Abstract
Staphylococcus aureus is the leading cause of hospital-acquired infections in the United States. The emergence of multi-drug resistant strains of S. aureus has created an urgent need for new antibiotics. S. aureus uses the sortase A (SrtA) enzyme to display surface virulence factors suggesting that compounds that inhibit its activity will function as potent anti-infective agents. Here we report the identification of several inhibitors of SrtA using virtual screening methods that employ the relaxed complex scheme, an advanced computer-docking methodology that accounts for protein receptor flexibility. Experimental testing validates that several compounds identified in the screen inhibit the activity of SrtA. A lead compound based on the 2-phenyl-2,3-dihydro-1H-perimidine scaffold is particularly promising and its binding mechanism was further investigated using molecular dynamics simulations and by conducting preliminary structure activity relationship studies.
Keywords: Staphylococcus aureus, MRSA, sortase, SrtA, transpeptidation, Gram-positive, drug discovery, virtual screening, relaxed complex scheme, molecular dynamics, docking
Introduction
Staphylococcus aureus is a leading cause of hospital- and community-acquired infections in the United States and produces a wide spectrum of diseases, ranging from minor skin infections to osteomyelitis, meningitis, endocarditis, septicemia, and toxic shock syndrome (1, 2). The widespread occurrence of methicillin-resistant S. aureus (MRSA), which is often resistant to many commonly used antibiotics in addition to methicillin (3), makes treatment difficult. In 2011, there were 80,000 cases of invasive MRSA infection in the United States, which resulted in more than 11,000 deathsa. The effectiveness of vancomycin, which was once regarded as a drug of last resort to treat MRSA infections, has been marginalized by the emergence of vancomycin-resistant strains (4). Moreover, S. aureus resistance to newer generation drugs such as linezolid and daptomycin has also now been reported (5, 6). This creates an urgent need for new therapeutic agents to treat MRSA infections, preferably ones that do not lead to rapid emergence of drug-resistant strains.
One potential attractive approach to treat infections caused by S. aureus and other pathogens is to use small molecules that effectively strip the bacteria of their surface proteins, which frequently function as virulence factors (7). S. aureus and many other Gram-positive pathogens use sortase enzymes to anchor surface proteins to their cell walls (8–10). In S. aureus, 21 distinct surface proteins are anchored to the cell wall by the extracellular sortase A (SrtA) enzyme (11). This cysteine transpeptidase catalyzes the formation of a peptide bond between a cell wall sorting signal located at the C-terminal end of the precursor surface protein and the cell wall precursor molecule lipid-II (9). The lipid-II linked protein product is then incorporated into the peptidoglycan by the transglycosylation and transpeptidation reactions that synthesize the cell wall (9). Many surface proteins attached to the cell wall by SrtA are virulence factors that play key roles in the infection process by promoting nutrient acquisition from the host, bacterial adhesion and immune evasion (11). Disrupting the display of these proteins by blocking the activity of SrtA using a small molecule could therefore effectively reduce bacterial virulence and thus promote bacterial clearance by the host. Indeed, numerous animal model studies of S. aureus infection have shown that srtA− strains of S. aureus are significantly attenuated in their virulence, underscoring the therapeutic potential of a small molecule SrtA inhibitor (12–16). Attractively, SrtA inhibitors may also be less likely to induce selective pressures that lead to drug resistance, as srtA− strains do not exhibit impaired growth outside of their human host in culture medium (17).
A number of different strategies have been employed to search for sortase inhibitors (7, 18). These include screening natural products (19–31) and small compound libraries (32–35), as well as synthesizing rationally designed peptidomimetics and small molecules (36–41). Structures of SrtA in its apo- and substrate-bound forms (42–44) have now been determined enabling pharmacophore and three dimensional quantitative structure-activity relationships to be established for a select number of inhibitors (45, 46). Currently this structural information has been employed in one virtual screen for sortase inhibitors, which made use of the crystal structure of SrtA determined in its unbound state (47). However, virtual docking efforts were hindered because the structure used in this study exhibited significant conformational heterogeneity and mobility, presumably because the protein was not co-crystallized with its sorting signal substrate. In subsequent work, our group determined the three-dimensional structure of SrtA bound to its sorting signal substrate. This new structure may be better suited for virtual screening approaches as its active site becomes conformationally ordered, and undergoes substantial changes in its structure, upon binding the substrate (43, 48–50). We therefore used it as a starting point for virtual screening effort in which the relaxed complex scheme (RCS) method was used to account for receptor and ligand flexibility during docking. Experimental testing of compounds identified in this analysis revealed that (2-(2,3-dihydro-1H-perimidin-2-yl)-phenoxy)-acetic acid inhibits SrtA with an IC50 value of 47 ± 5.9 µM. Molecular dynamics simulations and a preliminary structure activity relationship study of this lead compound provide insight into its binding mechanism, and strategies to improve its activity.
Methods and Materials
Initial screen against the NMR structure
A 70% cluster of the clean lead-like library was obtained from the ZINC databaseb and consists of 33,161 small molecules (51). The LigPrep programc in Schrödinger Suite 2011 was used to prepare the ligands. Protonation states were assigned at pH 7.0 ± 2.0 with Epikd (52, 53). A total of 55,789 ligands were generated that had distinct structures, stereochemistries, and charge and tautomerisation states. All 55,789 ligands were docked into the lowest energy NMR structure of SrtA bound to a substrate analog (holo-SrtA, PDB ID: 2KID). The receptor was processed using the default Protein Preparation Wizarde, which employs a restrained, partial energy minimization. Grids were generated by Glidef (54–56) with the grid box set around the substrate analog using default settings. The substrate analog was excluded in grid calculations. Docking was done with Glide using SP settings.
Molecular dynamics (MD) simulations and clustering
The MD simulations used in the current study have been described previously (48). Briefly, six 100-ns conventional MD simulations were performed on holo-SrtA using the AMBER99SB-ILDN force field with the simulation package NAMD (57, 58). In three of these simulations the sorting signal remained in the active site, whereas in the other three the sorting signal adopted metastable states outside of the active site. The three simulations in which the sorting signal remained in the active site were chosen for clustering, as conformations from these simulations are likely to be more representative of the bound state than when the sorting signal was not bound near the catalytic triad. From the last 80 ns of each of these MD simulations, 1,600 frames at regularly spaced intervals were extracted, which yielded a total of 4,800 frames. These frames were aligned by the protein Cα atoms in the active site (residues 90–112, 120–130, 161–176 and 183–196) and clustered by root mean square deviation (RMSD) conformational clustering using the gromos algorithm as implemented in GROMACS 4.5 (59). Twenty-one clusters were obtained with an RMSD cutoff of 1.35 Å. The centroid member of each cluster was presumed to best represent members of the ensemble and was selected for subsequent docking studies.
