of an eye or hand from individual cell types. Outside biomedicine, the field of synthetic biology must also move beyond metabolic circuits in soups of individual cells toward the bioengineering of complex autonomous structures. All of these goals require that we learn to reprogram not just single cells but the information-processing and communication pathways that orchestrate individual cell behaviours towards the patterning needs of a host organism (Fig. 1).
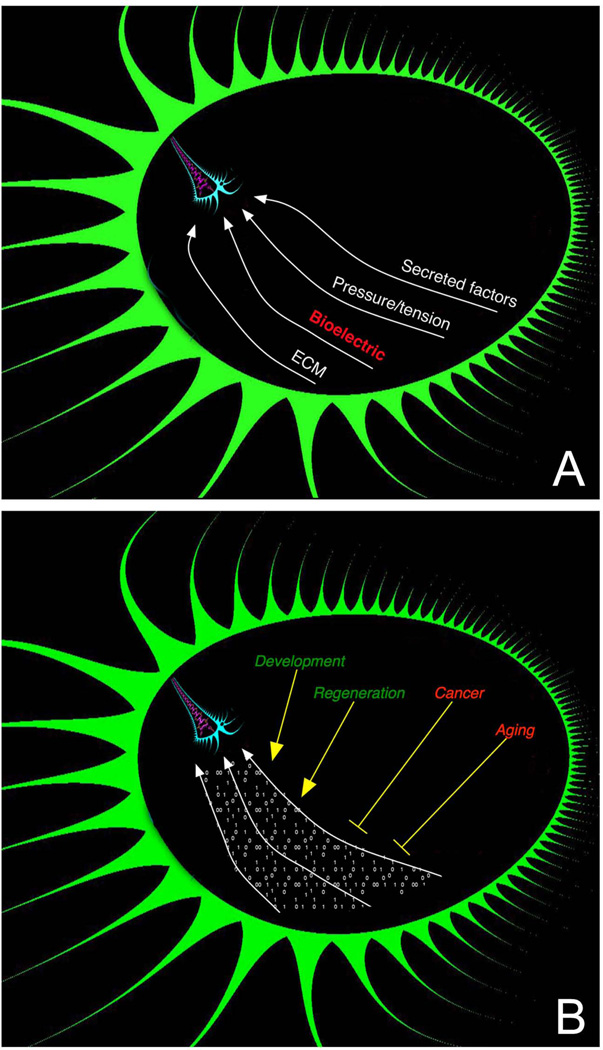
Figure 1. the morphogenetic field.
(A) Cell activity is guided by a complex, spatially-distributed set of signals from the host organism mediated by diffusing chemical, extracellular matrix, tension/pressure, and bioelectrical properties. (B) This morphogenetic field orchestrates cell behaviour towards large-scale anatomical programs during development and regeneration; its influence is subverted during oncogenic transformation and aging. Mastery of the information stored in this field, and of the mechanisms by which cells interact with it, will result in the ability to reprogram large-scale tissue and organ shape, with transformative implications for the fields of birth defects, regenerative medicine, cancer, and synthetic bioengineering.
Examples of large-scale tissue reprogramming abound in nature. Planarian flatworms can regenerate the entire animal after almost any kind of amputation, and continuously remodel their entire body to match available cell numbers to their target morphology and correct proportions among all organs5. Salamanders can regenerate limbs, eyes, jaws, hearts, and portions of the brain6 throughout life. Deer – large adult mammals – regenerate meters of bone and innervation each year7, and even human children can regenerate their fingertips completely8, reminding us that regeneration is not only the province of non-mammalian species. Remarkably, embryonic9, 10 and regenerative11, 12 environments can reprogram cancerous growth into normal anatomy13, 14, revealing the importance of context and patterning information in regulating individual cell behaviour. While the current focus on unravelling this control circuitry has been on transcriptional networks and gradients of secreted biochemicals, recent work has highlighted the importance of another powerful control system: endogenous bioelectricity15, 16.
All cells, not just nerve and muscle, utilize ion channels and pumps to establish specific voltage gradients across plasma and intracellular membranes (resting potential Vmem, Figures 2–3). While solid functional physiology over the last 40 years has implicated transepithelial electric fields as providing cues for cell migration, cell orientation, wound healing, and even limb regeneration17–24, it is now beginning to be appreciated that Vmem itself is a powerful regulator of cell proliferation, differentiation, and apoptosis25–27. Interestingly, recent work has demonstrated that spatio-temporal patterns of Vmem distribution in vivo contain instructive information for specifying organ identity and large-scale anatomical order (Fig. 4). Distinct from effects of applied electromagnetic field exposure28, 29 and from ultraweak electromagnetic radiations from cells30–32, this exciting new field is “Molecular Bioelectricity” - the investigation of instructive patterning and cell:cell signalling roles of voltage gradients in non-excitable cells. The modification and interpretation of these gradients during embryogenesis, regeneration, and cancer underlies endogenous and experimentally-induced cell and tissue reprogramming. This Opinion essay discusses the current state of the field of Bioelectricity, its major open questions and opportunities for new discoveries, and implications for basic biology and biomedicine.
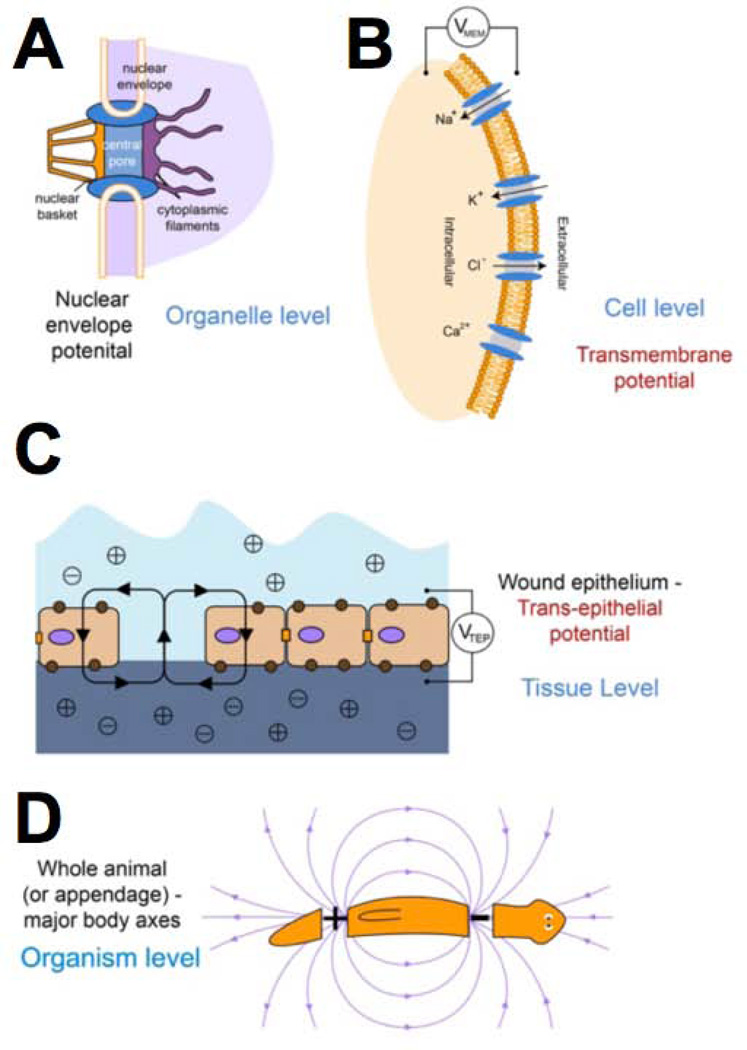
Figure 2. bioelectric gradients exist at multiple scales of size and levels of biological organization.
Organelles (A) and whole cells (B) are bound by membranes containing ion channel, pump, and transporter proteins. The activity of these ion translocators give rise to differences in resting potential (Vmem) across the membrane. Stacked in parallel, cells also give rise to a trans-epithelial potential (C), and electric fields have been characterized that correspond to appendages (limbs) or entire body axes (D). This overlapping set of cues provides positional information, organ identity, and other cues for cell behaviour and morphogenesis. This figure was drawn by Maria Lobikin, and is used with permission.
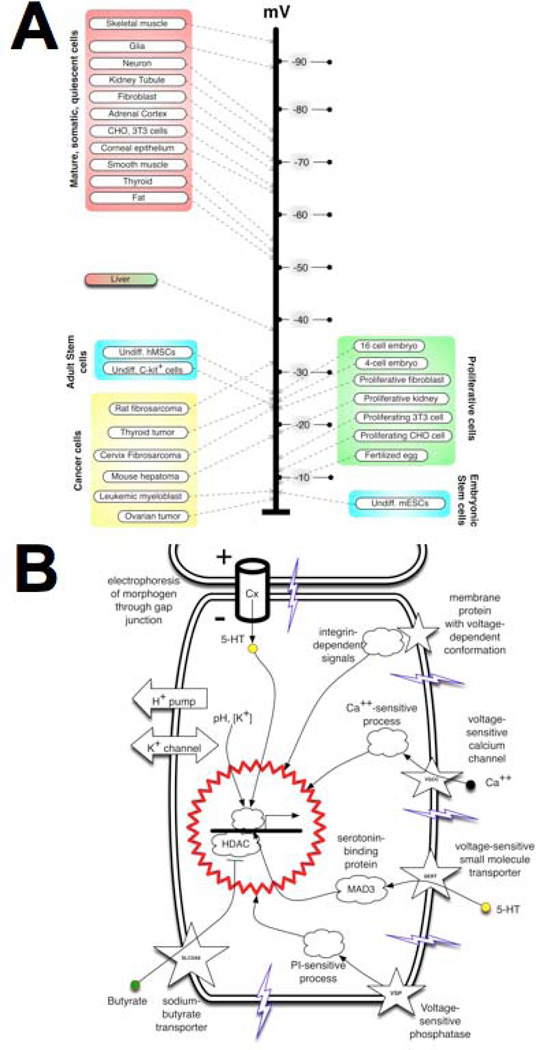
Figure 3. Vmem at the level of single cells: its transduction impacts cell states.
(A) A sample survey of many cell types (modified and updated after102), and recent functional data25, 103, 104, 171–174, reveals that at the level of single cells, Vmem determines cell plasticity and proliferation potential. Depolarized cells tend to be rapidly proliferating and undifferentiated (e.g., embryonic, stem, or tumor cells) while terminally-differentiated somatic cells tend to be highly polarized. Importantly, cell state can be functionally altered (switched between these two classes, in either direction) by artificial change of Vmem. This panel is modified after Fig. 1 of15. (B) A range of mechanisms have now been characterized that transduce alterations of Vmem into downstream effector cascades (transcriptional changes). These include signalling proteins with a voltage-sensitive conformation (e.g., integrins and voltage-sensitive phosphatases) and transporters of small signalling molecules whose activity is regulated by Vmem (such as gap junctions, voltage-gated calcium channels, and solute carriers, which allow Vmem changes to signal via serotonin, Ca++, butyrate, and likely many other yet-to-be-discovered compounds). This figure is modified after Fig. 1B of113.
