Abstract
The nuclear factor-κB (NF-κB) signaling pathway plays a critical role in a multitude of cellular processes. Activation of the NF-κB transcription factor family is essential for the initiation of inflammation, immunity, cell proliferation and apoptosis through a list of responsive genes. In hepatic tissue, activation of the NF-κB pathway has been implicated in a number of pathological conditions. Here we described a mouse model for noninvasive quantification of NF-κB activation in the hepatic tissues. Mice were subjected to hydrodynamic delivery with a mixture of pattB-NF-κB-Fluc reporter and φC31o integrase vector. Hepatic expression of φC31o integrase mediated chromosomal integration of the pattB-NF-κB-Fluc reporter, resulting in stable luciferase expression at 300 days post transfection. We applied noninvasive imaging and were able to detect NF-κB activation under acute liver injury and hepatitis conditions. During hepatectomy-induced liver regeneration, NF-κB activation was detected locally in the tissues at the surgery site. Treatment with Sorafenib suppressed NF-κB activation, accompanied with perturbation of liver regeneration. In conclusion, we established a method for stable transfection of the hepatic tissues and applied the transfected mice to longitudinal monitoring of NF-κB activity under pathological conditions. Further exploration of this methodology for establishment of other disease models and for evaluation of novel pharmaceuticals is likely to be fruitful.
Keywords: Nuclear factor-κB, noninvasive molecular imaging, φC31o integrase, Sorafenib, animal model.
Introduction
Nuclear factor-κB was first described in 1986 as a nuclear transcription factor required for immunoglobulin kappa light chain transcription in B cells 1. The NF-κB transcription factor consists of subunits that associate in dimmers 2. The commonly described dimer forms are p50 (NF-κB1) with RelA (p65) and p52 (NF-κB2) with RelB. The NF-κB1/RelA complex remains in the cytoplasm in an inactive form in association with I-κB. Upon stimulation with LPS or proinflammatory cytokines, IKK is activated and causes I-κB phosphorylation and degradation 2. This releases the NF-κB/RelA complex from I-κB inhibition and allows its translocation to the nucleus where it transcriptionally activates key genes involved in inflammation and chemotaxis 3-5.
NF-κB activation has been shown as a pivotal process in the regulation of viral replication, tumorigenesis, apoptosis and autoimmune diseases 6, 7. Under pathological hepatic conditions, such as hepatitis, fibrosis, hepatocarcinoma, ischemia-reperfusion injury and liver regeneration, involvement of the NF-κB signaling pathway has been reported 8-10. However, there has been little study on the impact of therapeutic intervention of the NF-κB pathway on hepatic disease initiation and progression. This is largely due to the lack of a modeling system for assessing NF-κB activity under in vivo conditions 11, 12.
Recently, bioluminescent imaging has been applied to noninvasive assessment of cell signaling pathways under in vivo conditions 13. Carlsen and colleagues reported a NF-κB-luciferase transgenic mouse model for monitoring the NF-κB signaling pathway 14. However, detection of NF-κB activation with this model is subjected to perturbance by luciferase expression in extra-hepatic tissues. Hyoudou and colleagues reported a mouse model for monitoring NF-κB activation specifically in the liver through i.v. delivery of a NF-κB reporter to the hepatic tissues 15. However, transient NF-κB reporter expression precludes further application for monitoring NF-κB activation under chronic pathological conditions. In this report, we generated a pattB-NF-κB-Fluc reporter plasmid that contains an attB integration site and the firefly luciferase gene under the transcriptional regulation of the NF-κB consensus sequences fused to a TATA-like promoter. Using a hydrodynamic transfection method 16, 17, we transfected the liver tissues with the pattB-NF-κB-Fluc reporter and a φC31o integrase 18, 19 vector. Expression of φC31o integrase was expected to mediate recombination between the attB site of the pattB-NF-κB-Fluc reporter and attP site of the chromosome, allowing stable chromosomal integration of the pattB-NF-κB-Fluc reporter cassette in the transfected cells. Using this novel approach, we established stable transfection of the hepatic tissues with the pattB-NF-κB-Fluc reporter and were able to monitor NF-κB activation longitudinally. We further investigated the kinetics of NF-κB activation following partial hepatectomy (PHx) and revealed that suppression of the NF-κB activation with Sorafenib was associated with impairment of liver regeneration.
Materials and Methods
Reagents
Bacterial lipopolysaccharide (LPS), Concanavalin A (ConA) and Pyrrolidinedithiocarbamate (PDTC) were obtained from Sigma-Aldrich Chemical Co (St. Louis, MO). D-Luciferin was purchased from Promega (Madison, Wisconsin). Sorafenib tosylate was obtained from Bayer HealthCare (Pharmaceuticals, Montville, NJ). The dosages, route of administration and dosage volume are listed in Table 1.
Table 1.
The dosages used, route of administration and dosages volume of the reagents used in this report.

Mice
BALB/c mice (male, 6-8 weeks) were purchased from the National Beijing Center for Drug Safety Evaluation and Research (NBCDSER). Mice were housed under specific pathogen-free (SPF) conditions. All animals received human care and in vivo experiments were approved by the ethics committee of the NBCDSER (Permit No.11-1166-3).
Plasmids
M. P. Calos (Department of Genetics, Stanford University, USA) kindly provided the pTA-attB plasmid 20. Plasmid pattB-NF-κB-Fluc was generated by cloning a 297-bp attB containing fragment of pTA-attB into the Kpn I site of pNF-κB-Fluc (Takara, Japan). A mouse codon-optimized φC31 (φC31o) was obtained from Addgene, Cambridge, USA (PphiC31o) (Supplementary Material: Fig. S1). Plasmid DNA was purified using an Endotoxin Free Maxi Kit (Qiagen, Hilden, Germany).
Hydrodynamics-based liver transfection
Hepatic tissues were transfected following a hydrodynamics-based DNA delivery technique 16, 21. Briefly, the plasmid DNA was dissolved in saline and rapidly injected intravenously within 5 seconds to mice in a volume equivalent to 10% of the mouse body weight (i.e. 2 ml for a 20 g mouse).
Partial hepatectomy
Two-thirds PHx was performed according to a method of Higgins and Andersen 22.
Bioluminescence imaging
Bioluminescence imaging was performed using an IVIS imaging system (Xenogen, Alameda, CA). Mice were i.p. injected with 150 mg/kg of D-luciferin. After 10 minutes, mice were anesthetized with 1-3% isoflurane and imaged for luciferase expression. The regions of interest from displayed images were quantified as photons/s/cm2/sr using the Living Image software 4.2 (Xenogen, Alameda, CA).