Relaxed complex screen
Five hundred top scoring ligands from the initial screen using the NMR structure of holo-SrtA were docked into each of the 21 representative centroid structures. Procedures used for receptor preparation, grid generation and docking are the same as those described for the initial screen using the NMR structure. The compounds were ranked according to three ensemble-based criteria. First, the compounds were ranked by computing the average of the scores obtained from docking to the 21 centroid conformers (ensemble-average). Second, the compounds were ranked by the population-weighted ensemble-average scores, which were calculated according to Eq. 1:
| (1) |
, where Ē is the weighted ensemble-average score, wi is the size of cluster i, and Ei is the docking score of the compound docked into the centroid of cluster i. Third, the compounds were ranked by the best score they obtained from any of the docking calculations to the 21 centroid conformers (ensemble-best).
Compounds and reagents
Select lead compounds identified from the docking calculations were purchased from ChemBridge Corp. (San Diego, CA, USA), Enamine Ltd. (Ukraine), Sigma-Aldrich Co. LLC (St. Louis, MO, USA) and Vitas-M Laboratory Ltd. (the Netherlands), or synthesized in house. The fluorogenic substrate used in the enzyme assays (Abz-LPETG-Dap(Dnp)-NH2) was purchased from Pepnome Ltd. (China). All other reagents that were used were purchased from Sigma-Aldrich Co. LLC (St. Louis, MO, USA) or Thermo Fisher Scientific Inc. (Waltham, MA, USA), unless noted otherwise.
Enzymatic assays
Compounds were tested for SrtA enzymatic inhibition using an established Förster resonance energy transfer (FRET) assay. Work made use of SrtAΔN59, which consists of residue 60–206. The purification and FRET assay protocols have been described previously (32, 42). Briefly, 20 µL of SrtA (final assay concentration of 1 µM in FRET buffer: 20 mM HEPES, 5 mM CaCl2, 0.05% v/v Tween-20, pH 7.5) was incubated with 1 µL of test compound solution (dissolved in Me2SO, final assay concentration of 0.08–400 µM) for 1 hour at room temperature. Subsequently, 30 µL of substrate solution in FRET buffer (37.5 µM final assay concentration) was added to the mixture and the fluorescence was monitored using excitation and emission wavelengths of 335 and 420 nm, respectively. IC50 values were calculated by fitting three independent sets of data to Eq. 2 using SigmaPlot 6.0g:
| (2) |
, where νi and ν0 are initial velocity of the reaction in the presence and absence of inhibitor at concentration , respectively. The term h is Hill’s coefficient.
The activities of fluorescent compounds that could not be reliably assayed by FRET were tested using a high performance liquid chromatography (HPLC) assay as previously described (37). Briefly, 1 µM SrtA was pre-incubated with inhibitors for 30 min at 37°C to account for any time-dependent inactivation. Reactions were performed in a total volume of 100 µL with all reagents dissolved in FRET buffer. The assay was started by adding to the enzyme a mixture containing 1 mM Abz-LPETG-Dap(Dnp)-NH2 and 1 mM NH2-Gly3-OH (Sigma). After one hour, the reaction was quenched by adding 50 µL of 1 M HCl. A 100 µL of the quenched reaction mixture was then injected onto a reverse phase XSELECT™ HSS C18 5 µM 3.0 × 50 mm HPLC column (Waters Corp., Milford, MA) and its components separated using a linear gradient from 3–45% acetonitrile/0.1% trifluoroacetic acid applied over a period of 25 min. For each inhibitor the fractional activity remaining relative to uninhibited controls was calculated by measuring the difference in percent product formation (Abz-LPETGGG-OH) measured at 215 nm. IC50 values were calculated as described previously and are the average of three measurements.
Induced Fit Docking and Molecular Dynamics Simulations
The compound determined experimentally to have the lowest IC50 value (hereafter called “Compound 1”) was computationally redocked to the NMR structure using the Schrödinger Induced Fit protocolh (60, 61). This protocol accounts for receptor flexibility using a three-step method that includes an initial docking calculation with Glide, refinement of residues within 5 Å of the small molecule’s docked pose using Primei (62, 63), and a redocking stage that uses Glide. In this protocol, Glide was used as previously described, and the default parameters were used for Prime.
Molecular dynamics simulations of compound 1 were performed using a combination of the AMBER99SB-ILDN force field for the protein, and GAFF for the small molecule (64, 65). Partial charges for GAFF were determined from a RESP fit to quantum calculations at the HF/6-31G* level of theory. The complex was solvated in a triclinic box of TIP3P water molecules with sufficient sodium and calcium ions to create a neutral simulation box of approximately 150 mM NaCl. Following relaxation with the default protocol in Maestroj, a 50 ns MD simulation was performed with Desmondk (66). Hydrogen bond analysis was performed with the Hydrogen Bonds plugin in VMD using a donor-acceptor distance of 3.5 Å and an angle cutoff of 30° (67).
Results and Discussion
Virtual screening approaches are increasingly being used to identify lead molecules in drug discovery efforts (68). Typically, these campaigns make use of a single experimentally determined protein structure which is used by computational docking algorithms to predict the relative binding affinities and poses of a large number of small molecules (47). However, in solution, proteins are thought to adopt an ensemble of interchanging conformers (metastates), with the experimentally determined structure presumably representing an average of the low energy conformers sampled experimentally (69, 70). In principle, small molecule binding to any one of the conformers in the ensemble might stabilize it and thereby shift the population equilibrium towards this conformation (71). Therefore, using only one, or a few, static experimentally determined protein structures in virtual screening may fail to discover high affinity binding small molecules that could be developed further into drugs. To account for protein flexibility in virtual screening, a number of techniques have been developed that in many instances allow for protein-side chain movement during the docking process (72). However, to account for full protein motion, docking to multiple structures obtained from x-ray crystallography, NMR, or molecular dynamics (MD) simulations is necessary (72, 73). In this study we make use of the relaxed complex scheme (RCS), a virtual screening approach that combines the dynamic structural information afforded by MD simulations with docking algorithms. This method uses receptor snapshots generated from MD simulations to search for small molecule binders via docking, therefore explicitly accounting for the flexibility of both the receptor and the ligands (74, 75). A number of high affinity binders have been discovered through the RCS (76–78), including the FDA approved drug raltegravir which targets HIV-1 integrase (79–81).