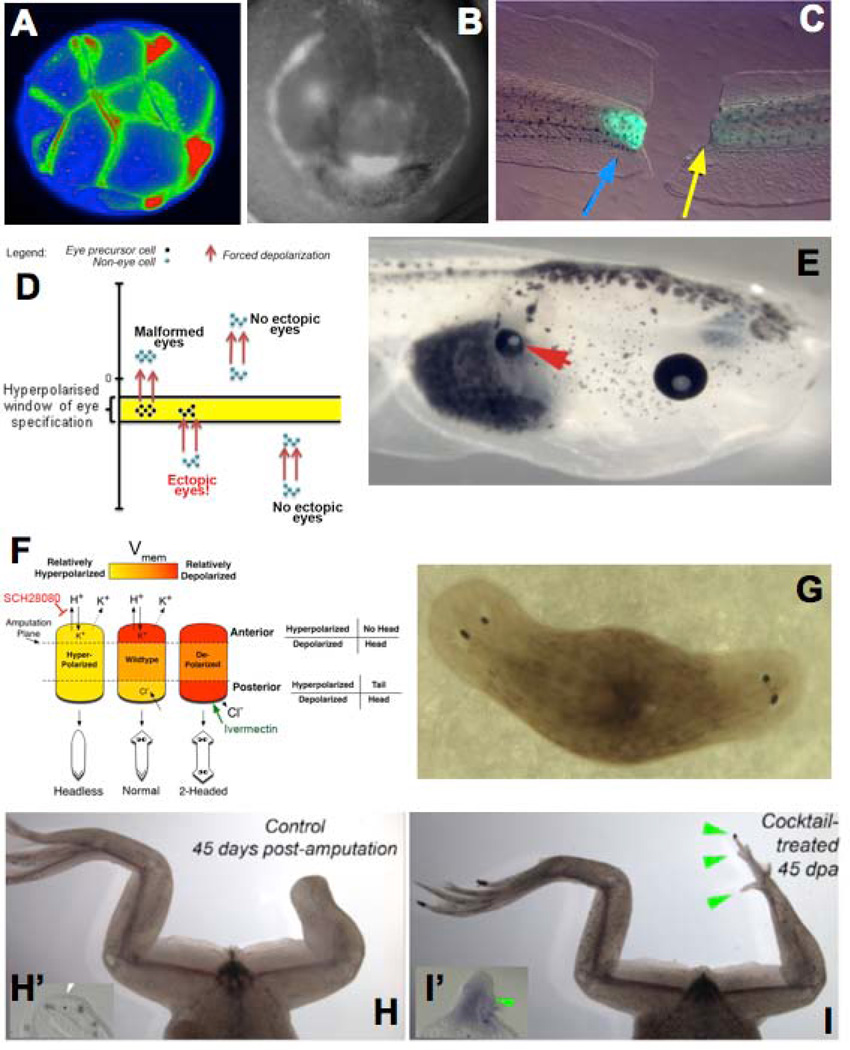
Figure 4. Large-scale tissue and organ reprogramming by Vmem gradients.
Voltage-sensitive fluorescent dyes reveal spatio-temporal patterns of bioelectrical gradients in vivo. Examples of gradients in the Xenopus laevis (frog) model include cleavage stages (A), craniofacial patterning (B, showing the hyperpolarizations in tissues that will become eye, branchial arches, and cement gland), and tail regeneration (C, regenerating tail on the left, and one that is prevented from regeneration by inhibition of V-ATPase activity on the right; green fluorescence signal indicates the normal repolarization of the wound (blue arrow), and when the repolarization is experimentally prevented (yellow arrow), regeneration is blocked). Functional data reveal a model in which a narrow range of Vmem (D) forces somatic cells, such as gut cells, to form a whole complete eye (E, arrowhead). A similar bioelectric circuit model describes the pathway regulating head vs. tail identity of regenerating tissue in planaria (F); experimental control of Vmem in this model results in 2-head animals after amputation (G), revealing the ability to control the shape of organs constructed by adult stem cells by bioelectric signalling. Brief treatment with sodium ionophore cocktail induces froglet hindlegs, which normally do not regenerate (H, showing wound region and lack of expression of the blastema gene MSX1 in inset panel H’) to regenerate legs with toes and toenails (I, showing blastema and induction of MSX1 expression in inset panel I’). This figure is modified after figures from references16, 50, 61, 146.
State-of-the-art tools of molecular bioelectricity
A number of tools have now been developed to facilitate analysis of bioelectric regulatory pathways in many model species33. The system in question (explanted tissue, in vitro organ culture, or entire developing embryo) can be interrogated with respect to the spatio-temporal profile of bioelectric properties. For example, the Vmem gradients that were observed in the early Xenopus face34 preceded and co-localized with specific inductions of genes such as frizzled, suggesting the hypothesis that the observed voltage distribution played a functional role in craniofacial morphogenesis. Characterization of ion flux is best performed using ion-selective vibrating probes35; trans-epithelial potentials can be mapped out using standard electrophysiology23, 36, while distributions of Vmemare detected non-invasively by soaking samples in fluorescent voltage reporter dyes coupled with quantitative microscopy37–39. Both vibrating probes and voltage dyes offer the opportunity of discovering multiple distinct voltage domains on the surface of individual cells15, 33 – a potentially important combinatorial code that is obscured by traditional electrophysiology which reports only one Vmem value for any given cell.
Alternatively, a number of screens can be used to first ask whether bioelectricity is functionally relevant in one’s favourite system (and if the result is positive, then followed up with characterization of the bioelectric profile in the native state). The basic loss-of-function screen, first performed in frog and chick embryos in a left-right asymmetry patterning assay to determine whether ion flow is important40, 41, is used to simultaneously identify the molecular-genetic origin of the signal. In this scheme42, 43, a panel of ion channel/pump drugs with known targets is used to build a pharmacological profile of the ion conductance whose inhibition results in a specific patterning or cell behaviour change. Remarkably, in many such screens it has been possible to adjust levels of inhibition so as to dissociate housekeeping functions of Vmem from subtle instructive patterning roles, revealing functions in development34, 40, 44–50, regeneration18, 51–53, and cancerous disorganization54–59 in the absence of generalized toxicity. The power of such inverse screens resides in the hierarchical organization of the reagents: first relatively non-specific compounds are used to test the involvement of large families of translocators (e.g., barium chloride to probe all K+ channels). Any negative result rules out the entire family, while positive results are followed by increasingly more-specific reagents to narrow down to a single class. Such screens usually only require the use of ~10–20 compounds total; in the case of left-right patterning, the screen identified two potassium channels and two ion pumps which were subsequently validated by molecular-genetic dominant negative approaches. Because of the logarithmic reduction in the number of candidates at each step, the relatively inexpensive nature of drug experiments, and the ability of compounds to target multiple family members at once, inverse screens are a very accessible, rapid way to determine which ion translocator proteins are involved in any process of interest, and a wide variety of reagents including morpholinos, RNAi, and genetic deletion can be used for molecular validation. Moreover, unlike molecular targeting of individual genes, such drug screens overcome the high compensation (redundancy) among channel members and allow the interrogation of maternal proteins (which do not rely on zygotic transcription for their presence and are thus hard to detect in standard knockdown screens). Nevertheless, despite the drawbacks of traditional genetic screens for uncovering bioelectrical regulators of patterning, a number of channelopathies have now been uncovered as underlying specific morphological phenotypes (Table 1).
Table 1.
Channelopathies with patterning phenotypes
| Protein | Morphogenetic role | Species | Reference |
|---|---|---|---|
| TMEM16A chloride channel | Tracheal morphogenesis | Mouse | 175 |
| Kir7.1 potassium channel | Melanosome development | Zebrafish | 176 |
| Cx41.8 gap junction | Pigmentation pattern | Zebrafish | 177 |
| Cx43 gap junction | Fin regeneration | Zebrafish | 178 |
| Kir2.1 potassium channel | Wing patterning | Drosophila | 44 |
| Cx43 gap junction | Fin size regulation, Craniofrontonasal syndrome | Zebrafish Mouse | 179, 180 |
| Cx43 gap junction | Osteoblast differentiation | Mouse | 181 |
| Kir2.1 potassium channel | Craniofacial (Andersen-Tawil syndrome) and limb patterning | Mouse | 44, 127 |
| CFTR chloride channel | Bilateral absence of vas deferens | Human | 182, 183 |
| Girk2 potassium channel | Cerebellar development | Mouse | 184–187 |
| GABA-A receptor (chloride channel) | Craniofacial patterning, (Angelman Syndrome) | Mouse | 188 |
| KCNH2 K+ channel | Cardiac patterning | Mouse | 189 |
| NHE2 sodium/proton exchanger | Epithelial patterning | Drosophila | 190 |
| V-ATPase proton pump | Wing hair patterning | Drosophila | 191 |
| KCNQ1 potassium channel | Hypertrophy of tongue, liver, spleen, pancreas, kidneys, adrenals, genitalia – Beckwith-Wiedemann syndrome; craniofacial defects | Human, Mouse | 132, 192, 193 |
| Kir6.2 potassium channel | Craniofacial defects | Human | 134 |
Gain-of-function strategies rely on misexpression of specific channels and pumps to perturb Vmem gradients in vivo, to test specific hypotheses generated by screens or physiomic profiling. For example, the observation that the left side of the 4-cell frog embryo pumped half as many protons per unit time as the right side led to a misexpression experiment in which a yeast proton pump mRNA was introduced into left-side cells to equalize the H+ efflux; this resulted in specific randomization of the position of the asymmetric internal organs40, 41, supporting a functional role for voltage gradients in embryonic left-right asymmetry. A large panel of well-characterized channels and pumps now exists that can be used to rationally modulate the bioelectric state of specific cells, and misexpression of a variety of ion channels in early frog embryos revealed the surprising reprogramming of many somatic areas into eyes50. These can be introduced into cells using transfection, microinjection of mRNA or DNA vectors, or viral infection. Pharmacological manipulation of natively-expressed channels or pumps (e.g., using channel opener drugs) is the method of choice in biomedical applications where gene therapy is to be avoided; for example, a recent study showed how a cocktail based on sodium flux could be used to initiate regeneration of spinal cord and muscle in a tadpole model of tail regeneration52. An important strategy using misexpression of wild-type and mutant channels concerns dissecting the mechanism of action. For example, a pore mutant can be used to determine whether a channel’s role is scaffolding/binding or truly related to its ion translocation role. Further, by changing the Vmem to the same overall level using channels for different ions (K+, Na+, Cl−), it is possible to determine whether a particular ion’s concentration is what matters or whether it really is the Vmem level that carries instructive information in the given system.
With functional and physiomic (ion flux or Vmem gradient) data in hand, quantitative modelling is then used to synthesize the ion conductance dynamics into a circuit with predicable Vmem control properties60–62. The last remaining piece of the puzzle is to connect Vmem changes to downstream transcriptional cascades. This is done through a suppression screen for transduction machinery. Given an assay in which a Vmem change produces a specific cell- or tissue- outcome, each of the known transduction pathways is inhibited in turn to determine which one prevents the Vmem change from being sensed by cells. Known transduction mechanisms that allow cells to convert bioelectrical signals into gene expression changes include voltage-sensitive phosphatases63, gap junctions64–66, transporters of serotonin41, 67–69 and butyrate54, 70, 71, and integrin signaling72. By identifying the transduction mechanism linking Vmem to transcription in a given system, the circle is closed, providing mechanistic details of every step from the expression of the channels that drive a Vmem change, to the mechanism that converts that change into 2nd messenger activity, to the genes (in many cases, ion channel genes!) whose activity is modified. Patterning thus occurs as a cycling, dynamical system implemented by the continuous interplay of genetics and biophysics.