Histological and immunohistochemical evaluations
The livers were fixed in 10% neutrally buffered formalin for 24 hours and then embedded in paraffin. The sections (5 μm) were affixed to slides, deparaffinized and stained with hematoxylin-eosin (HE). Immunohistochemical staining with anti-PCNA antibody (1:100 dilution; Epitomics, California) was performed to determine the morphological change and proliferation property of the hepatic tissues.
Analysis of genomic integration by nested PCR
Mice were sacrificed at 30 days after transfection and the liver tissues were excised. Genomic DNA of the liver tissues was isolated using the Wizard Genomic DNA Purification Kit (Promega, Madison, Wisconsin) according to the manufacturer's instructions. Nested PCR was performed to detect site-specific integration at mpsL1 (mice pseudo-site from liver). The first round of PCR with primers mspL1 (reverse) and attB-1 was performed with the isolated genomic DNA. Under similar conditions, the products were used as templates for the second round of PCR with primers mspL1 (reverse) and attB-2. The second round of PCR products were cloned into pGEM-T (Promega, Madison, Wisconsin) and sequenced. The following primers were used: mspL1 (reverse): 5'-TGAGGAGGAGCCTTAGCAAC-3', attB-1: 5'-GTAGGTCACGGTCTCGAAGC-3', and attB-2: 5'-CGAAGCCGCGGTGCGGGTGCCA-3'.
Western blot analysis
Hepatic cell extract containing 50 micrograms of total protein was subjected to electrophoresis with a 10% polyacrylamide gel (SDS-PAGE). Proteins were transferred to polyvinylidenedifluoride membranes (Millipore, Billerica, MA). Luciferase expression was evaluated with an anti-luciferase mAb (1:1000 dilution; Promega, Madison, Wisconsin). An anti-GAPDH mAb (1:1000 dilution; Santa Cruz, CA) was used as an internal control. After incubation with the primary antibodies, detection was made with horseradish peroxidase-conjugated secondary rabbit antibodies against either goat or mouse IgG (1:5000 dilution; Beijing CoWin Biotech, China). The western blot analysis was performed using the standard techniques of the Immobilon Western (Millipore, Billerica, MA).
Electrophoretic mobility shift assay (EMSA)
Hepatic nuclear extracts were applied to DNA binding assays using a Nuclear and Cytoplasmic Extraction Kit (Beijing CoWin Biotech, China) according to the manufacturer's protocol. Biotin-labeled NF-κB oligonucleotides (5'-AGTTGAGGGGACTTTCCCAGGC-3') were used for the EMSA 23. The reaction mixture was loaded on a 4% native polyacrylamide gel in 0.5% Tris-borate-EDTA and blotted onto nylon membranes (Millipore, Billerica, MA). After UV-cross-linking, interaction between the biotin-labeled NF-κB oligonucleotides and NF-κB protein was detected using a Chemiluminescent EMSA kit (Beyotime, China) with a streptavidin-HRP conjugate and a chemiluminescent substrate (Millipore, Billerica, MA).
Statistical analysis
All experiments were performed at least twice, with a minimum of three samples for each study. Data were shown as the means ± SD. The statistical significance was determined using the two-tailed unpaired Student's t test when 2 groups were compared. A probability value of <0.05 was considered as statistically significant.
Results
Detection of luciferase expression in the mouse liver after hydrodynamics-based gene delivery
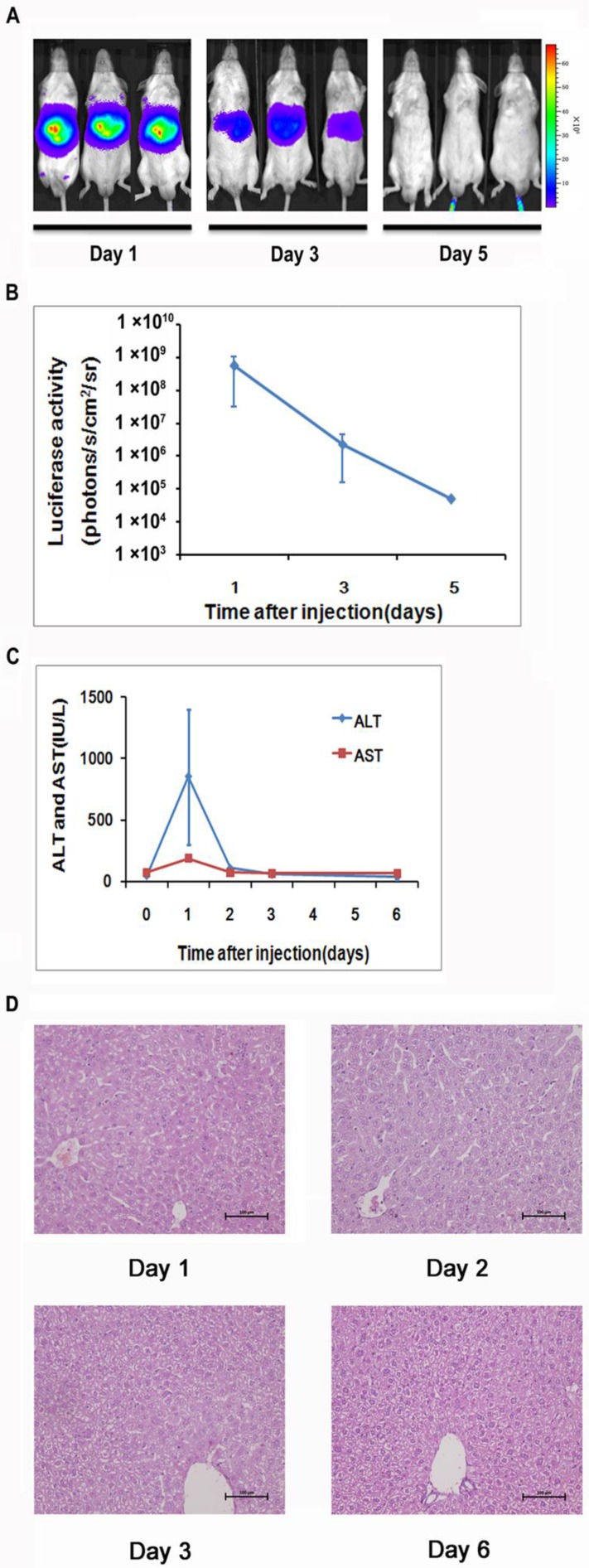
To generate a mouse model for evaluating the NF-κB transcriptional activity in the hepatic tissues, we delivered 10 μg of pattB-NF-κB-Fluc plasmid DNA to each mouse through a hydrodynamic injection method as described in the materials and methods. And before the formal experiment, the success rate, the survival rate and stability of hydrodynamic injection method were studied (Supplementary Material: Fig. S2). As shown in Fig. 1A and 1B, we detected strong bioluminescent signals (approximately107-108 p/s/cm2/sr) in the livers one day after the injection. However, the signals rapidly diminished in the following imaging sessions and became undetectable on day 5. We also observed that alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels significantly increased in the serum at day 1 and followed the similar clearance kinetics as luciferase (Fig. 1C). These results suggested that the hydrodynamic DNA delivery procedure elicited an acute hepatic inflammatory process which involved NF-κB activation.