NMR and MD studies have revealed that S. aureus SrtA is a highly dynamic protein suggesting that virtual screening approaches would benefit from the application of the RCS (43, 48, 49, 82). In particular, two of its active site loops, the β6/β7 and β7/β8 loops, undergo major conformational changes when SrtA binds to its sorting signal substrate. The largest changes occur in the β6/β7 loop, which is unstructured and flexible in the apo-state, and transitions into a structured loop containing a 310 helix when bound to the substrate analog (Figure S1a). Because structures generated from MD simulations are particularly well suited for improving the predictive power of docking results to flexible proteins (83), we therefore used the RCS method to conduct a virtual screen of compound libraries to identify inhibitors of the SrtA enzyme.
Virtual screening using the RCS
The procedures used for virtual screening are summarized in Figure 1. A total of 33,161 compounds were downloaded from the ZINC database. 55,789 ligands were then obtained after accounting for their different charge states, stereoisomers and tautomerization states. We performed the screen in two stages, since it was computationally intractable to dock all 55,789 ligands to the NMR structure, as well as to numerous structures generated from MD calculations. In the first stage, all 55,789 ligands were docked to the substrate bound form of the enzyme determined by NMR (hereafter, called holo-SrtA). This structure was chosen for docking because it presumably represents the enzymatically active form of the protein, and the atomic positions of the active site residues are well defined unlike structures of the enzyme determined in its apo-state. Moreover, previous studies have shown that the ligand bound structures of proteins which are generally less flexible, are more amenable to predictive docking experiments as compared to unliganded protein structures (84). Small molecules were docked to holo-SrtA after the in silico removal of the bound sorting signal. The top 500 compounds based on their docking scores were then chosen for the second round of screening using the RCS approach.
Figure 1.
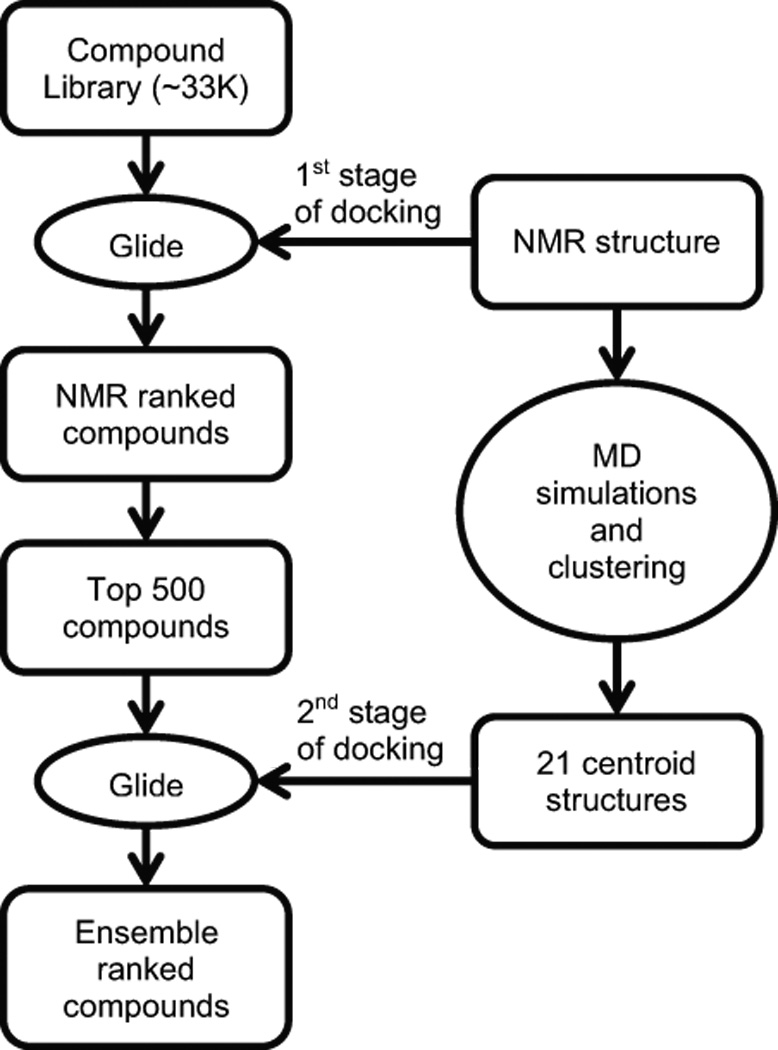
Overview of the two-staged virtual screening procedure that used the Relaxed Complex Scheme. In the first stage, small molecules from the ZINC compound library are docked using the program Glide to the NMR structure of substrate bound form of SrtA (PDB ID: 2KID). In the second stage, six 100 ns MD simulations of the NMR structure are performed and their snapshots are clustered by an RMSD-based algorithm, generating 21 clusters. The top 500 compounds obtained from the first screen are then docked using Glide to 21 centroid structures that represent each of the 21 clusters. Finally, the compounds are ranked by three different methods and the top 15 compounds in each ranking category are selected for experimental testing.
To prepare for the second round of screening that made use of the RCS, six 100 ns MD simulations of the NMR derived structure of the holo-SrtA were performed (48). A total of 4,800 snapshots from these calculations were clustered into 21 groups of related conformers using an RMSD-based clustering algorithm. The centroid member for each cluster was considered to be the best representative of each group and was used in subsequent analyses. As expected, an overlay of the 21 centroid structures reveals that most of the structural differences between the centroids occur in the β6/β7 loop (Figure S1b). In the second round of screening, each of the 21 centroid structures was docked to the top 500 ligands derived from the first screen. To evaluate the docking results, three approaches were used. First, the compounds were ranked by computing the average of the scores obtained from docking to the 21 centroid conformers (ensemble-average). Second, the compounds were ranked by their modified ensemble-average scores such that the number of conformers each centroid structure represents was taken into account (population-weighted ensemble-average). Third, the compounds were ranked by the best score they obtained from any of the docking calculations to the 21 centroid conformers (ensemble-best). For further analysis, the top 15 ligands in each ranking category were considered for experimental testing, which after accounting for redundancy corresponded to 24 unique compounds.