Bioelectrical determinants of individual cell behavior
It has long been known that endogenous electric fields provide spatial cues for orientation, outgrowth, and migration for a broad range of cell types36, 73–79. Growth cone pathfinding and cell orientation guided by bioelectrical cues involves integrins, cAMP, rac, and cdc4224, 80–86. A particularly thorough combination of biochemistry, transgenic mouse technology, and electric field perturbation18 dissected the mechanisms of electrotaxis showing that mammalian wound healing requires cells to sense endogenous fields (generated by trans-epithelial potential) by a PTEN - and PI(3)K-γ-dependent pathway. Cell differentiation is also controlled by changes in Vmem, as has been shown in human mesenchymal stem cells26, 87, embryonic stem cells88, 89, myoblasts (in which hyperpolarization driven by the Kir2.1 channel plays a crucial role)90, 91, the specification of neurotransmitter types92, and the control of precursor differentiation93–97 in the developing nervous system and heart. Tissue engineers have also begun to take advantage of this pathway using applied electric field stimulation98. Given the known roles of Vmem in regulating normal migration, differentiation, and proliferation99, it is not surprising that control of ion flux58, 100 and membrane voltage56, 57 are also increasingly implicated in cancer (Table 2). Interestingly, electric cues often dominate competing biochemical signals; for example, depolarization trumps the induction of differentiation by insulin+dexamethasone in human mesenchymal stem cells26, while physiological-strength electric fields override opposing chemical trophic factors, contact inhibition release, and population pressure101.
Table 2.
Ion translocators as oncogenes
A useful heuristic (Fig. 3A) is that depolarized cells tend to be plastic, undifferentiated, and highly proliferative (stem cells, cancer cells, and embryonic cells), while strongly polarized cells tend to be the mature, somatic terminally-differentiated cells: Vmem is a strong regulator of cell fate plasticity and mitotic rates102–104. However, bioelectric cues also provide spatially-patterned signals. The differential activation of voltage-responsive transduction mechanisms on opposite sides of a cell allows bioelectric signals to regulate cell polarity. This was first discovered in the algae Fucus105, and has been recently shown in yeast106 and pollen tubes107. During left-right patterning of the Xenopus embryo, voltage gradients set up at the first cleavages link individual cell dynamics to axial patterning of the entire bodyplan by redistributing a long-range morphogen108, 109,110, 111. The dissection and synthesis of such systems, at the genetic and physiological levels, is helping to understand the properties of biophysical pathways by which individual cell polarity is integrated into large-scale patterning outcomes112 - advances that are required for applications in tissue and organ (re)programming.
A fundamentally physical event such as ion flow or Vmem change needs to be transduced into genetic responses (Fig. 3B). Several distinct mechanisms convert slow changes in resting Vmem levels into second-messenger cascades in non-excitable cells that ultimately drive transcriptional responses (reviewed in detail in113). These include: electrophoretic redistribution of small signalling molecules through gap junctional paths, voltage-based regulation of membrane transporters of signalling molecules like calcium and various neurotransmitters, and electrophoretic separation or clustering of protein complex subunits within the plane of the cell membrane.
The most recent transduction mechanism to be described involves the SLC5A8. This protein, already implicated by genetic data in colon cancer114–117, converts ion levels into the movements of butyrate, which in turn is an important regulator of histone deacetylases and thus of epigenetic chromatin state118. Recent attempts to reprogram cancer in a frog model showed that Vmem is not only a promising diagnostic modality for non-invasively revealing tumour sites and margins, but is also a functional parameter regulating oncogene-mediated tumorigenesis. Cells artificially hyperpolarized by genetic or pharmacological means resisted forming tumours despite high levels of oncogene expression54, 55. This suppression effect was mediated by SLC5A8, which linked voltage change to HDAC activity modulation by butyrate. A similar pathway has been found to occur in limb and tail regeneration119, 120. Thus, as has been seen with voltage regulation of serotonin movement and signalling in non-neural cells during left-right patterning41, 67–69, Vmem control of small molecule flow and signalling may be a conserved module for the regulation of programs that mediate cell activities into anatomical structures and away from tumorigenesis.
Bioelectric gradients determine large-scale pattern
Large-scale morphogenesis emerges from the orchestrated interactions of individual cells. Recent data has shown that contact-dependent depolarization regulates the interactions of distinct cell types121, as well as regulates the paths taken through the body by migratory cell types57. Screens have also revealed new roles for bioelectric signals in endogenous regulation of complex organ patterning programs in vivo (Fig. 4). In regenerating planaria, a circuit driven by the H,K-ATPase has been described that regulates whether a head or tail appears at an amputation site61, 122. By experimentally regulating the Vmem at the wound and the physiological communication among cells via gap junctions, 0-, 2-, or even 4-headed (rhombus-shaped) worms can be created – a remarkable control over large-scale shape123. This is an example of bioelectric cues directly regulating what structure is built by the adult stem cells (neoblasts), and serves as an important model for the use of bioelectric signalling to coax stem cell-mediated growth in biomedical contexts.
Recent data in Drosophila embryos linked the Kir2.1 channel to the important TGF-β patterning pathway in wing patterning34. In Xenopus embryogenesis, regionalization of the anterior field by patterns of hyperpolarized and depolarized cells specifies a prepattern for gene expression and subsequent anatomy of the face. Specific endogenous patterns of differential Vmem in the naïve tissue preceded and controlled the position of eyes50, 124 and many components of the face34, while experimental alterations of this native pattern produced predictable craniofacial defects. Bioelectric regulation of the embryonic face is likely to be highly relevant to human medicine, as several channels have now been implicated in craniofacial dysmorphias125–134, including Kir2.1 (Andersen-Tawil syndrome), GABA-A (Angelman syndrome), and KCNQ1 (Beckwith-Wiedemann syndrome). Reprogramming tissue for applications addressing these birth defects will require increased understanding of the origin and role of the patterns of Vmem during craniofacial patterning.
One important area for future work in synthetic biology and bioengineering is to understand the links between Vmem and mechanical properties of tissues135. It is now clear that electrical stimulation can alter biomechanical outcomes136–138, and future work must distinguish the individual roles of electric fields, resting potential, and the roles of proteins that transduce bioelectricity to and from mechanical deformation, such as prestins139 and stretch-activated ion channels140.
Manipulation of Vmem gradients: applications
In addition to the endogenous roles of bioelectric gradients that have been discovered by loss-of-function approaches, the most exciting data for reprogramming applications come from gain-of-function studies that ask what shape changes are possible to make by appropriate modulation of Vmem16, 113, 141, 142. Such data were first derived from screens in which ion channels and pumps were randomly misexpressed in frog embryos to gauge the range of phenotypes that might be obtained by perturbation of endogenous bioelectric gradients, followed by focused experiments designed to induce specific physiological states (e.g., ones associated with regenerative response53, 143).
The Xenopus larva regenerates its tail – a highly patterned appendage containing a spinal cord, muscle, peripheral innervation, vasculature, and connective tissues144, 145. Interestingly, tadpoles undergo age-dependent decline of regenerative ability, as do human beings. The non-regenerative (refractory) state in the Xenopus tail can be overcome by transgenes driving strong proton efflux53 or by a cocktail that modulates sodium content52. In either case, the downstream sequellae of the regeneration-specific physiological state are induction of regenerative genes (such as Notch and BMP4), a strong increase in cell proliferation at the wound, and extensive innervation towards the outgrowth. There are several important aspects to this pathway. First, the whole complex cascade of organ regeneration can be triggered by an extremely simple event (proton pumping “on” – likely only 1 bit of information since no attempt was made to tune the strength, pattern, or timing of the flux). Indeed in the case of the sodium-based cocktail, an exposure of just 1 hour was sufficient to kick-start the whole regenerative process. Moreover, what formed was a normal tail of the right size, shape and orientation – not a small tail or tumour, suggesting that in this case (unlike in the case of the craniofacial prepattern) what is encoded by Vmem here is a “master regulator” or top network node signal – a sort of subroutine call that activates a complex, self-limiting downstream developmental module that builds the tail. More recently, a preliminary study showed that the same cocktail initiates leg regeneration after amputation146, suggesting that this system interacts with positional information cues to specify a “build whatever structure normally goes here” signal, not a set of cues that directly micromanage the morphogenetic process.
A considerably different picture of the role of voltage gradients is painted by another recent finding. By modulating Vmem states in vivo in frog embryos, any location in the tadpole could be turned into eye tissue –in some cases, a complete eye with all of the normal tissues arranged in proper morphology50. The effect takes place via a feedback loop in which a hyperpolarized voltage state activates the Pax6 eye gene and vice versa. However, the mechanism must involve much more than upregulation of Pax6 because Pax6 itself only makes eyes when it is misexpressed in the anterior neuroectoderm but not outside the head. Appropriate misexpression of ion channels was able to induce eyes anywhere, including in the gut, tail, and lateral plate mesoderm. Since it was previously thought that only neurectoderm was competent to make eye, these data suggest that bioelectric pathways may necessitate a revision to current lineage restriction maps, and may be a powerful way to control differentiation of iPS cells, embryonic stem cells, and somatic cells that need to be reprogrammed. As in tail regeneration, the whole eye was induced without having to specify the details of its construction (a desirable property for applications in regenerative medicine); however, because eyes could be formed anywhere (in ectopic, inappropriate locations) simply by achieving a specific range of Vmem (about 15 mV wide), the authors hypothesized that individual organs could be encoded by specific ranges of Vmem values. In addition to “form whatever normally goes here” (the kind of effect observed in the induction of tail/limb regeneration), it might be possible to coax cells to build appropriate organs as needed on demand. The mapping between Vmem ranges and other structures besides eye remains to be elucidated. Thus, bioelectricity offers opportunities for control at multiple levels: reprogramming of cell behaviors such as de-differentiation and mitotic control, and regulating large-scale morphogenesis at the level of anatomical polarity, growth control, and organ specification. Future work must delineate the precise methodology and bioelectrical parameters that induce the former or latter mode of signalling from Vmemchange.