Fig 1.
Detection of luciferase expression in the mouse liver after hydrodynamics-based gene delivery. NF-κB reporter gene expression in the livers (n=5) was monitored longitudinally through noninvasive molecular imaging (A, B). Serum ALT and AST levels at indicated time points were shown. n=5 in each group (C). Histological examination of tissue damage through HE staining of liver sections from mice at 1, 2, 3, and 6 days after the hydrodynamics-based gene delivery was shown. The results are representative of 3 experiments. Original magnification ×200 (D).
The extent of liver damage was evaluated by histology. As shown in Fig 1D, the liver tissues obtained at day 1 after hydrodynamic DNA delivery showed signs of severe damage, manifested by the presence of apoptotic and necrotic hepatocytes. In addition, infiltrating neutrophils were presented in the damaged tissue. These pathological changes abated at day 3 and completely diminished at day 6, suggesting that the damaged hepatic tissues underwent a rapid recovery process (Fig. 1D).
Monitoring NF-κB activation during LPS and ConA mediated acute inflammation
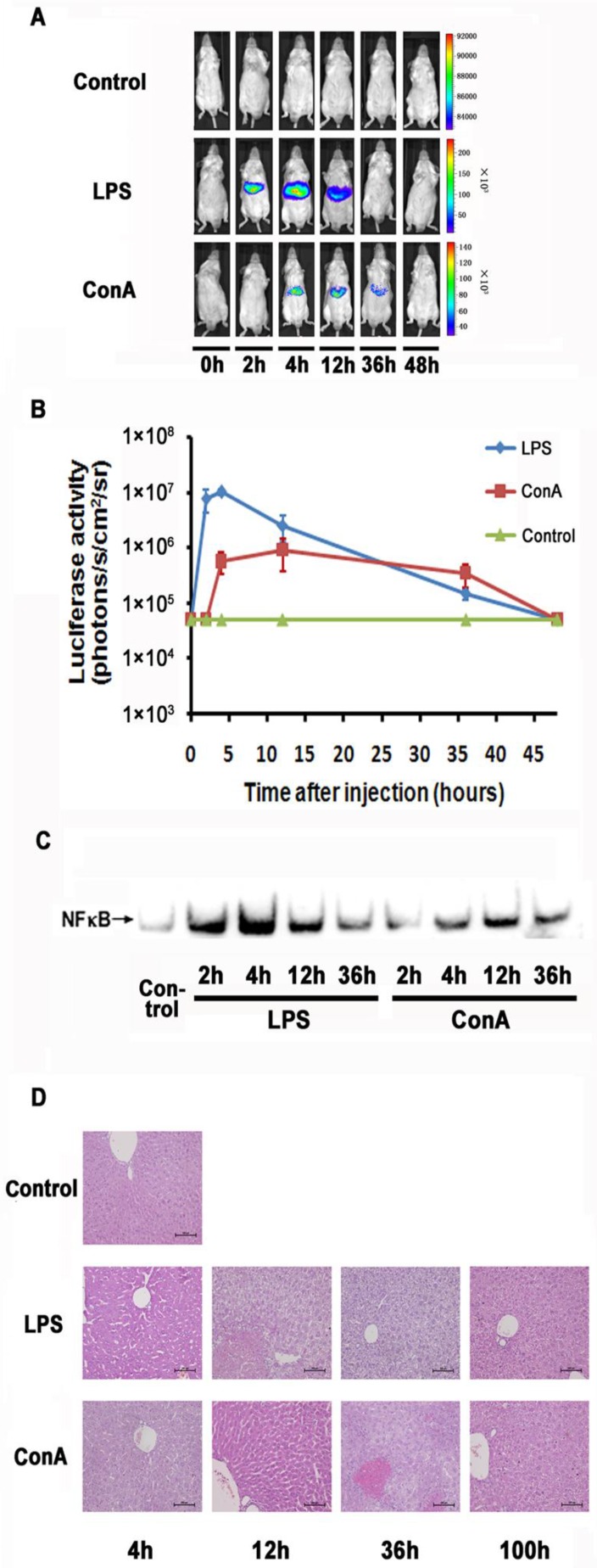
LPS and ConA were reported to induce acute inflammation in the liver. We examined the imaging detectibility of NF-κB activation in the NF-κB reporter transfected mice after treatment with these agents. As shown in Fig. 2A and 2B, we were able to detect luciferase activity in the livers of LPS or ConA treated mice. The kinetics of response in these mice showed a remarkable difference, with the LPS and ConA treated mice showing a peak activity at 2 and 12 hours respectively. The magnitude of induction was more robust in LPS treated mice than that in ConA treated mice. However, the duration of induction was much longer in ConA treated mice than that in the LPS treated mice. At 36 hours, the ConA treated mice still showed detectable luciferase activity while the activity of LPS-treated mice retreated to a basal level. Analysis with EMSA showed that the activated NF-κB levels in the hepatocytes reached their peaks at 4 and 12 hours in LPS- and ConA-treated mice respectively (Fig. 2C). The kinetics of NF-κB activation thus correlated nicely with imaging-detected luciferase activities in these mice. We further performed histological examination of the livers and showed that apoptosis and necrosis were more severe at 12 and 36 hours than at other time points, followed by recovery at 36 and 100 hours in LPS and ConA treated mice respectively (Fig. 2D).
Fig 2.