Experimental screening of SrtA inhibition
A FRET-based assay was used to experimentally evaluate the inhibitory activity of lead compounds identified in the virtual screen. Of the 24 unique molecules, a total of 16 compounds were tested experimentally that were either purchased (14 total) or synthesized in house (2 total). The remaining eight compounds were not characterized experimentally as they were deemed too expensive to purchase, as well as too difficult to synthesize in house. However, of these eight compounds, a total of three closely related analogs were purchased and tested. Thus, of the initial 24 lead molecules identified in the virtual screen, a total of 19 lead molecules or closely related compounds were tested experimentally for their ability to inhibit SrtA. The FRET assay was used to evaluate 17 of the 19 compounds, while the remaining 2 molecules were fluorescent and needed to be tested with an HPLC assay.
Eight out of nineteen compounds tested had an IC50 between 47 and 368 µM (see Table 1). The most active compound identified from this screening is compound 1, which had an IC50 value of 47.2 ± 5.9 µM. It is interesting to note that most of the experimentally determined inhibitory compounds that were deemed the best molecules using the RCS approach did not rank highly in the first stage of the virtual screen when they were docked only to the NMR structure. For example, compound 1, which has the lowest IC50, ranked 77th when docked to the NMR structure, but it ranked 9th when docked to the ensemble using the ensemble-best ranking method. This result illustrates the utility of the RCS method, since given limited resources, without application of the RCS method the initial low ranking of compound using conventional approaches may have resulted in it not being tested experimentally. It is also interesting to note that each of the different ranking methods (ensemble-average, population-weighted ensemble-average or ensemble-best) produced a comparable number of experimentally verified hits, and that most of these verified potent molecules were detected by only one of the three ranking methods. This highlights the usefulness of using different methods to rank ligands docked to an ensemble of structures.
Table 1.
Compounds identified from the virtual screen that inhibit SrtA
| Compound | ZINC ID | NMR Rank |
Ensm- Avg Rank |
Weighted Ensm- Avg Rank |
Ensm- Best Rank |
IC50 (µM) |
|---|---|---|---|---|---|---|
| 1 | 406572 | 77 | 27 | 24 | 9 | 47.2 ± 5.9 |
| 2 | 33733644a | 145 | 8 | 4 | 13 | 98.9 ± 7.7 |
| 3 | 46093796 | 158 | 18 | 7 | 8 | 114 ± 13 |
| 4 | 41495051 | 468 | 4 | 10 | 19 | 132 ± 21 |
| 5 | 28294435b | 124 | 7 | 8 | 47 | 189 ± 31 |
| 6 | 6538309 | 440 | 16 | 26 | 11 | 256 ± 21 |
| 7 | 6598689 | 148 | 19 | 12 | 77 | 276 ± 20 |
| 8 | 13610765 | 161 | 11 | 35 | 26 | 368 ± 29 |
Ensm-Avg refers to Ensemble-Average; Weighted Ensm-Avg refers to Population-weighted Ensemble-Average; Ensm-Best refers to Ensemble-Best;
ZINC33733644 identified in the virtual screen was not available for purchase and was deemed too technically difficult to synthesize in house. Therefore, its close analog ChemBridge 7253325 was tested instead;
ZINC28294435 identified in the virtual screen was not available for purchase and was deemed too technically difficult to synthesize in house. Therefore, its close analog ChemBridge 5303268 was tested instead.
Our virtual screen using the RCS yielded a higher hit rate than previously reported virtual screen that made use of more traditional methods. Previously, a virtual screen for sortase inhibitors was reported that made use of the structure of apo-SrtA, the only structure that was available at that time. A total of ~150,000 compounds were virtually screened for binding (47). After experimental testing of the leads identified from the screen, 7.4% were inhibitory; a total of 8 out of 108 experimentally tested compounds in this study had IC50 values ranging between 75 to 400 µM (47). In contrast, 42.2% of the lead molecules we tested that were identified in our virtual screen were active; a total of 8 out of 19 molecules experimentally tested had IC50 values ranging between 47 to 368 µM. Because the virtual screens were performed by different research groups using different docking algorithms and virtual compound libraries, it is not possible to rigorously explain why we obtained a higher hit rate. However, there seems to be two likely reasons for this difference. First, we used the structure of holo-SrtA as the receptor in the initial docking calculations, which may yield better results than docking to apo-SrtA as its active site is more rigid and well-defined. Second, our analysis made use of the RCS, which accounts for protein motion by docking ligands to an ensemble of structures obtained from MD simulations.
Compound 1: Structure and dynamics of its predicted binding mode
Compound 1 was chosen for additional characterization as it has the lowest IC50 value and a number of derivatives of this molecule could readily be purchased. To further investigate the binding pose of compound 1, it was redocked into the NMR structure using the “Induced Fit” workflow in Maestro, which combines both docking and protein rearrangement stages (see Methods for more details) (60, 61). The structure of compound 1 is based on a 2-phenyl-2,3-dihydro-1H-perimidine scaffold. It contains a dihydroperimidine (DHP) group and a phenyl ring with an oxyacetic acid group attached at the ortho position. In the docking pose the molecule is positioned in the active site with the DHP group placed underneath the β6/β7 loop, and the phenyl ring projected towards the active site H120, C184, and R197 (Figure 2a). Specificity for this orientation is achieved by interactions that originate from the carboxyl group of the small molecule, which simultaneously forms hydrogen bonds to the catalytically important residues R197 and H120 within the active site (Figure 2a). A predicted hydrogen bond between the backbone of P163 and the amine of the DHP group in the small molecule also presumably stabilizes ligand binding (Figure 2a). In the binding pose the naphthalene ring of the DHP group is wedged into a hydrophobic pocket formed by V166, I182, A118 and V161. This positioning orients the phenyl ring towards several potential hydrogen bonding groups within the enzyme’s active site (e.g. the side chains of T183, C184, and the backbone of G119) suggesting that molecules in which this ring are appropriately modified could exhibit improved binding selectivity and affinity.
Figure 2.