Unique features of bioelectrical signalling
A major area of inquiry remains the cracking of the bioelectric code – detailing the mapping between Vmem state distributions and the resulting anatomy. It is necessary to more clearly understand the role of Vmem gradients in kick-starting complex developmental modules that build whole organs vs. serving as direct templates for tissue patterning. To fully capitalize on the power of bioelectrical cues for cell and tissue reprogramming, it is important to exploit several major distinctions between bioelectrical controls and the biochemical/genetic signals for which modern molecular and cell biology tools are optimized. First, there is not always a simple one-to-one correspondence between genetics and bioelectricity. The post-translational gating of channels means that it is impossible to determine the Vmem state of a cell from expression data describing which channel proteins are present. Thus, the real-time physiological prepatterns of Vmem in tissue will be invisible to high-resolution mRNA or protein profiling technologies. Vmem should be thought of as an aggregate, higher-level property akin to “pressure” or “temperature”, resulting from the ensemble properties of many individual gene products. The fact that Vmem is determined by a contribution of all of the ion conductances in any cell means that single channel knockouts will fall prey to false negatives (due to compensation and redundancy). Understanding and controlling the bioelectrical gradients that encode the programs of pattern formation will require deep physiomic profiling data, and continued development of tools for regulating the Vmem of tissue regions at will. One such technology is optogenetics147; although so far these reagents have been used largely in neural and excitable cells, the application of light-based control of ion flows148, 149 to non-neuronal, somatic cells will enable the construction of true computational tissues into which bioelectric patterning information can be dynamically written and read from, for guided self-assembly of physiological and thus genetic and anatomical patterns. The first steps have been taken, as a recent report showed the induction of tail regeneration by optical modulation of bioelectric state after amputation150.
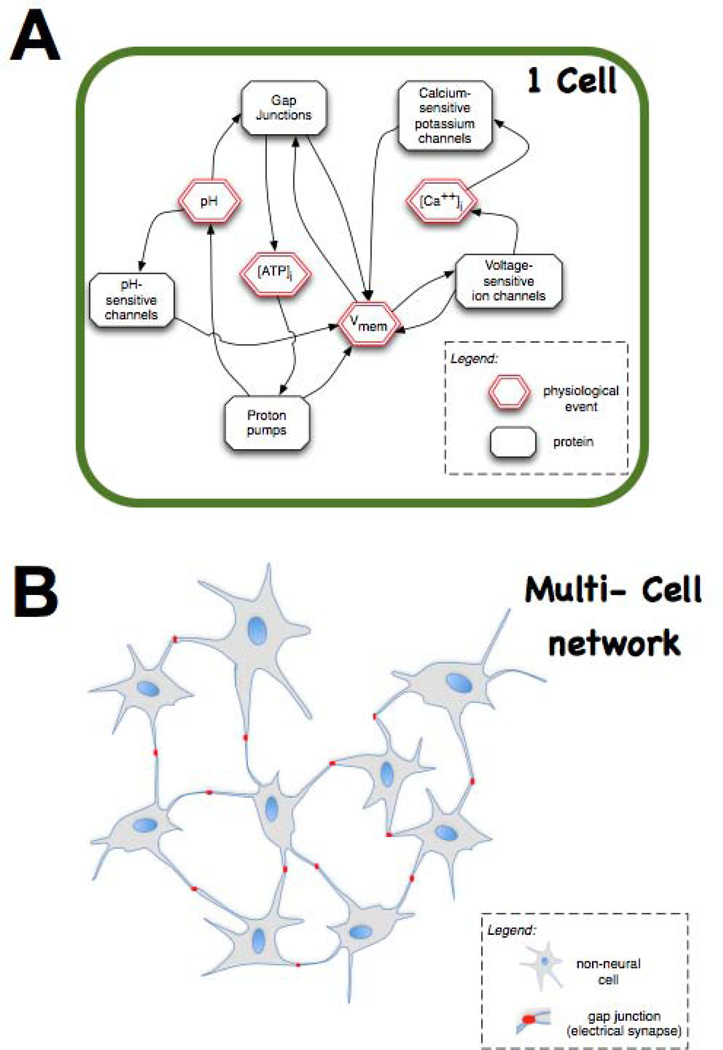
One exciting yet speculative hypothesis is suggested by the recent advances in bioelectricity (Fig. 5). The complex feedback dynamics between physiological parameters and proteins that are both regulated by, and regulate, those bioelectric states (e.g., voltage-sensitive potassium channels) can result in multiple stable attractors for cell physiology. This in turn means that cells could store information in their bioelectrical states151. Moreover, since non-neural cells are able to participate in electrical communication with their neighbours using electrical synapses (gap junctions152, 153), there may be no fundamental difference between electrically-communicating somatic tissues and neural networks. It has been suggested that synapses are an evolutionary innovation based on much more primitive cell:cell signalling modes using gap junctions, voltage regulation, and neurotransmitter molecule movement154; thus it is tempting to speculate that the information processing abilities of neural networks are also not restricted to brains. Indeed, non-neural cells may be able to store immense amounts of information encoded in the stable many-valued Vmem levels (at each of many domains across the cell surface). The development of new technologies for tracking and modulating the bioelectrical communication among actively patterning tissues will allow the testing of this hypothesis, and may reveal memory, decision-making, and other functions formerly reserved for neural systems implemented in somatic tissues155, 156 – a finding that would have important implications not only for strategies to reprogram morphogenesis but also the design of novel architectures for computer technology157, 158.
Figure 5. Bioelectric signals enable non-neural cell fields to function as a computational medium.
(A) At the level of single cells, elements such as voltage-sensitive ion channels result in feedback loops between the activity of ion translocator proteins and physiological parameters such as Vmem. These feedback loops ensure that physiological networks have non-obvious (emergent) behaviour dynamics, which can display hysteresis and multiple attractor states – thus able to store information encoded in stable Vmem states (e.g., depolarized = 1, hyperpolarized = 0) that would be invisible to genetic or proteomic profiling. (B) Even more interestingly, multiple (non-neural) cells communicating electrically via gap junctions (electrical synapses) could potentially store information and make decisions in the same way as do neural networks. The testing of this speculative hypothesis (using paradigms well-developed in computational neuroscience) may reveal entirely novel ways to understand and manipulate tissue-wide information that directs morphogenesis, and new approaches for the development of new (biologically-embedded) computational platforms. This figure is modified after figure 5 of reference146.
Conclusion
The true potential of biological reprogramming will only be realized when we gain control of shape at the level of large-scale multicellular structures166. The next phase of synthetic bioengineering will usher in the development of computational tissues: multicellular biological constructs that can be programmed as an excitable medium158, 167. The output of such programs will be complex 3-dimensional structures such as eyes, limbs, and synthetic constructs with shapes never before seen in the evolutionary history of Earth. The “master regulator” properties of bioelectric and other signalling modalities will allow bioengineers to offload some of the incredible computational complexity onto the organism itself, which is already ideally suited to carry out the assembly and repair process. When integrated with new bioreactors168, 169 and delivery methodology that can tweak the time evolution of complex systems by providing instructive cues at key decision times, such guided self-assembly processes will greatly expand our ability to produce required structures for regeneration in situ or for transplantation. This will revolutionize not only regenerative approaches but also the creation of hybrid biological robotic devices170.
Such advances require the development of new theory and methodology. On the theoretic front, both analysis and synthesis of complex shapes will benefit from the development of new models for understanding how information can be stored and manipulated in physiological networks, and of next-generation artificial intelligence tools supporting a bioinformatics of shape and pattern control. In technique development, advances in fields such as optogenetics will greatly facilitate hypothesis testing by allowing the bioelectric signals within complex tissues to be altered as needed. Next frontiers in molecular bioelectricity include the functional relevance of nuclear envelope voltage potentials, and the significance of multiple Vmem domains across the surface of cells. The development of mature bioelectric technology will revolutionize the field of molecular physiology and reprogramming as the development of restriction enzymes did for molecular cell biology. Just as the understanding of cell function was transformed by the ability to construct desired DNA sequences, the cracking of the bioelectric code will propel the field to exciting new vistas in the control of growth and form, with biomedical applications in birth defects, regenerative repair, cancer, and synthetic biology.
Sidebar: A new bioinformatics of shape.
“There is an obvious discrepancy between the single-cell genetic input and the multicellular geometrical output. To bridge the gap, a mathematical proof, usually in the form of computer simulations, becomes necessary”159. While the tools of bioinformatics have revolutionized molecular, cell, and evolutionary biology, they are largely limited to operating with genomic, transcriptional, or proteomic data – at the level of molecules. Likewise, recent work in the robot scientist field160, 161 is most suited to biochemistry, drug discovery, and metabolism. Computational efforts in synthetic biology162 have begun to develop frameworks for guiding top-down design of complex pathways and control circuitry. However, in the fields of developmental and regenerative biology, scientists are still required to manually attempt to derive testable models from functional data. Organisms such as salamanders, planaria, and deer provide striking proof-of-principle that complex organ repair in adulthood is an achievable goal. As we probe these model systems to uncover the rules guiding dynamic remodelling (reprogramming) of cell behaviour and organ shape, an ever-increasing deluge of functional data floods the literature. What are needed are constructive (algorithmic) models that specify the actions and decisions made at each step163, 164. A true explanation of pattern regulation must specify the information and energy flow at step that is sufficient to produce the self-repairing systems we seek to emulate – not only outline gene-regulatory networks based on loss-of-function data that reveal what components are necessary for the system to work correctly. However, the mountain of information on experimental perturbations that result in specific changes of anatomical shape is already so large and complex that scientists can rarely produce algorithmic models that fit those data. Additional data inhibit, not help, the efforts of human scientists to keep up with the facts and try to mentally construct a model whose behaviour fits all of the data. A fundamental piece of this problem is that feedback, emergence, and complexity (in the dynamical systems sense) render it extremely difficult to know in advance what dynamic behaviour will result from a model specified in terms of components and rules. Thus, one key pillar of continuing advances past the current plateau is the development of a new Bioinformatics of Shape. This marriage of bench biology and true computer science is beginning to give rise to artificial intelligence tools165 to help mine the database of functional experiments on pattern formation and suggest testable, mechanistic, algorithmic models of biological systems that acquire, modify, and repair their shape.
Acknowledgements
The author thanks Douglas J. Blackiston and Laura N. Vandenberg for helpful comments on a draft of this manuscript, and members of the Levin lab and the bioelectricity community for many helpful discussions. This work was supported by National Institutes of Health (EY018168, AR061988, GM078484, AR055993), the G. Harold and Leila Y. Mathers Charitable Foundation, and the Telemedicine and Advanced Technology Research Center at the US Army Medical Research and Materiel Command through award W81XWH-10–2-0058.
Footnotes
No conflict of interest.
- Pullar CE. The physiology of bioelectricity in development, tissue regeneration, and cancer. Biological effects of electromagnetics series 2011. Available at: http://www.crcnetbase.com/isbn/978-1-4398-3724-5.
- Nuccitelli R. Ionic currents in development. New York: Alan R. Liss; 1986.
- Borgens R, Robinson K, Vanable J, McGinnis M. Electric Fields in Vertebrate Repair. New York: Alan R. Liss; 1989.