Monitoring NF-κB activation during LPS and ConA mediated acute inflammation. NF-κB in the mouse liver was activated through LPS and ConA treatment (A). The level of luciferase expression (n=4) was quantified as photons/s/cm2/sr using the Living Image software 4.2. The results are representative of 2 experiments (B). NF-κB activation in the hepatocytes at different time points was detected using EMSA. Each blot represents at least three independent experiments (C). Histological examination of liver tissues harvested at indicated time points from LPS and ConA treated mice. The results are representative of 2 experiments. Original magnification ×200 (D).
Phage φC31ointegrase mediated stable integration of the NF-κB-luc reporter
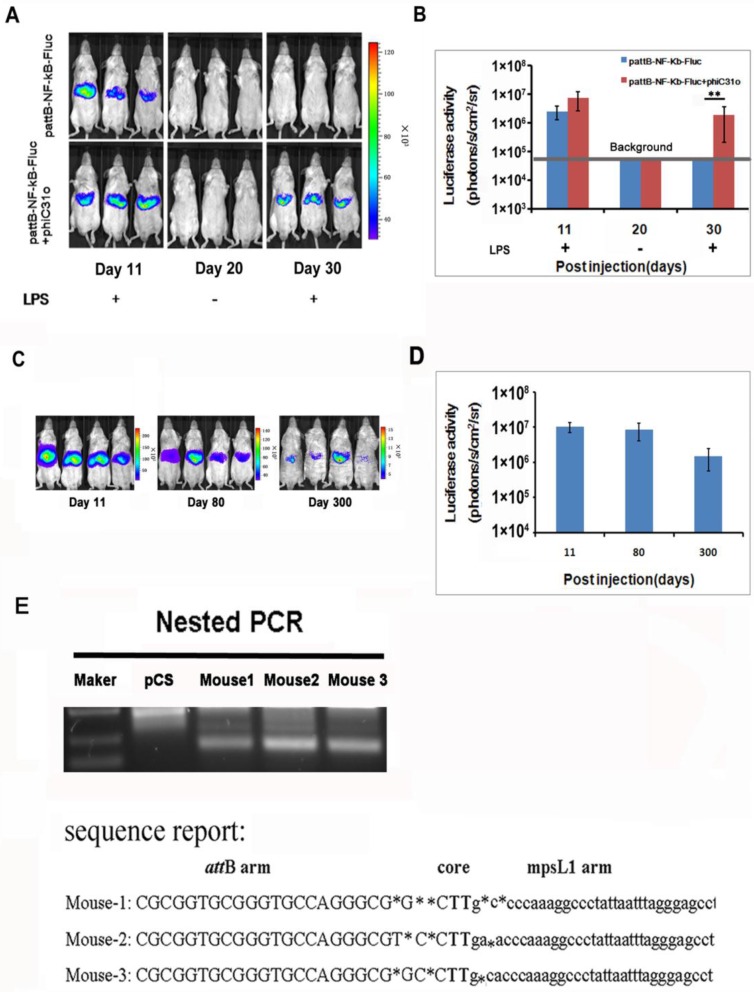
We examined the NF-κB activation after gene delivery and found that the luciferase activity was detectable on day 11 and became difficult to detect after day 30 (Fig. 3A, top panel), indicating that the gene delivery procedure only produced transient transfection of the hepatic tissues. To facilitate stable integration of the NF-κB-luc reporter, a mouse codon-optimized φC31, named φC31o integrase, was co-transfected to the hepatic tissues. The mouse codon-optimized φC31 integrase (PphiC31o) is a new system different from the originalφC31 integrase (pCMV-int) previously reported. Mice co-injected with PphiC31o displayed higher luciferase activity than mice co-injected with pCMV-int 60 days post-injection, indicating that φC31o improved recombination efficiency in mice livers and could be a feasible and viable alternative method to create genetically modified laboratory mice models (Supplementary Material: Fig. S1). As shown in Fig. 3A (bottom panel), we were able to detect luciferase induction by LPS in the co-transfected mice at 30 days after gene delivery. Expectedly, the luciferase induction by LPS was still detectable in the co-transfected mice at 80 and 300 days after the hydrodynamic gene delivery procedure (Fig. 3C and 3D). These results indicated that the φC31o integrase had mediated stable chromosomal integration of the NF-κB reporter in the transfected hepatic tissues.
Fig 3.
PhageφC31ointegrase mediated stable integration of the NF-κB-luc reporter. A total of 10 μg of pattB-NF-κB-Fluc was delivered to mice with and without 20 μg of PphiC31oby hydrodynamic injection. On day 11 and 30, the luciferase activity was monitored after LPS challenge. Imaging on day 20 was performed without LPS challenge and served as a baseline control for the day 30 imaging data. (A). Luciferase activities after LPS challenge of the transfected mice (n=4) on day 11, 30 (B) and day 80 and 300 (C, D) were quantified and shown. The results are representative of 2 experiments. Nested PCR was used to detect the junction between the attB site of pattB-NF-κB-Fluc and mpsL1 at 30 days after the DNA injection (E).
The DNA sequences at the integration sites are termed pseudo attP sites. Phage φC31o integrase has been shown to mediate efficient gene transfer of plasmid DNA into host cells through a recombination event between the attachment sequence attB and the pseudo attP sites present in the host genome. The most prevalent pseudo attP site (mpsL1) favored for integration into the mouse genome has been described previously 20. Here we determined whether integration into this site occurred through nested PCR. The resultant PCR products were sequenced and aligned with the murine genomic sequence. The switch from the attB site to the genomic sequence near the TG core and the sequence between the genomic sequence and the attP site support φC31o mediated integration at genomic pseudo-attP sites (Fig. 3E). These results confirmed that φC31o mediated chromosomal integration of the NF-κB reporter, which accounted for the long-term imaging detection of the NF-κB signaling pathway in the co-transfected mice.