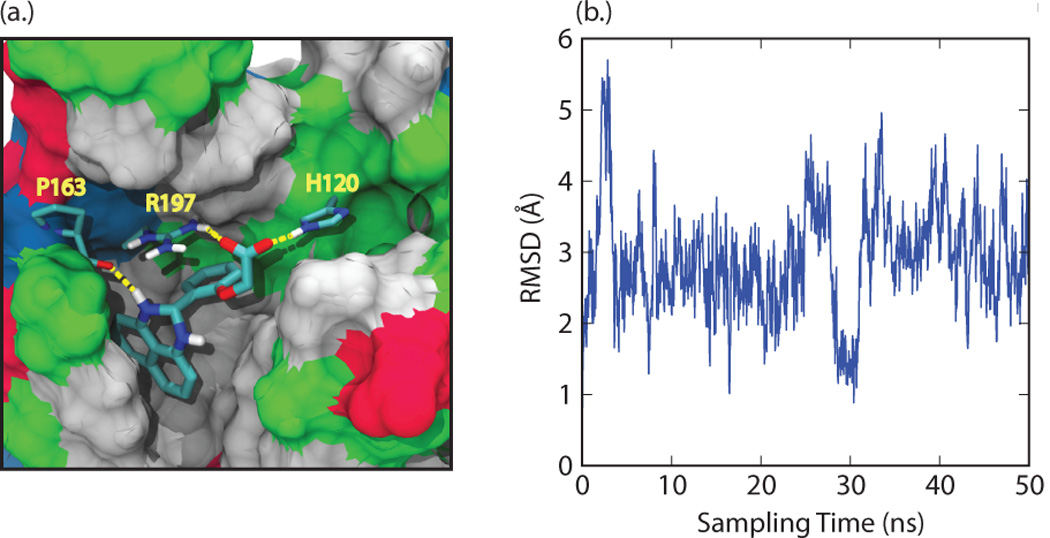
(a) Docking pose for compound 1 generated in "induced fit docking" calculations. Residues H120, P163, and R197 from SrtA are explicitly represented, along with their intermolecular hydrogen bonds to the small molecule (yellow lines). The remainder of SrtA is shown by a surface representation, with non-polar residues in grey, polar residues in green, acidic residues in red, and basic residues in blue. (b) Root-mean squared deviation (RMSD) of the position of the compound relative to its initial binding pose in SrtA at various time points during the 50 ns molecular dynamics simulation.
To gain insight into the dynamics of the bound state, a single, 50 ns MD simulation of SrtA:Compound 1 complex was performed. Over the course of the simulation, the structure of the protein resembled most closely several of the centroid structures, with the RMSD of the active site residues calculated to be as low as 1 Å. By the end of the simulation, the structure of the complex was structurally most similar to several of the centroid structures (active site RMSD ~ 1.5 Å) and less similar to the NMR structure (active site RMSD ~ 2.5 Å). RMSD calculations of the ligand relative to the protein show that the molecule experiences motions that result in atomic displacements on the order of 2–3 Å relative to the initial pose. Interestingly, a major excursion from the binding mode can occur transiently which causes a >5 Å displacement from the initial binding pose, as well as a return to conformation that is very similar to the initial binding pose (less than 1.5 Å from the induced fit docking results) (Figure 2b). This larger excursion is caused by movement of the naphthalene ring within the hydrophobic pocket formed underneath the β6/β7 loop. Presumably, the addition of nonpolar substituents to this ring to fill this pocket could further improve binding affinity. The side chain of R197 maintained hydrogen bond contacts with compound 1 for 53% of the simulation, primarily with atoms in the carboxylic acid group. Other contacts were more transient, with the most dominant interactions to compound 1 being between the NH in the DHP group and the backbone of A104 (11% of the simulation), the backbone of G167 (8% of the simulation), and the backbone of A92 (4% of the simulation). Early in the simulation the side chain of H120 flipped such that the hydrogen bonds between it and compound 1 were broken, and in the course of the simulation they did not reform. Overall, these results indicate that a reasonable strategy in lead development may be to create additional contacts to stabilize compound 1 in the binding site to increase the propensity of these hydrogen bonds.
Preliminary structure activity relationship study of compound 1
To develop compound 1 further, we performed a similarity search on the ChemBridge small molecule database. A total of 22 compounds with the 2-phenyl DHP scaffold were identified. Based on the docking and MD calculations, 10 of these compounds were purchased and their inhibitory activity determined experimentally. These molecules contain polar substituents in the phenyl ring to facilitate hydrogen bonding to the active site, and include a smaller compound that only contains the 2-phenyl DHP scaffold (summarized in Table 2). Compounds containing naphthalene ring substituents may also exhibit improved binding, but were not tested in this study because they are not available for purchase from ChemBridge. The scaffold compound 1–1 did not inhibit SrtA, which is probably the result of missing hydrogen bonds to the active site R197 and H120, which underscores the importance of having polar groups on the phenyl ring. Compounds containing a nitro group (1–5) or chloro group (1–6 and 1–7) at the para position are the most active with IC50 values close to, or less than, 100 µM. The retention of activity after modification of the phenyl ring is presumably because these polar groups form favorable interactions with the side chains of R197 or H120 within the active site. Interestingly, compounds containing a substituent at the meta position of the phenyl ring (1–2, 1–4, 1–8, 1–9 and 1–10) are less active or even inactive. This may be due to steric clashes at this site in the small molecules with residues projecting from the β2/α1 loop, or from the β7 and β8 strands. Lower activity was also observed when the nitro or chloro group at the para position was replaced by a smaller fluoro group (compare 1–3 with 1–5 and 1–6). Unfortunately, none of the compounds inhibited SrtA better than our lead, probably because the substituents in the phenyl ring are not long enough to interact with both the active site R197 and H120. Future work will focus on synthesizing compounds with phenyl rings containing longer polar groups, and will explore different substituents on the naphthalene ring to increase contacts to the hydrophobic pocket.
Table 2.