References
- 1.Levin M. Morphogenetic fields in embryogenesis, regeneration, and cancer: non-local control of complex patterning. Bio Systems. 2012;109:243–261. doi: 10.1016/j.biosystems.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin H. Cancer as a dynamic developmental disorder. Cancer Res. 1985;45:2935–2942. [PubMed] [Google Scholar]
- 3.Smithers DW. An attack on cytologism. Lancet. 1962;1:493–499. doi: 10.1016/s0140-6736(62)91475-7. [DOI] [PubMed] [Google Scholar]
- 4.Waddington CH. Cancer and the theory of organisers. Nature. 1935;135:606–608. [Google Scholar]
- 5.Gentile L, Cebria F, Bartscherer K. The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration. Dis Model Mech. 2011;4:12–19. doi: 10.1242/dmm.006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCusker C, Gardiner DM. The axolotl model for regeneration and aging research: a mini-review. Gerontology. 2011;57:565–571. doi: 10.1159/000323761. [DOI] [PubMed] [Google Scholar]
- 7.Li C. Deer antler regeneration: A stem cell-based epimorphic process. Birth defects research. Part C, Embryo today : reviews. 2012;96:51–62. doi: 10.1002/bdrc.21000. [DOI] [PubMed] [Google Scholar]
- 8.Illingworth CM. Trapped fingers and amputated finger tips in children. J Pediatr Surg. 1974;9:853–858. doi: 10.1016/s0022-3468(74)80220-4. [DOI] [PubMed] [Google Scholar]
- 9.Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer. 2007;7:246–255. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 10.Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci U S A. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose SM, Wallingford HM. Transformation of renal tumors of frogs to normal tissues in regenerating limbs of salamanders. Science. 1948;107:457. [PubMed] [Google Scholar]
- 12.Tsonis PA. Effects of carcinogens on regenerating and non-regenerating limbs in amphibia (review) Anticancer Research. 1983;3:195–202. [PubMed] [Google Scholar]
- 13.Ingber DE. Can cancer be reversed by engineering the tumor microenvironment? Semin Cancer Biol. 2008;18:356–364. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bizzarri M, Cucina A, Biava PM, Proietti S, D'Anselmi F, Dinicola S, Pasqualato A, Lisi E. Embryonic morphogenetic field induces phenotypic reversion in cancer cells. Review article. Curr Pharm Biotechnol. 2011;12:243–253. doi: 10.2174/138920111794295701. [DOI] [PubMed] [Google Scholar]
- 15.Levin M. Molecular bioelectricity in developmental biology: new tools and recent discoveries: control of cell behavior and pattern formation by transmembrane potential gradients. BioEssays : news and reviews in molecular, cellular and developmental biology. 2012;34:205–217. doi: 10.1002/bies.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin M, Stevenson CG. Regulation of cell behavior and tissue patterning by bioelectrical signals: challenges and opportunities for biomedical engineering. Annu Rev Biomed Eng. 2012;14:295–323. doi: 10.1146/annurev-bioeng-071811-150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCaig CD, Rajnicek AM, Song B, Zhao M. Controlling cell behavior electrically: current views and future potential. Physiol Rev. 2005;85:943–978. doi: 10.1152/physrev.00020.2004. [DOI] [PubMed] [Google Scholar]
- 18.Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–460. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]
- 19.Shi R, Borgens RB. Three-dimensional gradients of voltage during development of the nervous system as invisible coordinates for the establishment of embryonic pattern. Dev Dyn. 1995;202:101–114. doi: 10.1002/aja.1002020202. [DOI] [PubMed] [Google Scholar]
- 20.Borgens RB, Shi R. Uncoupling histogenesis from morphogenesis in the vertebrate embryo by collapse of the transneural tube potential. Developmental Dynamics. 1995;203:456–467. doi: 10.1002/aja.1002030408. [DOI] [PubMed] [Google Scholar]
- 21.Nuccitelli R. A role for endogenous electric fields in wound healing. Curr Top Dev Biol. 2003;58:1–26. doi: 10.1016/s0070-2153(03)58001-2. [DOI] [PubMed] [Google Scholar]
- 22.Robinson KR, Messerli MA. Electric embryos: the embryonic epithelium as a generator of developmental information. In: McCaig CD, editor. Nerve Growth and Guidance. London: Portland Press; 1996. pp. 131–150. [Google Scholar]
- 23.Yao L, Pandit A, Yao S, McCaig CD. Electric field-guided neuron migration: a novel approach in neurogenesis. Tissue Eng Part B Rev. 2011;17:143–153. doi: 10.1089/ten.TEB.2010.0561. [DOI] [PubMed] [Google Scholar]
- 24.Rajnicek AM, Foubister LE, McCaig CD. Prioritising guidance cues: directional migration induced by substratum contours and electrical gradients is controlled by a rho/cdc42 switch. Dev Biol. 2007;312:448–460. doi: 10.1016/j.ydbio.2007.09.051. [DOI] [PubMed] [Google Scholar]
- 25.Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. 2009;8:3519–3528. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundelacruz S, Levin M, Kaplan DL. Membrane potential controls adipogenic and osteogenic differentiation of mesenchymal stem cells. PLoS One. 2008;3:e3737. doi: 10.1371/journal.pone.0003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 28.Funk RH, Monsees T, Ozkucur N. Electromagnetic effects - From cell biology to medicine. Prog Histochem Cytochem. 2009;43:177–264. doi: 10.1016/j.proghi.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Balint R, Cassidy NJ, Cartmell SH. Electrical stimulation: a novel tool for tissue engineering. Tissue engineering. Part B, Reviews. 2013;19:48–57. doi: 10.1089/ten.TEB.2012.0183. [DOI] [PubMed] [Google Scholar]
- 30.Trushin MV. Distant non-chemical communication in various biological systems. Riv Biol. 2004;97:409–442. [PubMed] [Google Scholar]
- 31.Sun Y, Wang C, Dai J. Biophotons as neural communication signals demonstrated by in situ biophoton autography. Photochem Photobiol Sci. 2010;9:315–322. doi: 10.1039/b9pp00125e. [DOI] [PubMed] [Google Scholar]
- 32.Fels D. Cellular communication through light. PLoS One. 2009;4:e5086. doi: 10.1371/journal.pone.0005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams DS, Levin M. Endogenous voltage gradients as mediators of cell-cell communication: strategies for investigating bioelectrical signals during pattern formation. Cell Tissue Res. 2013;352:95–122. doi: 10.1007/s00441-012-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandenberg LN, Morrie RD, Adams DS. V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev Dyn. 2011;240:1889–1904. doi: 10.1002/dvdy.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid B, Zhao M. Ion-selective self-referencing probes for measuring specific ion flux. Communicative & integrative biology. 2011;4:524–527. doi: 10.4161/cib.4.5.16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita M. Electric axon guidance in embryonic retina: galvanotropism revisited. Biochemical and biophysical research communications. 2013;431:280–283. doi: 10.1016/j.bbrc.2012.12.115. [DOI] [PubMed] [Google Scholar]
- 37.Adams DS, Levin M. General principles for measuring resting membrane potential and ion concentration using fluorescent bioelectricity reporters. Cold Spring Harbor protocols. 2012;2012:385–397. doi: 10.1101/pdb.top067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams DS, Levin M. Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4(3) and CC2-DMPE. Cold Spring Harbor Protocols. 2012 doi: 10.1101/pdb.prot067702. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oviedo NJ, Nicolas CL, Adams DS, Levin M. Live Imaging of Planarian Membrane Potential Using DiBAC4(3) Cold Spring Harb Protoc. 2008;2008 doi: 10.1101/pdb.prot5055. pdb.prot5055-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell. 2002;111:77–89. doi: 10.1016/s0092-8674(02)00939-x. [DOI] [PubMed] [Google Scholar]
- 41.Adams DS, Robinson KR, Fukumoto T, Yuan S, Albertson RC, Yelick P, Kuo L, McSweeney M, Levin M. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development. 2006;133:1657–1671. doi: 10.1242/dev.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams DS, Levin M. Strategies and techniques for investigation of biophysical signals in patterning. In: Whitman M, Sater AK, editors. Analysis of Growth Factor Signaling in Embryos. Taylor and Francis Books; 2006. pp. 177–262. [Google Scholar]
- 43.Adams DS, Levin M. Inverse drug screens: a rapid and inexpensive method for implicating molecular targets. Genesis. 2006;44:530–540. doi: 10.1002/dvg.20246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahal GR, Rawson J, Gassaway B, Kwok B, Tong Y, Ptacek LJ, Bates E. An inwardly rectifying K+ channel is required for patterning. Development. 2012;139:3653–3664. doi: 10.1242/dev.078592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA, Izpisua Belmonte JC. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- 46.Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- 47.Shimeld SM, Levin M. Evidence for the regulation of left-right asymmetry in Ciona intestinalis by ion flux. Dev Dyn. 2006;235:1543–1553. doi: 10.1002/dvdy.20792. [DOI] [PubMed] [Google Scholar]
- 48.Hibino T, Ishii Y, Levin M, Nishino A. Ion flow regulates left-right asymmetry in sea urchin development. Dev Genes Evol. 2006;216:265–276. doi: 10.1007/s00427-005-0051-6. [DOI] [PubMed] [Google Scholar]
- 49.Pineda RH, Svoboda KR, Wright MA, Taylor AD, Novak AE, Gamse JT, Eisen JS, Ribera AB. Knockdown of Nav1.6a Na+ channels affects zebrafish motoneuron development. Development. 2006;133:3827–3836. doi: 10.1242/dev.02559. [DOI] [PubMed] [Google Scholar]
- 50.Pai VP, Aw S, Shomrat T, Lemire JM, Levin M. Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development. 2012;139:313–323. doi: 10.1242/dev.073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozkucur N, Epperlein HH, Funk RH. Ion imaging during axolotl tail regeneration in vivo. Dev Dyn. 2010;239:2048–2057. doi: 10.1002/dvdy.22323. [DOI] [PubMed] [Google Scholar]
- 52.Tseng AS, Beane WS, Lemire JM, Masi A, Levin M. Induction of vertebrate regeneration by a transient sodium current. J Neurosci. 2010;30:13192–13200. doi: 10.1523/JNEUROSCI.3315-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams DS, Masi A, Levin M. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development. 2007;134:1323–1335. doi: 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- 54.Chernet BT, Levin M. Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumor development in a Xenopus model. Disease Models & Mechanisms. 2013 doi: 10.1242/dmm.010835. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lobikin M, Chernet B, Lobo D, Levin M. Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo. Physical Biology. 2012;9:065002. doi: 10.1088/1478-3975/9/6/065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morokuma J, Blackiston D, Adams DS, Seebohm G, Trimmer B, Levin M. Modulation of potassium channel function confers a hyperproliferative invasive phenotype on embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:16608–16613. doi: 10.1073/pnas.0808328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackiston D, Adams DS, Lemire JM, Lobikin M, Levin M. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis Model Mech. 2011;4:67–85. doi: 10.1242/dmm.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.House CD, Vaske CJ, Schwartz AM, Obias V, Frank B, Luu T, Sarvazyan N, Irby R, Strausberg RL, Hales TG, et al. Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion. Cancer Res. 2010;70:6957–6967. doi: 10.1158/0008-5472.CAN-10-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fraser SP, Diss JK, Chioni AM, Mycielska ME, Pan H, Yamaci RF, Pani F, Siwy Z, Krasowska M, Grzywna Z, et al. Voltage-gated sodium channel expression and potentiation of human breast cancer metastasis. Clin Cancer Res. 2005;11:5381–5389. doi: 10.1158/1078-0432.CCR-05-0327. [DOI] [PubMed] [Google Scholar]