Monitoring the suppression effect of PDTC on NF-κB activation
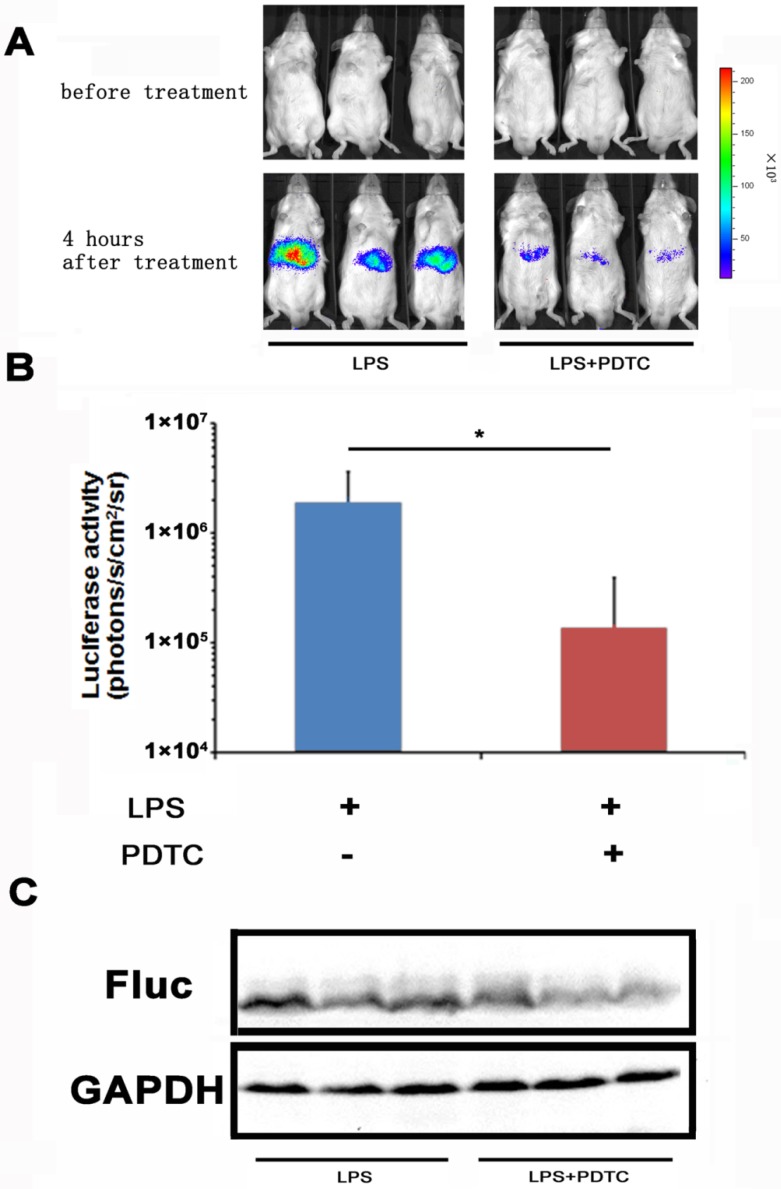
With the NF-κB-luc reporter transfected mice, we evaluated the efficacy of PDTC on LPS induced NF-κB activation in the hepatic tissues. PDTC has been reported as a potent and specific inhibitor of NF-κB 24, 25. In our study, we pre-dosed the NF-κB-luc reporter transfected mice with PDTC at 30 minutes before LPS challenge. In vivo imaging was performed at 4 hours and the results were shown in Fig. 4A. We observed a remarkable inhibition of LPS induced luciferase activity in the PDTC pre-treated mice. Quantitation analysis showed that the inhibition of luciferase activity by PDTC was statistically significant (p<0.05) (Fig. 4B). Furthermore, we confirmed by western blot that the hepatic tissues of PDTC treated mice had less luciferase protein expression than that of LPS treated mice (Fig. 4C).
Fig 4.
Monitoring the suppression effect of PDTC on NF-κB activation. PDTC was i.p. injected to mice at 30 min before LPS treatment. NF-κB activation (n=3) was measured at 4 hours through bioluminescence imaging. The results are representative of 2 experiments (A, B). The expression of the firefly luciferase was determined through western blot analysis (C).
Taken together, these results demonstrated a correlation between NF-κB activation and luciferase activity in the NF-κB-luc reporter transfected mice.
Sorafenib suppressed partial hepatectomy-induced liver regeneration via down-regulating NF-κB activation
Clinically, Sorafenib has been approved by the FDA for treatment of unresectable hepatocellular carcinoma (HCC). A previous report showed that Sorafenib affected the late stage liver mass restoration after partial hepatectomy 26. We hypothesize that Sorafenib mediated suppression of liver regeneration involves in the NF-κB pathway.
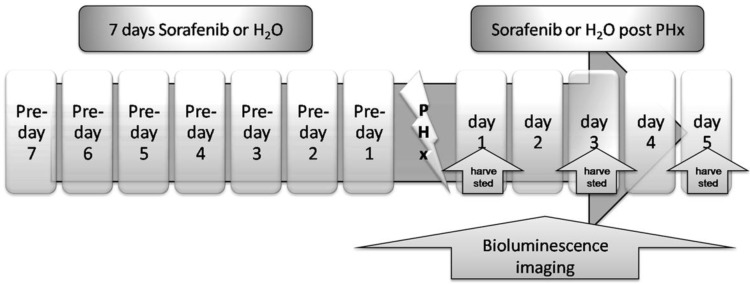
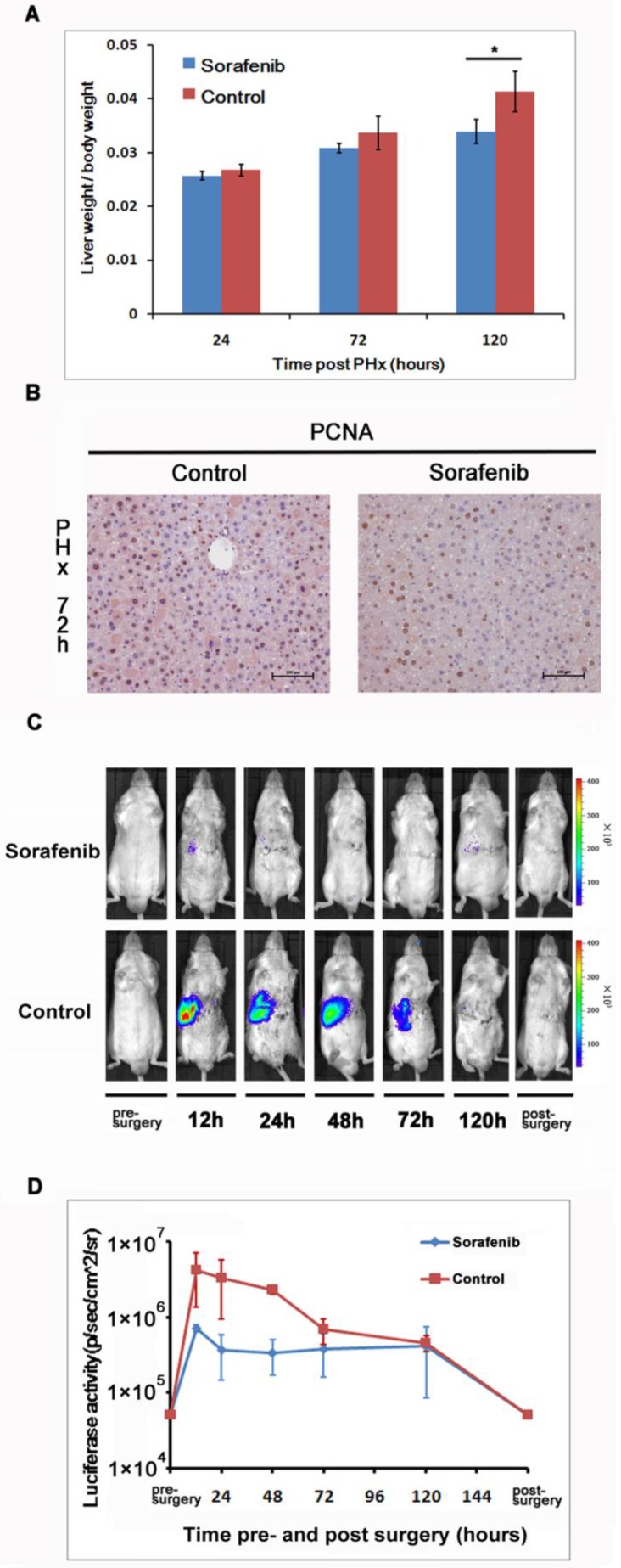
We investigated the impact of Sorafenib treatment on liver regeneration. The experimental scheme was delineated in Fig. 5. Mice were pre-treated with Sorafenib daily for 7 days before and 5 days after the partial hepatectomy procedure. Following the surgical procedure, mice were sacrificed at day 1, 3, and 5 respectively for measurement of the liver weight. In addition, bioluminescent imaging was performed at indicated time points. As shown in Fig. 6A, we observed a retarded restoration of the liver mass in Sorafenib treated mice in comparison to the vehicle treated control mice. Expression of the proliferating cell nuclear antigen (PCNA) in the hepatic tissues of Sorafenib-treated mice was remarkably lower than that in the control mice (Fig. 6B). Analysis of NF-κB activity showed that the PHx procedure induced NF-κB activation in the liver when the mice were imaged at 12 hours. The activity remained at a plateau level until 48 hours after surgery and declined dramatically from 72 hours to a basal level at 120 hours (Fig. 6C, bottom panel). These results indicated that the liver regeneration process is accompanied with NF-κB activation. In the Sorafenib treated mice, we observed a dramatic suppression of the NF-κB activity in the liver (Fig. 6C, top panel), which was further confirmed through quantitation of the luciferase signal Fig. 6D. Thus, our results suggested that Sorafenib may have exerted its inhibitory effect on liver regeneration through suppression of the NF-κB signaling pathway.
Fig 5.
Treatment scheme. Sorafenib or vehicle (distilled water) was p.o. dosed to mice daily for 7 days before PHx. Treatment was continued for 5 days post the PHx procedure. The bioluminescence imaging was performed at 1 day before PHx and continued for 5 days after PHx. The livers from the two animal groups were harvested at 24, 72, or 120 hours after PHx.
