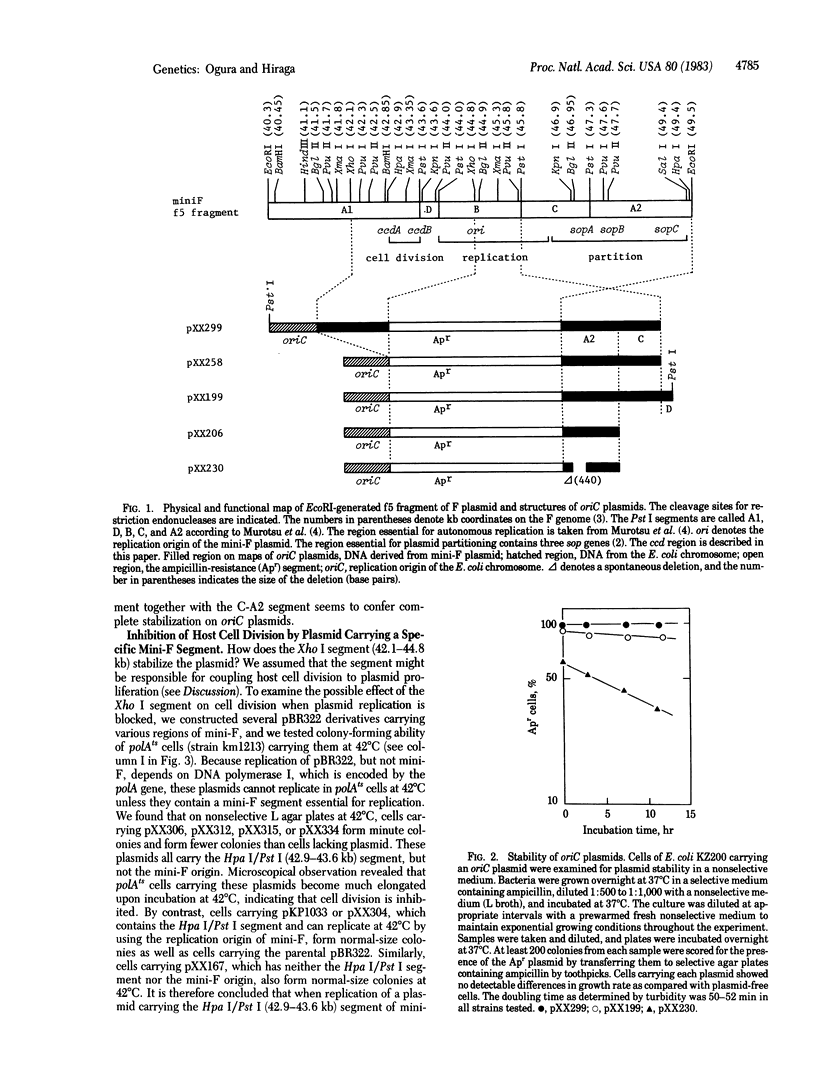
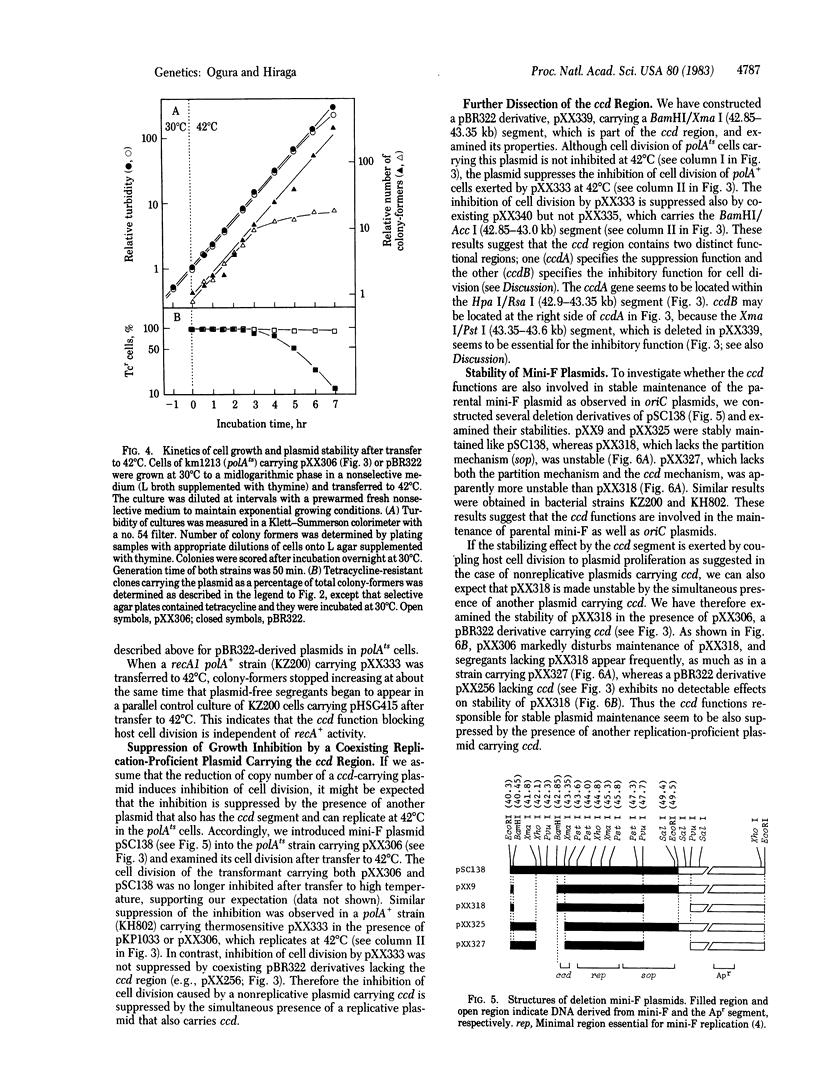
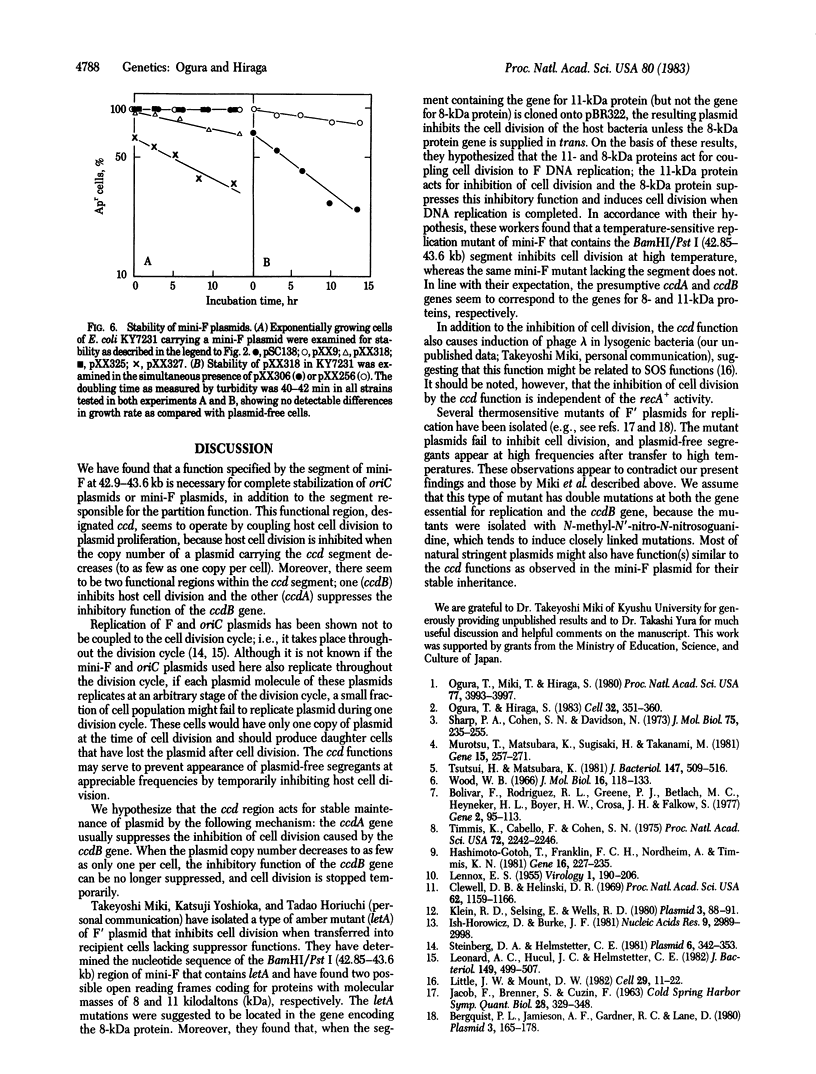
Abstract
A mechanism for stable maintenance of plasmids, besides the replication and partition mechanisms, has been found to be specified by genes of a mini-F plasmid. An oriC plasmid carrying both a mini-F segment necessary for partition [coordinates 46.4-49.4 kilobase pairs (kb) on the F map] and another segment (42.9-43.6 kb), designated ccd (coupled cell division), is more stably maintained than are oriC plasmids carrying only the partition segment; the stability is comparable to that of the parental mini-F plasmid. When replication of a plasmid carrying ccd is prevented and the plasmid copy number decreases, to as few as one per cell, host cell division is inhibited, but not increase of turbidity or chromosome replication. Appearance of plasmid-free segregants is therefore effectively prevented under such conditions. Experimental results suggest that reduction of the copy number of plasmids carrying the ccd region causes an inhibition of cell division and that the ccd region can be dissected into two functional regions; one (ccdB) inhibits cell division and the other (ccdA) releases the inhibition. The interplay of the ccdA and ccdB genes promotes stable plasmid maintenance by coupling host cell division to plasmid proliferation.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergquist P. L., Jamieson A. F., Gardner R. C., Lane D. Replication mutants of the F-plasmid of Escherichia coli K-12. I. Isolation and characterization of temperature-sensitive replication mutants of F'-gal+. Plasmid. 1980 Mar;3(2):165–178. doi: 10.1016/0147-619x(80)90107-9. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Franklin F. C., Nordheim A., Timmis K. N. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981 Dec;16(1-3):227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Leonard A. C., Hucul J. A., Helmstetter C. E. Kinetics of minichromosome replication in Escherichia coli B/r. J Bacteriol. 1982 Feb;149(2):499–507. doi: 10.1128/jb.149.2.499-507.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Matsubara K., Sugisaki H., Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981 Nov;15(2-3):257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Ogura T., Miki T., Hiraga S. Copy-number mutants of the plasmid carrying the replication origin of the Escherichia coli chromosome: evidence for a control region of replication. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3993–3997. doi: 10.1073/pnas.77.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Steinberg D. A., Helmstetter C. E. F plasmid replication and the division cycle of Escherichia coli B/r. Plasmid. 1981 Nov;6(3):342–353. doi: 10.1016/0147-619x(81)90042-1. [DOI] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Matsubara K. Replication control and switch-off function as observed with a mini-F factor plasmid. J Bacteriol. 1981 Aug;147(2):509–516. doi: 10.1128/jb.147.2.509-516.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]