- 60.Levin M. Is the early left-right axis like a plant, a kidney, or a neuron? The integration of physiological signals in embryonic asymmetry. Birth Defects Res C Embryo Today. 2006;78:191–223. doi: 10.1002/bdrc.20078. [DOI] [PubMed] [Google Scholar]
- 61.Beane WS, Morokuma J, Adams DS, Levin M. A Chemical Genetics Approach Reveals H,K-ATPase-Mediated Membrane Voltage Is Required for Planarian Head Regeneration. Chemistry & Biology. 2011;18:77–89. doi: 10.1016/j.chembiol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morokuma J, Blackiston D, Levin M. KCNQ1 and KCNE1 K+ channel components are involved in early left-right patterning in Xenopus laevis embryos. Cell Physiol Biochem. 2008;21:357–372. doi: 10.1159/000129628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamura Y, Dixon JE. Voltage-sensing phosphatase: its molecular relationship with PTEN. Physiology (Bethesda) 2011;26:6–13. doi: 10.1152/physiol.00035.2010. [DOI] [PubMed] [Google Scholar]
- 64.Levin M, Mercola M. Gap junction-mediated transfer of left-right patterning signals in the early chick blastoderm is upstream of Shh asymmetry in the node. Development. 1999;126:4703–4714. doi: 10.1242/dev.126.21.4703. [DOI] [PubMed] [Google Scholar]
- 65.Levin M, Mercola M. Gap junctions are involved in the early generation of left-right asymmetry. Dev Biol. 1998;203:90–105. doi: 10.1006/dbio.1998.9024. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Levin M. Particle tracking model of electrophoretic morphogen movement reveals stochastic dynamics of embryonic gradient. Dev Dyn. 2009;238:1923–1935. doi: 10.1002/dvdy.22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fukumoto T, Kema IP, Levin M. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Curr Biol. 2005;15:794–803. doi: 10.1016/j.cub.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 68.Carneiro K, Donnet C, Rejtar T, Karger BL, Barisone GA, Diaz E, Kortagere S, Lemire JM, Levin M. Histone deacetylase activity is necessary for left-right patterning during vertebrate development. BMC Dev Biol. 2011;11:29. doi: 10.1186/1471-213X-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukumoto T, Blakely R, Levin M. Serotonin transporter function is an early step in left-right patterning in chick and frog embryos. Dev Neurosci. 2005;27:349–363. doi: 10.1159/000088451. [DOI] [PubMed] [Google Scholar]
- 70.Tseng AS, Levin M. Transducing bioelectric signals into epigenetic pathways during tadpole tail regeneration. Anatomical record. 2012;295:1541–1551. doi: 10.1002/ar.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta N, Martin PM, Prasad PD, Ganapathy V. SLC5A8 (SMCT1)-mediated transport of butyrate forms the basis for the tumor suppressive function of the transporter. Life Sci. 2006;78:2419–2425. doi: 10.1016/j.lfs.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 72.Arcangeli A. Ion channels and transporters in cancer. 3. Ion channels in the tumor cell-microenvironment cross talk. American journal of physiology. Cell physiology. 2011;301:C762–C771. doi: 10.1152/ajpcell.00113.2011. [DOI] [PubMed] [Google Scholar]
- 73.Jaffe LF, Poo MM. Neurites grow faster towards the cathode than the anode in a steady field. Journal of Experimental Zoology. 1979;209:115–128. doi: 10.1002/jez.1402090114. [DOI] [PubMed] [Google Scholar]
- 74.Patel N, Poo MM. Orientation of neurite growth by extracellular electric fields. Journal of Neuroscience. 1982;2:483–496. doi: 10.1523/JNEUROSCI.02-04-00483.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borgens RB, Blight AR, McGinnis ME. Behavioral recovery induced by applied electric fields after spinal cord hemisection in guinea pig. Science. 1987;238:366–369. doi: 10.1126/science.3659920. [DOI] [PubMed] [Google Scholar]
- 76.Nishiyama M, von Schimmelmann MJ, Togashi K, Findley WM, Hong K. Membrane potential shifts caused by diffusible guidance signals direct growth-cone turning. Nat Neurosci. 2008;11:762–771. doi: 10.1038/nn.2130. [DOI] [PubMed] [Google Scholar]
- 77.Ozkucur N, Perike S, Sharma P, Funk RH. Persistent directional cell migration requires ion transport proteins as direction sensors and membrane potential differences in order to maintain directedness. BMC Cell Biol. 2011;12:4. doi: 10.1186/1471-2121-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao L, Wei D, Reid B, Zhao S, Pu J, Pan T, Yamoah E, Zhao M. Endogenous electric currents might guide rostral migration of neuroblasts. EMBO reports. 2013;14:184–190. doi: 10.1038/embor.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan L, Borgens RB. Strict perpendicular orientation of neural crest-derived neurons in vitro is dependent on an extracellular gradient of voltage. J Neurosci Res. 2012;90:1335–1346. doi: 10.1002/jnr.22809. [DOI] [PubMed] [Google Scholar]
- 80.Yao L, McCaig CD, Zhao M. Electrical signals polarize neuronal organelles, direct neuron migration, and orient cell division. Hippocampus. 2009;19:855–868. doi: 10.1002/hipo.20569. [DOI] [PubMed] [Google Scholar]
- 81.Pan L, Borgens RB. Perpendicular organization of sympathetic neurons within a required physiological voltage. Exp Neurol. 2010;222:161–164. doi: 10.1016/j.expneurol.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Gao RC, Zhang XD, Sun YH, Kamimura Y, Mogilner A, Devreotes PN, Zhao M. Different Roles of Membrane Potentials in Electrotaxis and Chemotaxis of Dictyostelium Cells. Eukaryot Cell. 2011 doi: 10.1128/EC.05066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajnicek AM, Foubister LE, McCaig CD. Growth cone steering by a physiological electric field requires dynamic microtubules, microfilaments and Rac-mediated filopodial asymmetry. J Cell Sci. 2006;119:1736–1745. doi: 10.1242/jcs.02897. [DOI] [PubMed] [Google Scholar]
- 84.Pullar CE, Zhao M, Song B, Pu J, Reid B, Ghoghawala S, McCaig C, Isseroff RR. Beta-adrenergic receptor agonists delay while antagonists accelerate epithelial wound healing: evidence of an endogenous adrenergic network within the corneal epithelium. J Cell Physiol. 2007;211:261–272. doi: 10.1002/jcp.20934. [DOI] [PubMed] [Google Scholar]
- 85.Pullar CE, Baier BS, Kariya Y, Russell AJ, Horst BA, Marinkovich MP, Isseroff RR. beta4 integrin and epidermal growth factor coordinately regulate electric field-mediated directional migration via Rac1. Mol Biol Cell. 2006;17:4925–4935. doi: 10.1091/mbc.E06-05-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pullar CE, Isseroff RR. Cyclic AMP mediates keratinocyte directional migration in an electric field. J Cell Sci. 2005;118:2023–2034. doi: 10.1242/jcs.02330. [DOI] [PubMed] [Google Scholar]
- 87.You MH, Song MS, Lee SK, Ryu PD, Lee SY, Kim DY. Voltage-gated K(+) channels in adipogenic differentiation of bone marrow-derived human mesenchymal stem cells. Acta pharmacologica Sinica. 2012 doi: 10.1038/aps.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng SY, Chin CH, Lau YT, Luo J, Wong CK, Bian ZX, Tsang SY. Role of voltage-gated potassium channels in the fate determination of embryonic stem cells. J Cell Physiol. 2010 doi: 10.1002/jcp.22113. [DOI] [PubMed] [Google Scholar]
- 89.Du Y, Du Z, Zheng H, Wang D, Li S, Yan Y, Li Y. GABA exists as a negative regulator of cell proliferation in spermaogonial stem cells. Cellular & molecular biology letters. 2013 doi: 10.2478/s11658-013-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hinard V, Belin D, Konig S, Bader CR, Bernheim L. Initiation of human myoblast differentiation via dephosphorylation of Kir2.1 K+ channels at tyrosine 242. Development. 2008;135:859–867. doi: 10.1242/dev.011387. [DOI] [PubMed] [Google Scholar]
- 91.Li F, Yin J, Yue T, Liu L, Zhang H. The chloride intracellular channel 5 (CLIC5) involved in C2C12 myoblasts proliferation and differentiation. Cell Biol Int. 2010 doi: 10.1042/CBI20090334. [DOI] [PubMed] [Google Scholar]
- 92.Root CM, Velazquez-Ulloa NA, Monsalve GC, Minakova E, Spitzer NC. Embryonically expressed GABA and glutamate drive electrical activity regulating neurotransmitter specification. J Neurosci. 2008;28:4777–4784. doi: 10.1523/JNEUROSCI.4873-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yasuda T, Adams DJ. Physiological roles of ion channels in adult neural stem cells and their progeny. J Neurochem. 2010;114:946–959. doi: 10.1111/j.1471-4159.2010.06822.x. [DOI] [PubMed] [Google Scholar]
- 94.Lange C, Prenninger S, Knuckles P, Taylor V, Levin M, Calegari F. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem cells and development. 2011;20:843–850. doi: 10.1089/scd.2010.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Vliet P, de Boer TP, van der Heyden MA, El Tamer MK, Sluijter JP, Doevendans PA, Goumans MJ. Hyperpolarization induces differentiation in human cardiomyocyte progenitor cells. Stem Cell Rev. 2010;6:178–185. doi: 10.1007/s12015-010-9142-5. [DOI] [PubMed] [Google Scholar]
- 96.Ring H, Mendu SK, Shirazi-Fard S, Birnir B, Hallbook F. GABA maintains the proliferation of progenitors in the developing chick ciliary marginal zone and non-pigmented ciliary epithelium. PloS one. 2012;7:e36874. doi: 10.1371/journal.pone.0036874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liebau S, Tischendorf M, Ansorge D, Linta L, Stockmann M, Weidgang C, Iacovino M, Boeckers T, von Wichert G, Kyba M, et al. An inducible expression system of the calcium-activated potassium channel 4 to study the differential impact on embryonic stem cells. Stem cells international. 2011;2011:456815. doi: 10.4061/2011/456815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sauer H, Rahimi G, Hescheler J, Wartenberg M. Effects of electrical fields on cardiomyocyte differentiation of embryonic stem cells. J Cell Biochem. 1999;75:710–723. doi: 10.1002/(sici)1097-4644(19991215)75:4<710::aid-jcb16>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 99.Aprea J, Calegari F. Bioelectric state and cell cycle control of mammalian neural stem cells. Stem cells international. 2012;2012:816049. doi: 10.1155/2012/816049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park JY, Helm JF, Zheng W, Ly QP, Hodul PJ, Centeno BA, Malafa MP. Silencing of the candidate tumor suppressor gene solute carrier family 5 member 8 (SLC5A8) in human pancreatic cancer. Pancreas. 2008;36:e32–e39. doi: 10.1097/MPA.0b013e3181630ffe. [DOI] [PubMed] [Google Scholar]
- 101.Zhao M. Electrical fields in wound healing-An overriding signal that directs cell migration. Semin Cell Dev Biol. 2009;20:674–682. doi: 10.1016/j.semcdb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 102.Binggeli R, Weinstein R. Membrane potentials and sodium channels: hypotheses for growth regulation and cancer formation based on changes in sodium channels and gap junctions. Journal of Theoretical Biology. 1986;123:377–401. doi: 10.1016/s0022-5193(86)80209-0. [DOI] [PubMed] [Google Scholar]