Fig 6.
Sorafenib suppressed partial hepatectomy-induced liver regeneration via down-regulating NF-κB activation. The livers were harvested at 24, 72, and 120 hours after PHx and weighted. The ratios of liver weight over the whole body weight were shown. The values represents means±SD, with n=4 for each group. (A). Expression of PCNA in the livers was evaluated with immunohistochemical staining. Original magnification ×200 (B).Sorafenib or vehicle treated mice were imaged for luciferase expression at indicated time points following PHx (C). Quantitation of luciferase signal from the liver was shown (n=4).The results are representative of 2 experiments. (D).
Discussion
A growing body of evidence suggested that the NF-κB-dependent pathway participated in the development of liver failure, chronic liver diseases, and hepatocellular carcinoma. Thus, NF-κB is an attractive target for pharmacological intervention of these pathological conditions 27-29. Being able to monitor the NF-κB-dependent pathway under various pathological states would facilitate the validation process of novel therapeutic agents and approaches. Our development of the NF-κB-luc stable transfection model fits right to this purpose.
Bioluminescence imaging has been proven to be a highly sensitive method for monitoring gene expression in luciferase reporter manifested transgenic mice 30, 31. The noninvasive nature of this technology enables convenient longitudinal studies, which leads to the generation of statistically preferable data sets with fewer animals 32-34. Generation of transgenic mice for imaging the NF-κB signaling pathway was reported previously 14. However, development of transgenic animal model is time consuming. Transient transfection with luciferase reporters has been used for monitoring NF-κB activation in the liver 15. However, the reporter expression lasted for only a few days, thus was not suitable for monitoring NF-κB activity during chronic pathological conditions.
φC31o integrase has been exploited to facilitate stable chromosomal integration of the transfected DNA. Within the transfected cells, stable chromosomal integration was achieved though φC31o integrase catalyzed recombination between the attachment attB sequence of the transfected DNA and the pseudo attP sites of the host genome. In our study, we experimented the φC31o integrase mediated integration approach and achieved efficient and stable integration of the NF-κB reporter in the transfected hepatic tissues. Chromosomal integration of the reporter was verified through nested PCR analysis of DNA sequences adjacent to the mpsL1 site, which was previously identified as one of the frequently targeted attP site in the murine genome 18-20. We demonstrated that the transfected mice expressed the reporter nearly one year after the gene delivery. Induction of the NF-κB pathway was non-invasively monitored during LPS and ConA mediated inflammatory processes. Response to LPS challenge occurred more rapidly than that to the ConA treatment. It has been established that acute liver injury by LPS involved NF-κB activation in Kupffer cells and secretion of proinflammatory cytokines, which orchestrated a cascade of pathological events towards the liver failure 35, 36. ConA treatment was used to establish T cell-mediated acute hepatitis through cytokine induction that involved NF-κB activation, which was conducive to hepatic apoptosis and necrosis. The difference of NF-κB activation kinetics after LPS and ConA treatments could be due to that the Kupffer cells and T cells were the predominant cell populations in the responses following the respective challenges 37, 38.