Preliminary structure activity relationship study of compound 1
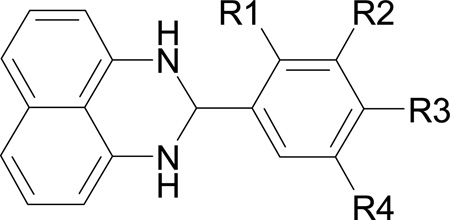 | |||||
|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | R4 | IC50 (µM) |
| 1 (lead) | OCH2COOH | H | H | H | 47.2 ± 5.9 |
| 1–1 | H | H | H | H | >400 |
| 1–2 | H | Cl | H | H | >400 |
| 1–3 | H | H | F | H | 276 ± 29 |
| 1–4 | H | OH | OCH3 | H | >400 |
| 1–5 | H | H | NO2 | H | 80 ± 5 |
| 1–6 | H | H | Cl | H | 89 ± 12 |
| 1–7 | Cl | H | Cl | H | 111 ± 9 |
| 1–8 | H | OCH3 | OH | H | 231 ± 60 |
| 1–9 | H | OCH2CH3 | OH | H | >400 |
| 1–10 | H | OCH3 | OH | Br | 136 ± 20 |
Conclusions and Future Directions
A virtual screen identified molecules containing the 2-phenyl-2,3-dihydro-1H-perimidine scaffold as possible inhibitors of the S. aureus SrtA enzyme. A structure activity relationship analysis indicates that the best molecule in this class, (2-(2,3-dihydro-1H-perimidin-2-yl)-phenoxy)-acetic acid, inhibits the activity of SrtA with an IC50 value of 47.2 ± 5.9 µM. MD simulations of this molecule bound to SrtA provide insight into its binding mechanism and serve as the foundation for future structure guided studies to uncover analogs that might have increased potency. Our virtual screen made use of the RCS and had a significantly higher success rate in identifying inhibitor compounds of SrtA as compared to conventional methods, highlighting the improved predictive power of the ensemble docking approach (85, 86). The 2-phenyl-2,3-dihydro-1H-perimidine-based lead compound discovered in this study is a promising candidate for further development into a therapeutically useful anti-infective agent that can be used to treat infections caused by MRSA, and other multi-drug resistant bacterial pathogens.
Supplementary Material
Acknowledgments
We thank Dr. Sara E. Nichols for her help with the docking calculations. This research was supported by funding from the National Institutes of Health (AI52217 to R.T.C. and M.E.J.; and GM31749 to J.A.M.). A.H.C. was supported by the UCLA Chemistry Biology Interface Training program (NIH T32GM008496). Additional support at UCSD has been provided by the National Science Foundation, the Howard Hughes Medical Institute, the Center for Theoretical Biological Physics, the National Biomedical Computation Resource, and the NSF Supercomputer Centers.
Abbreviations
- SrtA
sortase A
- MRSA
methicillin-resistant Staphylococcus aureus
- HIV
human immunodeficiency virus
- MD
molecular dynamics
- RCS
relaxed complex scheme
- RMSD
root mean square deviation
- NMR
nuclear magnetic resonance
- FRET
Förster resonance energy transfer
- HPLC
high performance liquid chromatography
- DHP
dihydroperimidine
Footnotes
Conflict of Interest
There are no conflicts of interest.
Centers for Disease Control and Prevention. 2011. Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus aureus, 2011. http://www.cdc.gov/abcs/reports-findings/survreports/mrsa11.html.
ZINC clean lead-like subset, 70% cluster. Downloaded on Oct 26, 2011 from http://zinc.docking.org.
LigPrep, version 2.5, Schrödinger, LLC, New York, NY, 2011.
Epik, version 2.2, Schrödinger, LLC, New York, NY, 2011.
Schrödinger Suite 2011 Protein Preparation Wizard; Epik version 2.2, Schrödinger, LLC, New York, NY, 2011; Impact version 5.7, Schrödinger, LLC, New York, NY, 2011; Prime version 3.0, Schrödinger, LLC, New York, NY, 2011.
Glide, version 5.7, Schrödinger, LLC, New York, NY, 2011.
SigmaPlot 6.0, SPSS, Inc., Chicago, IL, 2000.
Schrödinger Suite 2011 Induced Fit Docking protocol; Glide version 5.7, Schrödinger, LLC, New York, NY, 2011; Prime version 3.0, Schrödinger, LLC, New York, NY, 2011.
Prime, version 3.0, Schrödinger, LLC, New York, NY, 2011.
Maestro, version 9.2, Schrödinger, LLC, New York, NY, 2011.
Desmond Molecular Dynamics System, version 3.0, D. E. Shaw Research, New York, NY, 2011. Maestro-Desmond Interoperability Tools, version 3.0, Schrödinger, New York, NY, 2011.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Ippolito G, Leone S, Lauria FN, Nicastri E, Wenzel RP. Methicillin-resistant Staphylococcus aureus: the superbug. Int J Infect Dis. 2010;14(Suppl 4):S7–S11. doi: 10.1016/j.ijid.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Otto M. MRSA virulence and spread. Cell Microbiol. 2012;14:1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welsh KJ, Abbott AN, Lewis EM, Gardiner JM, Kruzel MC, Lewis CT, et al. Clinical characteristics, outcomes, and microbiologic features associated with methicillin-resistant Staphylococcus aureus bacteremia in pediatric patients treated with vancomycin. J Clin Microbiol. 2010;48:894–899. doi: 10.1128/JCM.01949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marty FM, Yeh WW, Wennersten CB, Venkataraman L, Albano E, Alyea EP, et al. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J Clin Microbiol. 2006;44:595–597. doi: 10.1128/JCM.44.2.595-597.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda-Dantsuji Y, Hanaki H, Sakai F, Tomono K, Takesue Y, Honda J, et al. Linezolid-resistant Staphylococcus aureus isolated from 2006 through 2008 at six hospitals in Japan. J Infect Chemother. 2011;17:45–51. doi: 10.1007/s10156-010-0085-1. [DOI] [PubMed] [Google Scholar]
- 7.Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 8.Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers. 2010;94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paterson GK, Mitchell TJ. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004;12:89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson IM, Mazmanian SK, Schneewind O, Verdrengh M, Bremell T, Tarkowski A. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J Infect Dis. 2002;185:1417–1424. doi: 10.1086/340503. [DOI] [PubMed] [Google Scholar]
- 15.Weiss WJ, Lenoy E, Murphy T, Tardio L, Burgio P, Projan SJ, et al. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal models of infection. J Antimicrob Chemother. 2004;53:480–486. doi: 10.1093/jac/dkh078. [DOI] [PubMed] [Google Scholar]
- 16.Miyazaki S, Matsumoto Y, Sekimizu K, Kaito C. Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol Lett. 2012;326:116–124. doi: 10.1111/j.1574-6968.2011.02439.x. [DOI] [PubMed] [Google Scholar]