- 103.Cone CD, Tongier M. Control of somatic cell mitosis by simulated changes in the transmembrane potential level. Oncology. 1971;25:168–182. doi: 10.1159/000224567. [DOI] [PubMed] [Google Scholar]
- 104.Cone CD. Unified theory on the basic mechanism of normal mitotic control and oncogenesis. Journal of Theoretical Biology. 1971;30:151–181. doi: 10.1016/0022-5193(71)90042-7. [DOI] [PubMed] [Google Scholar]
- 105.Jaffe LF. Electrical currents through the developing fucus egg. Proc Natl Acad Sci U S A. 1966;56:1102–1109. doi: 10.1073/pnas.56.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Minc N, Chang F. Electrical control of cell polarization in the fission yeast Schizosaccharomyces pombe. Curr Biol. 2010;20:710–716. doi: 10.1016/j.cub.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Certal AC, Almeida RB, Carvalho LM, Wong E, Moreno N, Michard E, Carneiro J, Rodriguez-Leon J, Wu HM, Cheung AY, et al. Exclusion of a proton ATPase from the apical membrane is associated with cell polarity and tip growth in Nicotiana tabacum pollen tubes. Plant Cell. 2008;20:614–634. doi: 10.1105/tpc.106.047423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aw S, Levin M. Is left-right asymmetry a form of planar cell polarity? Development. 2009;136:355–366. doi: 10.1242/dev.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Levin M, Palmer AR. Left-right patterning from the inside out: widespread evidence for intracellular control. Bioessays. 2007;29:271–287. doi: 10.1002/bies.20545. [DOI] [PubMed] [Google Scholar]
- 110.Vandenberg LN, Morrie RD, Seebohm G, Lemire JM, Levin M. Rab GTPases are required for early orientation of the left-right axis in Xenopus. Mechanisms of development. 2013;130:254–271. doi: 10.1016/j.mod.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vandenberg LN, Lemire JM, Levin M. Serotonin has early, cilia-independent roles in Xenopus left-right patterning. Disease models & mechanisms. 2012 doi: 10.1242/dmm.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marshall WF. Origins of cellular geometry. BMC biology. 2011;9:57. doi: 10.1186/1741-7007-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Levin M. Large-scale biophysics: ion flows and regeneration. Trends in Cell Biology. 2007;17:262–271. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 114.Ganapathy V, Thangaraju M, Gopal E, Martin PM, Itagaki S, Miyauchi S, Prasad PD. Sodium-coupled monocarboxylate transporters in normal tissues and in cancer. AAPS J. 2008;10:193–199. doi: 10.1208/s12248-008-9022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paroder V, Spencer SR, Paroder M, Arango D, Schwartz S, Jr, Mariadason JM, Augenlicht LH, Eskandari S, Carrasco N. Na(+)/monocarboxylate transport (SMCT) protein expression correlates with survival in colon cancer: molecular characterization of SMCT. Proc Natl Acad Sci U S A. 2006;103:7270–7275. doi: 10.1073/pnas.0602365103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miyauchi S, Gopal E, Fei YJ, Ganapathy V. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na(+)-coupled transporter for short-chain fatty acids. J Biol Chem. 2004;279:13293–13296. doi: 10.1074/jbc.C400059200. [DOI] [PubMed] [Google Scholar]
- 117.Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L, Lutterbaugh J, Rerko RM, Casey G, Issa JP, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A. 2003;100:8412–8417. doi: 10.1073/pnas.1430846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Archer SY, Johnson J, Kim HJ, Ma Q, Mou H, Daesety V, Meng S, Hodin RA. The histone deacetylase inhibitor butyrate downregulates cyclin B1 gene expression via a p21/WAF-1-dependent mechanism in human colon cancer cells. American journal of physiology. Gastrointestinal and liver physiology. 2005;289:G696–G703. doi: 10.1152/ajpgi.00575.2004. [DOI] [PubMed] [Google Scholar]
- 119.Taylor AJ, Beck CW. Histone deacetylases are required for amphibian tail and limb regeneration but not development. Mechanisms of development. 2012 doi: 10.1016/j.mod.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 120.Tseng AS, Carneiro K, Lemire JM, Levin M. HDAC activity is required during Xenopus tail regeneration. PloS one. 2011;6:e26382. doi: 10.1371/journal.pone.0026382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Inaba M, Yamanaka H, Kondo S. Pigment pattern formation by contact-dependent depolarization. Science. 2012;335:677. doi: 10.1126/science.1212821. [DOI] [PubMed] [Google Scholar]
- 122.Beane WS, Morokuma J, Lemire JM, Levin M. Bioelectric signaling regulates head and organ size during planarian regeneration. Development. 2013;140:313–322. doi: 10.1242/dev.086900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oviedo NJ, Morokuma J, Walentek P, Kema IP, Gu MB, Ahn JM, Hwang JS, Gojobori T, Levin M. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol. 2010;339:188–199. doi: 10.1016/j.ydbio.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pai VP, Vandenberg LN, Blackiston D, Levin M. Neurally-derived tissues in Xenopus laevis embryos exhibit a consistent bioelectrical left-right asymmetry. Stem cells international. 2012;2012:16. doi: 10.1155/2012/353491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tristani-Firouzi M, Etheridge SP. Kir 2.1 channelopathies: the Andersen-Tawil syndrome. Pflugers Arch. 2010;460:289–294. doi: 10.1007/s00424-010-0820-6. [DOI] [PubMed] [Google Scholar]
- 126.Choi BO, Kim J, Suh BC, Yu JS, Sunwoo IN, Kim SJ, Kim GH, Chung KW. Mutations of KCNJ2 gene associated with Andersen-Tawil syndrome in Korean families. Journal of human genetics. 2007;52:280–283. doi: 10.1007/s10038-006-0100-7. [DOI] [PubMed] [Google Scholar]
- 127.Bendahhou S, Donaldson MR, Plaster NM, Tristani-Firouzi M, Fu YH, Ptacek LJ. Defective potassium channel Kir2.1 trafficking underlies Andersen-Tawil syndrome. J Biol Chem. 2003;278:51779–51785. doi: 10.1074/jbc.M310278200. [DOI] [PubMed] [Google Scholar]
- 128.Roden WH, Peugh LD, Jansen LA. Altered GABA(A) receptor subunit expression and pharmacology in human Angelman syndrome cortex. Neuroscience letters. 2010;483:167–172. doi: 10.1016/j.neulet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Galanopoulou AS. Mutations affecting GABAergic signaling in seizures and epilepsy. Pflugers Archiv : European journal of physiology. 2010;460:505–523. doi: 10.1007/s00424-010-0816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liljelund P, Handforth A, Homanics GE, Olsen RW. GABAA receptor beta3 subunit gene-deficient heterozygous mice show parent-of-origin and gender-related differences in beta3 subunit levels, EEG, and behavior. Brain research. Developmental brain research. 2005;157:150–161. doi: 10.1016/j.devbrainres.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 131.Weksberg R, Shuman C, Caluseriu O, Smith AC, Fei YL, Nishikawa J, Stockley TL, Best L, Chitayat D, Olney A, et al. Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum Mol Genet. 2002;11:1317–1325. doi: 10.1093/hmg/11.11.1317. [DOI] [PubMed] [Google Scholar]
- 132.Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C, Steele L, Cameron J, Smith A, Ambus I, et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet. 2001;10:2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 133.Gaston V, Le Bouc Y, Soupre V, Burglen L, Donadieu J, Oro H, Audry G, Vazquez MP, Gicquel C. Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith-Wiedemann syndrome. European Journal of Human Genetics. 2001;9:409–418. doi: 10.1038/sj.ejhg.5200649. [DOI] [PubMed] [Google Scholar]
- 134.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. The New England journal of medicine. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 135.Ghosh K, Ingber DE. Micromechanical control of cell and tissue development: implications for tissue engineering. Adv Drug Deliv Rev. 2007;59:1306–1318. doi: 10.1016/j.addr.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 136.Joshi SD, von Dassow M, Davidson LA. Experimental control of excitable embryonic tissues: three stimuli induce rapid epithelial contraction. Experimental cell research. 2010;316:103–114. doi: 10.1016/j.yexcr.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Titushkin I, Cho M. Regulation of cell cytoskeleton and membrane mechanics by electric field: role of linker proteins. Biophysical journal. 2009;96:717–728. doi: 10.1016/j.bpj.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song G, Qin J, Yao C, Ju Y. Effect of steep pulsed electric field on proliferation, viscoelasticity and adhesion of human hepatoma SMMC-7721 cells. Anticancer research. 2008;28:2245–2251. [PubMed] [Google Scholar]
- 139.Zhang R, Qian F, Rajagopalan L, Pereira FA, Brownell WE, Anvari B. Prestin modulates mechanics and electromechanical force of the plasma membrane. Biophys J. 2007;93:L07–L09. doi: 10.1529/biophysj.107.107573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nilius B, Honore E. Sensing pressure with ion channels. Trends in neurosciences. 2012;35:477–486. doi: 10.1016/j.tins.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 141.Levin M. Endogenous Bioelectric Signals as Morphogenetic Controls of Development, Regeneration, and Neoplasm. In: Pullar CE, editor. The Physiology of Bioelectricity in Development, Tissue Regeneration, and Cancer. Boca Raton, FL: CRC Press; 2011. pp. 39–89. [Google Scholar]
- 142.Levin M. Bioelectric mechanisms in regeneration: Unique aspects and future perspectives. Semin Cell Dev Biol. 2009;20:543–556. doi: 10.1016/j.semcdb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reid B, Song B, Zhao M. Electric currents in Xenopus tadpole tail regeneration. Dev Biol. 2009;335:198–207. doi: 10.1016/j.ydbio.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 144.Slack JM, Lin G, Chen Y. The Xenopus tadpole: a new model for regeneration research. Cellular and molecular life sciences : CMLS. 2008;65:54–63. doi: 10.1007/s00018-007-7431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mochii M, Taniguchi Y, Shikata I. Tail regeneration in the Xenopus tadpole. Dev Growth Differ. 2007;49:155–161. doi: 10.1111/j.1440-169X.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- 146.Tseng A, Levin M. Cracking the bioelectric code: Probing endogenous ionic controls of pattern formation. Communicative & Integrative Biology. 2013;6:1–8. doi: 10.4161/cib.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Knopfel T, Lin MZ, Levskaya A, Tian L, Lin JY, Boyden ES. Toward the second generation of optogenetic tools. J Neurosci. 2010;30:14998–15004. doi: 10.1523/JNEUROSCI.4190-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annual review of neuroscience. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fortin DL, Dunn TW, Kramer RH. Engineering light-regulated ion channels. Cold Spring Harb Protoc. 2011;2011:579–585. doi: 10.1101/pdb.top112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Adams DS, Tseng AS, Levin M. Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration: sparking system-level controls in vivo. Biology open. 2013;2:306–313. doi: 10.1242/bio.20133665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gallaher J, Bier M, van Heukelom JS. First order phase transition and hysteresis in a cell's maintenance of the membrane potential-An essential role for the inward potassium rectifiers. Biosystems. 2010;101:149–155. doi: 10.1016/j.biosystems.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 152.Bennett MV. Gap junctions as electrical synapses. Journal of Neurocytology. 1997;26:349–366. doi: 10.1023/a:1018560803261. [DOI] [PubMed] [Google Scholar]