HCC is a common cause of cancer-related death, and the incidence has been increasing worldwide 39. Clinical, HCC patients often need to undergo partial hepatectomy to remove the primary lesion before chemotherapy. Sorafenib is a multi-kinase inhibitor and has been used to treat patients with HCC 40. However, researchers have shown that Sorafenib affected liver mass restoration after partial hepatectomy with unknown molecular mechanism 26. As NF-κB has been indicated to play a critical rule during liver regeneration, we hypothesized that Sorafenib treatment-induced delay of liver regeneration could be due to its modulation of the NF-κB pathway. Indeed, our results demonstrated that Sorafenib suppressed the NF-κB activation following hepatectomy and that the suppression correlated with a delay in liver regeneration (Fig. 6). This finding may serve as a cautionary guidance on the timing of Sorafenib treatment of HCC patients following hepatectomy.
In conclusion, we reported a method of stable transfection of the hepatic tissues and demonstrated that the NF-κB signaling pathway can be non-invasively monitored under pathological conditions such as acute liver injury, hepatitis and liver regeneration. We provided evidence that suppression of the NF-κB signaling pathway was the molecular mechanism of Sorafenib treatment-elicited retardation of liver regeneration. Further exploration of this stable transfection approach for establishment of other disease models will enable in vivo validation of novel pharmaceuticals.
Supplementary Material
Fig.S1 - Fig.S2.
Acknowledgments
The plasmid pT-attB was generously provided by M. P. Calos, Department of Genetics, Stanford University, USA. This work was supported by the Mega-projects of Science Research for the 12th Five-Year Plan of China (#2012ZX10004-502, #2011ZXJ092-031),the Natural Science Foundation of China (#31201757, #81102223, #30901812) and Beijing National Science Foundation (#7112104, #7122136).
Abbreviations
- NF-κB
nuclear factor-κB
- LPS
lipopolysaccharide
- φC31o
mouse-codon optimized φC31 integrase
- ConA
Concanavalin A
- PDTC
Pyrrolidinedithiocarbamate
- NBCDSER
National Beijing Center for Drug Safety Evaluation and Research
- SPF
specific pathogen-free
- PHx
partial hepatectomy
- HE
hematoxylin-eosin
- PCNA
proliferating cell nuclear antigen
- EMSA
electrophoretic mobility shift assay
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- HCC
hepatocellular carcinoma.
References
- 1.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–16. doi: 10.1016/0092-8674(86)90346-6. doi:0092-8674(86)90346-6 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. doi:10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 3.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. doi:10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baeuerle PA. IkappaB-NF-kappaB structures: at the interface of inflammation control. Cell. 1998;95:729–31. doi: 10.1016/s0092-8674(00)81694-3. doi:S0092-8674(00)81694-3 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Delhase M. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin Immunol. 2000;12:85–98. doi: 10.1006/smim.2000.0210. doi:S1044-5323(00)90210-0 [pii] 10.1006/smim.2000.0210. [DOI] [PubMed] [Google Scholar]
- 6.Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799:775–87. doi: 10.1016/j.bbagrm.2010.05.004. doi:10.1016/j.bbagrm.2010.05.004 S1874-9399(10)00065-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porcile C, Piccioli P, Stanzione S, Bajetto A, Bonavia R, Barbero S. et al. Proteasome inhibitors induce cerebellar granule cell death: inhibition of nuclear factor-kB activation. Ann N Y Acad Sci. 2002;973:402–13. doi: 10.1111/j.1749-6632.2002.tb04673.x. [DOI] [PubMed] [Google Scholar]
- 8.Suetsugu H, Iimuro Y, Uehara T, Nishio T, Harada N, Yoshida M. et al. Nuclear factor {kappa}B inactivation in the rat liver ameliorates short term total warm ischaemia/reperfusion injury. Gut. 2005;54:835–42. doi: 10.1136/gut.2004.043034. doi:54/6/835 [pii] 10.1136/gut.2004.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luedde T, Schwabe RF. NF-kappaB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–18. doi: 10.1038/nrgastro.2010.213. doi:10.1038/nrgastro.2010.213 nrgastro.2010.213 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seki E, Brenner DA. The role of NF-kappaB in hepatocarcinogenesis: promoter or suppressor? J Hepatol. 2007;47:307–9. doi: 10.1016/j.jhep.2007.05.006. doi:S0168-8278(07)00297-8 [pii] 10.1016/j.jhep.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottero V, Imbert V, Frelin C, Formento JL, Peyron JF. Monitoring NF-kappa B transactivation potential via real-time PCR quantification of I kappa B-alpha gene expression. Mol Diagn. 2003;7:187–94. doi: 10.1007/BF03260037. doi:738 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Badr CE, Niers JM, Tjon-Kon-Fat LA, Noske DP, Wurdinger T, Tannous BA. Real-time monitoring of nuclear factor kappaB activity in cultured cells and in animal models. Mol Imaging. 2009;8:278–90. [PMC free article] [PubMed] [Google Scholar]
- 13.Badr CE, Tannous BA. Bioluminescence imaging: progress and applications. Trends Biotechnol. 2011;29:624–33. doi: 10.1016/j.tibtech.2011.06.010. doi:10.1016/j.tibtech.2011.06.010 S0167-7799(11)00114-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. In vivo imaging of NF-kappa B activity. J Immunol. 2002;168:1441–6. doi: 10.4049/jimmunol.168.3.1441. [DOI] [PubMed] [Google Scholar]
- 15.Hyoudou K, Nishikawa M, Kobayashi Y, Kuramoto Y, Yamashita F, Hashida M. Analysis of in vivo nuclear factor-kappaB activation during liver inflammation in mice: prevention by catalase delivery. Mol Pharmacol. 2007;71:446–53. doi: 10.1124/mol.106.027169. doi:mol.106.027169 [pii] 10.1124/mol.106.027169. [DOI] [PubMed] [Google Scholar]