- 17.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 18.Suree N, Jung ME, Clubb RT. Recent advances towards new anti-infective agents that inhibit cell surface protein anchoring in Staphylococcus aureus and other gram-positive pathogens. Mini Rev Med Chem. 2007;7:991–1000. doi: 10.2174/138955707782110097. [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, Shin DS, Oh MN, Chung SC, Lee JS, Chang IM, et al. Inhibition of sortase, a bacterial surface protein anchoring transpeptidase, by beta-sitosterol-3-O-glucopyranoside from Fritillaria verticillata. Biosci Biotechnol Biochem. 2003;67:2477–2479. doi: 10.1271/bbb.67.2477. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Shin DS, Oh MN, Chung SC, Lee JS, Oh KB. Inhibition of the bacterial surface protein anchoring transpeptidase sortase by isoquinoline alkaloids. Biosci Biotechnol Biochem. 2004;68:421–424. doi: 10.1271/bbb.68.421. [DOI] [PubMed] [Google Scholar]
- 21.Kim SW, Chang IM, Oh KB. Inhibition of the bacterial surface protein anchoring transpeptidase sortase by medicinal plants. Biosci Biotechnol Biochem. 2002;66:2751–2754. doi: 10.1271/bbb.66.2751. [DOI] [PubMed] [Google Scholar]
- 22.Oh KB, Mar W, Kim S, Kim JY, Oh MN, Kim JG, et al. Bis(indole) alkaloids as sortase A inhibitors from the sponge Spongosorites sp. Bioorg Med Chem Lett. 2005;15:4927–4931. doi: 10.1016/j.bmcl.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Jang KH, Chung SC, Shin J, Lee SH, Kim TI, Lee HS, et al. Aaptamines as sortase A inhibitors from the tropical sponge Aaptos aaptos. Bioorg Med Chem Lett. 2007;17:5366–5369. doi: 10.1016/j.bmcl.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Kang SS, Kim JG, Lee TH, Oh KB. Flavonols inhibit sortases and sortase-mediated Staphylococcus aureus clumping to fibrinogen. Biol Pharm Bull. 2006;29:1751–1755. doi: 10.1248/bpb.29.1751. [DOI] [PubMed] [Google Scholar]
- 25.Park BS, Kim JG, Kim MR, Lee SE, Takeoka GR, Oh KB, et al. Curcuma longa L. constituents inhibit sortase A and Staphylococcus aureus cell adhesion to fibronectin. J Agric Food Chem. 2005;53:9005–9009. doi: 10.1021/jf051765z. [DOI] [PubMed] [Google Scholar]
- 26.Jeon JE, Na Z, Jung M, Lee HS, Sim CJ, Nahm K, et al. Discorhabdins from the Korean marine sponge Sceptrella sp. J Nat Prod. 2010;73:258–262. doi: 10.1021/np9005629. [DOI] [PubMed] [Google Scholar]
- 27.Lee YJ, Han YR, Park W, Nam SH, Oh KB, Lee HS. Synthetic analogs of indole-containing natural products as inhibitors of sortase A and isocitrate lyase. Bioorg Med Chem Lett. 2010;20:6882–6885. doi: 10.1016/j.bmcl.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 28.Oh I, Yang WY, Chung SC, Kim TY, Oh KB, Shin J. In vitro sortase A inhibitory and antimicrobial activity of flavonoids isolated from the roots of Sophora flavescens. Arch Pharm Res. 2011;34:217–222. doi: 10.1007/s12272-011-0206-0. [DOI] [PubMed] [Google Scholar]
- 29.Won TH, Jeon JE, Kim SH, Lee SH, Rho BJ, Oh DC, et al. Brominated Aromatic Furanones and Related Esters from the Ascidian Synoicum sp. J Nat Prod. 2012;75:2055–2061. doi: 10.1021/np3005562. [DOI] [PubMed] [Google Scholar]
- 30.Won TH, Jeon JE, Lee SH, Rho BJ, Oh KB, Shin J. Beta-carboline alkaloids derived from the ascidian Synoicum sp. Bioorg Med Chem. 2012;20:4082–4087. doi: 10.1016/j.bmc.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Bae J, Jeon JE, Lee YJ, Lee HS, Sim CJ, Oh KB, et al. Sesterterpenes from the tropical sponge Coscinoderma sp. J Nat Prod. 2011;74:1805–1811. doi: 10.1021/np200492k. [DOI] [PubMed] [Google Scholar]
- 32.Suree N, Yi SW, Thieu W, Marohn M, Damoiseaux R, Chan A, et al. Discovery and structure-activity relationship analysis of Staphylococcus aureus sortase A inhibitors. Bioorg Med Chem. 2009;17:7174–7185. doi: 10.1016/j.bmc.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maresso AW, Wu R, Kern JW, Zhang R, Janik D, Missiakas DM, et al. Activation of inhibitors by sortase triggers irreversible modification of the active site. J Biol Chem. 2007;282:23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh KB, Nam KW, Ahn H, Shin J, Kim S, Mar W. Therapeutic effect of (Z)-3-(2,5-dimethoxyphenyl)-2-(4-methoxyphenyl) acrylonitrile (DMMA) against Staphylococcus aureus infection in a murine model. Biochem Biophys Res Commun. 2010;396:440–444. doi: 10.1016/j.bbrc.2010.04.113. [DOI] [PubMed] [Google Scholar]
- 35.Oh KB, Kim SH, Lee J, Cho WJ, Lee T, Kim S. Discovery of diarylacrylonitriles as a novel series of small molecule sortase A inhibitors. J Med Chem. 2004;47:2418–2421. doi: 10.1021/jm0498708. [DOI] [PubMed] [Google Scholar]
- 36.Kruger RG, Barkallah S, Frankel BA, McCafferty DG. Inhibition of the Staphylococcus aureus sortase transpeptidase SrtA by phosphinic peptidomimetics. Bioorg Med Chem. 2004;12:3723–3729. doi: 10.1016/j.bmc.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 37.Kudryavtsev KV, Bentley ML, McCafferty DG. Probing of the cis-5-phenyl proline scaffold as a platform for the synthesis of mechanism-based inhibitors of the Staphylococcus aureus sortase SrtA isoform. Bioorg Med Chem. 2009;17:2886–2893. doi: 10.1016/j.bmc.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung ME, Clemens JJ, Suree N, Liew CK, Pilpa R, Campbell DO, et al. Synthesis of (2R,3S) 3-amino-4-mercapto-2-butanol, a threonine analogue for covalent inhibition of sortases. Bioorg Med Chem Lett. 2005;15:5076–5079. doi: 10.1016/j.bmcl.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 39.Liew CK, Smith BT, Pilpa R, Suree N, Ilangovan U, Connolly KM, et al. Localization and mutagenesis of the sorting signal binding site on sortase A from Staphylococcus aureus. FEBS Lett. 2004;571:221–226. doi: 10.1016/j.febslet.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 40.Connolly KM, Smith BT, Pilpa R, Ilangovan U, Jung ME, Clubb RT. Sortase from Staphylococcus aureus does not contain a thiolate-imidazolium ion pair in its active site. J Biol Chem. 2003;278:34061–34065. doi: 10.1074/jbc.M305245200. [DOI] [PubMed] [Google Scholar]