- 153.Bissiere S, Zelikowsky M, Ponnusamy R, Jacobs NS, Blair HT, Fanselow MS. Electrical synapses control hippocampal contributions to fear learning and memory. Science. 2011;331:87–91. doi: 10.1126/science.1193785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Buznikov GA, Shmukler YB. Possible role of "prenervous" neurotransmitters in cellular interactions of early embryogenesis: a hypothesis. Neurochem Res. 1981;6:55–68. doi: 10.1007/BF00963906. [DOI] [PubMed] [Google Scholar]
- 155.Turner CH, Robling AG, Duncan RL, Burr DB. Do bone cells behave like a neuronal network? Calcified Tissue International. 2002;70:435–442. doi: 10.1007/s00223-001-1024-z. [DOI] [PubMed] [Google Scholar]
- 156.Chakravarthy SV, Ghosh J. On Hebbian-like adaptation in heart muscle: a proposal for 'cardiac memory'. Biol Cybern. 1997;76:207–215. doi: 10.1007/s004220050333. [DOI] [PubMed] [Google Scholar]
- 157.Regot S, Macia J, Conde N, Furukawa K, Kjellen J, Peeters T, Hohmann S, de Nadal E, Posas F, Sole R. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 158.Adamatzky A, Costello BDL, Shirakawa T. Universal Computation with Limited Resources: Belousov-Zhabotinsky and Physarum Computers. International Journal of Bifurcation and Chaos. 2008;18:2373–2389. [Google Scholar]
- 159.Hamant O. Quantitative imaging strategies pave the way for testable biological concepts. BMC Biol. 2011;9:10. doi: 10.1186/1741-7007-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sparkes A, Aubrey W, Byrne E, Clare A, Khan MN, Liakata M, Markham M, Rowland J, Soldatova LN, Whelan KE, et al. Towards Robot Scientists for autonomous scientific discovery. Autom Exp. 2010;2:1. doi: 10.1186/1759-4499-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Soldatova LN, Clare A, Sparkes A, King RD. An ontology for a Robot Scientist. Bioinformatics. 2006;22:e464–e471. doi: 10.1093/bioinformatics/btl207. [DOI] [PubMed] [Google Scholar]
- 162.Beal J, Lu T, Weiss R. Automatic compilation from high-level biologically-oriented programming language to genetic regulatory networks. PloS one. 2011;6:e22490. doi: 10.1371/journal.pone.0022490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Slack JM. A serial threshold theory of regeneration. J Theor Biol. 1980;82:105–140. doi: 10.1016/0022-5193(80)90092-2. [DOI] [PubMed] [Google Scholar]
- 164.Barkai N, Ben-Zvi D. 'Big frog, small frog'--maintaining proportions in embryonic development. Febs J. 2009;276:1196–1207. doi: 10.1111/j.1742-4658.2008.06854.x. [DOI] [PubMed] [Google Scholar]
- 165.Lobo D, Malone TJ, Levin M. Planform: an application and database of graph-encoded planarian regenerative experiments. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Levin M. The wisdom of the body: future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer. Regenerative medicine. 2011;6:667–673. doi: 10.2217/rme.11.69. [DOI] [PubMed] [Google Scholar]
- 167.Schumann A, Adamatzky A. Toward semantical model of reaction-diffusion computing. Kybernetes. 2009;38:1518–1531. [Google Scholar]
- 168.Hechavarria D, Dewilde A, Braunhut S, Levin M, Kaplan DL. BioDome regenerative sleeve for biochemical and biophysical stimulation of tissue regeneration. Med Eng Phys. 2010;32:1065–1073. doi: 10.1016/j.medengphy.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cimetta E, Sirabella D, Yeager K, Davidson K, Simon J, Moon RT, Vunjak-Novakovic G. Microfluidic bioreactor for dynamic regulation of early mesodermal commitment in human pluripotent stem cells. Lab on a chip. 2013;13:355–364. doi: 10.1039/c2lc40836h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tsuda S, Zauner KP, Gunji YP. Robot control with biological cells. Bio Systems. 2007;87:215–223. doi: 10.1016/j.biosystems.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 171.Stillwell EF, Cone CM, Cone CD. Stimulation of DNA synthesis in CNS neurones by sustained depolarisation. Nature - New Biology. 1973;246:110–111. doi: 10.1038/newbio246110a0. [DOI] [PubMed] [Google Scholar]
- 172.Cone CD, Tongier M. Contact inhibition of division: involvement of the electrical transmembrane potential. Journal of Cellular Physiology. 1973;82:373–386. doi: 10.1002/jcp.1040820307. [DOI] [PubMed] [Google Scholar]
- 173.Cone CD., Jr Variation of the transmembrane potential level as a basic mechanism of mitosis control. Oncology. 1970;24:438–470. doi: 10.1159/000224545. [DOI] [PubMed] [Google Scholar]
- 174.Sundelacruz S, Levin M, Kaplan DL. Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev Rep. 2009;5:231–246. doi: 10.1007/s12015-009-9080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rock JR, Futtner CR, Harfe BD. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev Biol. 2008;321:141–149. doi: 10.1016/j.ydbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 176.Iwashita M, Watanabe M, Ishii M, Chen T, Johnson SL, Kurachi Y, Okada N, Kondo S. Pigment Pattern in jaguar/obelix Zebrafish Is Caused by a Kir7.1 Mutation: Implications for the Regulation of Melanosome Movement. PLoS Genet. 2006;2:e197. doi: 10.1371/journal.pgen.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Watanabe M, Iwashita M, Ishii M, Kurachi Y, Kawakami A, Kondo S, Okada N. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 2006;7:893–897. doi: 10.1038/sj.embor.7400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, Iovine MK. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol. 2008;317:541–548. doi: 10.1016/j.ydbio.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–219. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 180.Davy A, Bush JO, Soriano P. Inhibition of gap junction communication at ectopic Eph/ephrin boundaries underlies craniofrontonasal syndrome. PLoS Biol. 2006;4:e315. doi: 10.1371/journal.pbio.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Civitelli R. Cell-cell communication in the osteoblast/osteocyte lineage. Arch Biochem Biophys. 2008;473:188–192. doi: 10.1016/j.abb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Uzun S, Gokce S, Wagner K. Cystic fibrosis transmembrane conductance regulator gene mutations in infertile males with congenital bilateral absence of the vas deferens. The Tohoku journal of experimental medicine. 2005;207:279–285. doi: 10.1620/tjem.207.279. [DOI] [PubMed] [Google Scholar]
- 183.Wilschanski M, Dupuis A, Ellis L, Jarvi K, Zielenski J, Tullis E, Martin S, Corey M, Tsui LC, Durie P. Mutations in the cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. Am J Respir Crit Care Med. 2006;174:787–794. doi: 10.1164/rccm.200509-1377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rakic P, Sidman RL. Sequence of developmental abnormalities leading to granule cell deficit in cerebellar cortex of weaver mutant mice. The Journal of comparative neurology. 1973;152:103–132. doi: 10.1002/cne.901520202. [DOI] [PubMed] [Google Scholar]
- 185.Rakic P, Sidman RL. Weaver mutant mouse cerebellum: defective neuronal migration secondary to abnormality of Bergmann glia. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:240–244. doi: 10.1073/pnas.70.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hatten ME, Liem RK, Mason CA. Weaver mouse cerebellar granule neurons fail to migrate on wild-type astroglial processes in vitro. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1986;6:2676–2683. doi: 10.1523/JNEUROSCI.06-09-02676.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Patil N, Cox DR, Bhat D, Faham M, Myers RM, Peterson AS. A potassium channel mutation in weaver mice implicates membrane excitability in granule cell differentiation. Nature genetics. 1995;11:126–129. doi: 10.1038/ng1095-126. [DOI] [PubMed] [Google Scholar]
- 188.Culiat CT, Stubbs LJ, Woychik RP, Russell LB, Johnson DK, Rinchik EM. Deficiency of the beta 3 subunit of the type A gamma-aminobutyric acid receptor causes cleft palate in mice. Nature genetics. 1995;11:344–346. doi: 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- 189.Teng GQ, Zhao X, Lees-Miller JP, Quinn FR, Li P, Rancourt DE, London B, Cross JC, Duff HJ. Homozygous missense N629D hERG (KCNH2) potassium channel mutation causes developmental defects in the right ventricle and its outflow tract and embryonic lethality. Circulation research. 2008;103:1483–1491. doi: 10.1161/CIRCRESAHA.108.177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Simons M, Gault WJ, Gotthardt D, Rohatgi R, Klein TJ, Shao Y, Lee HJ, Wu AL, Fang Y, Satlin LM, et al. Electrochemical cues regulate assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nat Cell Biol. 2009;11:286–294. doi: 10.1038/ncb1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hermle T, Saltukoglu D, Grunewald J, Walz G, Simons M. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr Biol. 2010;20:1269–1276. doi: 10.1016/j.cub.2010.05.057. [DOI] [PubMed] [Google Scholar]
- 192.Lee MP, Ravenel JD, Hu RJ, Lustig LR, Tomaselli G, Berger RD, Brandenburg SA, Litzi TJ, Bunton TE, Limb C, et al. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. Journal of Clinical Investigation. 2000;106:1447–1455. doi: 10.1172/JCI10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Moore ES, Ward RE, Escobar LF, Carlin ME. Heterogeneity in Wiedemann-Beckwith syndrome: anthropometric evidence. American journal of medical genetics. 2000;90:283–290. [PubMed] [Google Scholar]
- 194.Onkal R, Djamgoz MB. Molecular pharmacology of voltage-gated sodium channel expression in metastatic disease: clinical potential of neonatal Nav1.5 in breast cancer. Eur J Pharmacol. 2009;625:206–219. doi: 10.1016/j.ejphar.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 195.Pardo LA, del Camino D, Sanchez A, Alves F, Bruggemann A, Beckh S, Stuhmer W. Oncogenic potential of EAG K(+) channels. Embo J. 1999;18:5540–5547. doi: 10.1093/emboj/18.20.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pei L, Wiser O, Slavin A, Mu D, Powers S, Jan LY, Hoey T. Oncogenic potential of TASK3 (Kcnk9) depends on K+ channel function. Proc Natl Acad Sci U S A. 2003;100:7803–7807. doi: 10.1073/pnas.1232448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Saito T, Schlegel R, Andresson T, Yuge L, Yamamoto M, Yamasaki H. Induction of cell transformation by mutated 16K vacuolar H+-atpase (ductin) is accompanied by down-regulation of gap junctional intercellular communication and translocation of connexin 43 in NIH3T3 cells. Oncogene. 1998;17:1673–1680. doi: 10.1038/sj.onc.1202092. [DOI] [PubMed] [Google Scholar]
- 198.Roepke TK, Purtell K, King EC, La Perle KM, Lerner DJ, Abbott GW. Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PLoS One. 2010;5:e11451. doi: 10.1371/journal.pone.0011451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lee MP, Hu RJ, Johnson LA, Feinberg AP. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]