- 16.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. doi:10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 17.Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. doi:3302891 [pii] 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- 18.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. doi:10.1073/pnas.090527097 090527097 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keravala A, Chavez CL, Hu G, Woodard LE, Monahan PE, Calos MP. Long-term phenotypic correction in factor IX knockout mice by using PhiC31 integrase-mediated gene therapy. Gene Ther. 2011;18:842–8. doi: 10.1038/gt.2011.31. doi:10.1038/gt.2011.31 gt201131 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA, Calos MP. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat Biotechnol. 2002;20:1124–8. doi: 10.1038/nbt753. doi:10.1038/nbt753 nbt753 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Yan S, Fu Q, Zhou Y, Wang J, Liu Y, Duan X. et al. High levels of gene expression in the hepatocytes of adult mice, neonatal mice and tree shrews via retro-orbital sinus hydrodynamic injections of naked plasmid DNA. J Control Release. 2012;161:763–71. doi: 10.1016/j.jconrel.2012.05.018. doi:10.1016/j.jconrel.2012.05.018 S0168-3659(12)00394-X [pii] [DOI] [PubMed] [Google Scholar]
- 22.Higgins GM, Anderson RM. Experimental pathology of the liver. I: restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 23.Naidu S, Wijayanti N, Santoso S, Kietzmann T, Immenschuh S. An atypical NF-kappa B-regulated pathway mediates phorbol ester-dependent heme oxygenase-1 gene activation in monocytes. J Immunol. 2008;181:4113–23. doi: 10.4049/jimmunol.181.6.4113. doi:181/6/4113 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Bessho R, Matsubara K, Kubota M, Kuwakado K, Hirota H, Wakazono Y. et al. Pyrrolidine dithiocarbamate, a potent inhibitor of nuclear factor kappa B (NF-kappa B) activation, prevents apoptosis in human promyelocytic leukemia HL-60 cells and thymocytes. Biochem Pharmacol. 1994;48:1883–9. doi: 10.1016/0006-2952(94)90586-x. [DOI] [PubMed] [Google Scholar]
- 25.Lauzurica P, Martinez-Martinez S, Marazuela M, Gomez del Arco P, Martinez C, Sanchez-Madrid F. et al. Pyrrolidine dithiocarbamate protects mice from lethal shock induced by LPS or TNF-alpha. Eur J Immunol. 1999;29:1890–900. doi: 10.1002/(SICI)1521-4141(199906)29:06<1890::AID-IMMU1890>3.0.CO;2-F. doi:10.1002/(SICI)1521-4141(199906)29:06<1890::AID-IMMU1890>3.0.CO;2-F [pii]10.1002/(SICI)1521-4141(199906)29:06<1890::AID-IMMU1890>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Hora C, Romanque P, Dufour JF. Effect of sorafenib on murine liver regeneration. Hepatology. 2011;53:577–86. doi: 10.1002/hep.24037. doi:10.1002/hep.24037. [DOI] [PubMed] [Google Scholar]
- 27.Kwak JH, Jung JK, Lee H. Nuclear factor-kappa B inhibitors; a patent review (2006-2010) Expert Opin Ther Pat. 2011;21:1897–910. doi: 10.1517/13543776.2011.638285. doi:10.1517/13543776.2011.638285. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore TD, Garbati MR. Inhibition of NF-kappaB signaling as a strategy in disease therapy. Curr Top Microbiol Immunol. 2011;349:245–63. doi: 10.1007/82_2010_105. doi:10.1007/82_2010_105. [DOI] [PubMed] [Google Scholar]
- 29.Sethi G, Tergaonkar V. Potential pharmacological control of the NF-kappaB pathway. Trends Pharmacol Sci. 2009;30:313–21. doi: 10.1016/j.tips.2009.03.004. doi:10.1016/j.tips.2009.03.004 S0165-6147(09)00068-6 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Zhang N, Weber A, Li B, Lyons R, Contag PR, Purchio AF. et al. An inducible nitric oxide synthase-luciferase reporter system for in vivo testing of anti-inflammatory compounds in transgenic mice. J Immunol. 2003;170:6307–19. doi: 10.4049/jimmunol.170.12.6307. [DOI] [PubMed] [Google Scholar]
- 31.Zhang N, Ahsan MH, Purchio AF, West DB. Serum amyloid A-luciferase transgenic mice: response to sepsis, acute arthritis, and contact hypersensitivity and the effects of proteasome inhibition. J Immunol. 2005;174:8125–34. doi: 10.4049/jimmunol.174.12.8125. doi:174/12/8125 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Contag CH, Jenkins D, Contag PR, Negrin RS. Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2000;2:41–52. doi: 10.1038/sj.neo.7900079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Feng JQ, Harris SE, Contag PR, Stevenson DK, Contag CH. Rapid in vivo functional analysis of transgenes in mice using whole body imaging of luciferase expression. Transgenic Res. 2001;10:423–34. doi: 10.1023/a:1012042506002. [DOI] [PubMed] [Google Scholar]
- 34.Niu G, Zhu L, Ho DN, Zhang F, Gao H, Quan Q. et al. Longitudinal bioluminescence imaging of the dynamics of Doxorubicin induced apoptosis. Theranostics. 2013;3:190–200. doi: 10.7150/thno.5825. doi:10.7150/thno.5825 thnov03p0190 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann F, Sass G, Zillies J, Zahler S, Tiegs G, Hartkorn A. et al. A novel technique for selective NF-kappaB inhibition in Kupffer cells: contrary effects in fulminant hepatitis and ischaemia-reperfusion. Gut. 2009;58:1670–8. doi: 10.1136/gut.2008.165647. doi:10.1136/gut.2008.165647 gut.2008.165647 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Sun D, Chen D, Du B, Pan J. Heat shock response inhibits NF-kappaB activation and cytokine production in murine Kupffer cells. J Surg Res. 2005;129:114–21. doi: 10.1016/j.jss.2005.05.028. doi:S0022-4804(05)00296-9 [pii] 10.1016/j.jss.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 37.Imose M, Nagaki M, Kimura K, Takai S, Imao M, Naiki T. et al. Leflunomide protects from T-cell-mediated liver injury in mice through inhibition of nuclear factor kappaB. Hepatology. 2004;40:1160–9. doi: 10.1002/hep.20438. doi:10.1002/hep.20438. [DOI] [PubMed] [Google Scholar]
- 38.Ke B, Shen XD, Ji H, Kamo N, Gao F, Freitas MC. et al. HO-1-STAT3 axis in mouse liver ischemia/reperfusion injury: regulation of TLR4 innate responses through PI3K/PTEN signaling. J Hepatol. 2012;56:359–66. doi: 10.1016/j.jhep.2011.05.023. doi:10.1016/j.jhep.2011.05.023 S0168-8278(11)00528-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabibbo G, Craxi A. Epidemiology, risk factors and surveillance of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2010;14:352–5. [PubMed] [Google Scholar]
- 40.Woo HY, Heo J. Sorafenib in liver cancer. Expert Opin Pharmacother. 2012;13:1059–67. doi: 10.1517/14656566.2012.679930. doi:10.1517/14656566.2012.679930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.S1 - Fig.S2.