- 41.Scott CJ, McDowell A, Martin SL, Lynas JF, Vandenbroeck K, Walker B. Irreversible inhibition of the bacterial cysteine protease-transpeptidase sortase (SrtA) by substrate-derived affinity labels. Biochem J. 2002;366:953–958. doi: 10.1042/BJ20020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suree N, Liew CK, Villareal VA, Thieu W, Fadeev EA, Clemens JJ, et al. The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J Biol Chem. 2009;284:24465–24477. doi: 10.1074/jbc.M109.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 45.Uddin R, Lodhi MU, Ul-Haq Z. Combined pharmacophore and 3D-QSAR study on a series of Staphylococcus aureus Sortase A inhibitors. Chem Biol Drug Des. 2012;80:300–314. doi: 10.1111/j.1747-0285.2012.01403.x. [DOI] [PubMed] [Google Scholar]
- 46.Mehta H, Khokra SL, Arora K, Kaushik P. Pharmacophore mapping and 3D-QSAR analysis of Staphylcoccus aureus Sortase a inhibitors. Der Pharma Chemica. 2012;4:1776–1784. [Google Scholar]
- 47.Chenna BC, Shinkre BA, King JR, Lucius AL, Narayana SV, Velu SE. Identification of novel inhibitors of bacterial surface enzyme Staphylococcus aureus Sortase A. Bioorg Med Chem Lett. 2008;18:380–385. doi: 10.1016/j.bmcl.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 48.Kappel K, Wereszczynski J, Clubb RT, McCammon JA. The binding mechanism, multiple binding modes, and allosteric regulation of Staphylococcus aureus Sortase A probed by molecular dynamics simulations. Protein Sci. 2012;21:1858–1871. doi: 10.1002/pro.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moritsugu K, Terada T, Kidera A. Disorder-to-order transition of an intrinsically disordered region of sortase revealed by multiscale enhanced sampling. J Am Chem Soc. 2012;134:7094–7101. doi: 10.1021/ja3008402. [DOI] [PubMed] [Google Scholar]
- 50.Naik MT, Suree N, Ilangovan U, Liew CK, Thieu W, Campbell DO, et al. Staphylococcus aureus Sortase A transpeptidase. Calcium promotes sorting signal binding by altering the mobility and structure of an active site loop. J Biol Chem. 2006;281:1817–1826. doi: 10.1074/jbc.M506123200. [DOI] [PubMed] [Google Scholar]
- 51.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: A Free Tool to Discover Chemistry for Biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 53.Greenwood JR, Calkins D, Sullivan AP, Shelley JC. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 54.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 55.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 56.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 57.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman W, Beard HS, Farid R. Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des. 2006;67:83–84. doi: 10.1111/j.1747-0285.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 61.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 62.Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 63.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, et al. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 66.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters; Proceedings of the ACM/IEEE Conference on Supercomputing (SC06); November 11–17; Tampa, Florida. 2006. [Google Scholar]
- 67.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 68.Tanrikulu Y, Kruger B, Proschak E. The holistic integration of virtual screening in drug discovery. Drug Discov Today. 2013;18:358–364. doi: 10.1016/j.drudis.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 69.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 70.Hardin C, Eastwood MP, Prentiss M, Luthey-Schulten Z, Wolynes PG. Folding funnels: the key to robust protein structure prediction. J Comput Chem. 2002;23:138–146. doi: 10.1002/jcc.1162. [DOI] [PubMed] [Google Scholar]
- 71.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durrant JD, McCammon JA. Computer-aided drug-discovery techniques that account for receptor flexibility. Curr Opin Pharmacol. 2010;10:770–774. doi: 10.1016/j.coph.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amaro RE, Li WW. Emerging methods for ensemble-based virtual screening. Curr Top Med Chem. 2010;10:3–13. doi: 10.2174/156802610790232279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin JH, Perryman AL, Schames JR, McCammon JA. The relaxed complex method: Accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers. 2003;68:47–62. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 75.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput Aided Mol Des. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Durrant JD, Hall L, Swift RV, Landon M, Schnaufer A, Amaro RE. Novel naphthalene-based inhibitors of Trypanosoma brucei RNA editing ligase 1. PLoS Negl Trop Dis. 2010;4:e803. doi: 10.1371/journal.pntd.0000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durrant JD, Urbaniak MD, Ferguson MA, McCammon JA. Computer-aided identification of Trypanosoma brucei uridine diphosphate galactose 4'-epimerase inhibitors: toward the development of novel therapies for African sleeping sickness. J Med Chem. 2010;53:5025–5032. doi: 10.1021/jm100456a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, et al. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc Natl Acad Sci U S A. 2004;101:11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schames JR, Henchman RH, Siegel JS, Sotriffer CA, Ni H, McCammon JA. Discovery of a novel binding trench in HIV integrase. J Med Chem. 2004;47:1879–1881. doi: 10.1021/jm0341913. [DOI] [PubMed] [Google Scholar]
- 81.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 82.Tian BX, Eriksson LA. Catalytic mechanism and roles of Arg197 and Thr183 in the Staphylococcus aureus sortase A enzyme. J Phys Chem B. 2011;115:13003–13011. doi: 10.1021/jp2058113. [DOI] [PubMed] [Google Scholar]
- 83.Nichols SE, Baron R, Ivetac A, McCammon JA. Predictive power of molecular dynamics receptor structures in virtual screening. J Chem Inf Model. 2011;51:1439–1446. doi: 10.1021/ci200117n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, modeled conformations of enzymes. J Med Chem. 2003;46:2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 85.Huang SY, Zou X. Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 86.Rao S, Sanschagrin PC, Greenwood JR, Repasky MP, Sherman W, Farid R. Improving database enrichment through ensemble docking. J Comput Aided Mol Des. 2008;22:621–627. doi: 10.1007/s10822-008-9182-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


