Abstract
Summary: At the rate that prokaryotic genomes can now be generated, comparative genomics studies require a flexible method for quickly and accurately predicting orthologs among the rapidly changing set of genomes available. SPOCS implements a graph-based ortholog prediction method to generate a simple tab-delimited table of orthologs and in addition, html files that provide a visualization of the predicted ortholog/paralog relationships to which gene/protein expression metadata may be overlaid.
Availability and Implementation: A SPOCS web application is freely available at http://cbb.pnnl.gov/portal/tools/spocs.html. Source code for Linux systems is also freely available under an open source license at http://cbb.pnnl.gov/portal/software/spocs.html; the Boost C++ libraries and BLAST are required.
Contact: leeann.mccue@pnnl.gov
1 INTRODUCTION
The prediction of orthologs and paralogs is a fundamental step in many comparative genomics studies. For example, orthology and paralogy relationships are used to identify the core and auxiliary genes among related organisms, assign protein function annotation to genes in a new genome and predict gene expansion events, such as those that give rise to protein families. Though the concepts of orthology and paralogy were defined many years ago, the advent of whole genome sequencing and the wide availability of these genomic data has been the catalyst for the development of numerous ortholog prediction programs [see Kristensen et al. (2011) for a recent review]. In large parts, these programs rely on the identification of homologous genes/proteins, and frequently employ BLAST (Basic Local Alignment Search Tool; Altschul et al., 1997) to identify these.
Many methods have focused on eukaryotic genomes, in which widespread gene duplication and domain rearrangement have made predicting orthologs particularly challenging (Sjölander et al., 2011). In prokaryotes, however, the challenge lies in keeping up with the rate of genomic data generation. As a bacterial genome can now be sequenced in a day or two, and many are often left in ‘high quality draft’ stage, comparative genomics studies on these data require the flexibility to quickly and accurately predict orthologs on-the-fly, when the genome data are updated frequently.
We have previously employed a graph-based ortholog/paralog prediction method (Callister et al., 2008; Konstantinidis et al., 2009), that combines the pairwise (two species) predictions from InParanoid (Remm et al., 2001), using custom Perl scripts to produce orthologs for groups of species. Here, we present a robust software tool called SPOCS (Species Paralogy and Orthology Clique Solver) that implements this approach, providing flexible and readily extensible ortholog predictions for groups of species defined by the user.
2 APPROACH
For every pair of species (i.e. pair of protein fasta files provided by the user), we use the ortholog/paralog definitions as described by Remm et al. (2001) and combine the pairwise predictions using a graph-based method to identify complete or near-complete cliques. SPOCS will generate a simple tab-delimited table of orthologs, and in addition, html files that provide a visualization of the predicted orthology/paralogy relationships. The html files link back to the protein sequence data and gene/protein expression metadata may be overlaid onto the visualization.
2.1 Implementation
SPOCS is written in C++ using the Boost libraries (http://www.boost.org/). For the identification of orthologs and paralogs between pairs of species, the InParanoid Perl code (Remm et al., 2001) was redesigned and implemented in C++. We used the MaxCliqueDyn algorithm (Konc and Janezic, 2007) to identify ortholog cliques, and the D3 Javascript library (http://d3js.org/) to generate SVG images of the ortholog graphs.
SPOCS is available both as source code and in a web application. For local installation on Unix/Linux machines, instructions are provided in a readme file within the downloadable source tar file. To run SPOCS locally, the BLAST executables must be in the user PATH. At the command line, SPOCS requires only a list of fasta formatted protein sequence files, a scratch output directory in which SPOCS will store intermediate files and a results directory for the final output files.
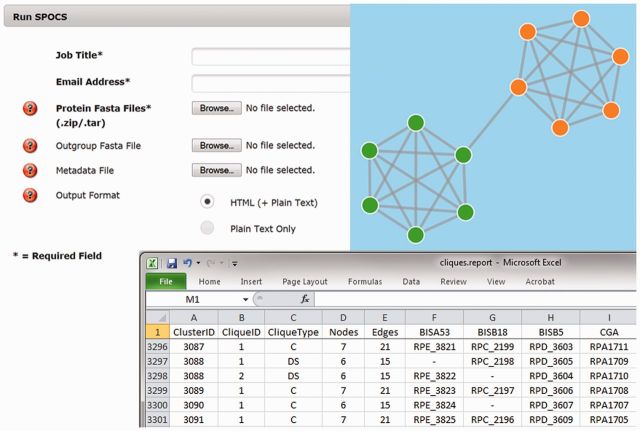
The web application requires only the protein fasta files, a job title and an email address (Fig. 1, upper left). The user is notified by email when results are available, because jobs may require hours to run, depending on the number of species involved. The stand-alone Linux implementation and the web application generate the same outputs: a tab-delimited report file in which each line represents a predicted set of orthologs (Fig. 1, lower right), and optionally, self-contained html output with visualizations of the ortholog relationships (Fig. 1, upper right). SPOCS also offers several options to control parameters associated with utilization of an outgroup species, identification of the reciprocal BLAST hits, generation of html files for visualization, and incorporation of metadata on gene/protein expression that can be overlaid on the html images.
Fig. 1.
Screen shots of the SPOCS web application home page, a sample of the text output (viewed in Excel) and an html page for a Degenerate group that will resolve two cliques (orange and green)
2.2 Application
Provided a set of n proteomes (fasta-formatted files of the predicted proteins from a species) and an optional proteome to serve as the outgroup, SPOCS proceeds through three main stages. First, SPOCS executes a series of BLAST (Altschul et al., 1997) runs between every pair of species to identify reciprocal best hits; this is the most compute-intensive stage, requiring n2 independent BLAST runs. When SPOCS is run locally, the BLAST results are stored in the scratch directory, allowing subsequent SPOCS runs that include some of the same n species to avoid performing BLAST runs if they already exist. In the second stage, SPOCS uses the BLAST results to generate an orthology/paralogy relationship graph based on merging the pairwise ortholog and in-paralog relationships [as defined by Remm et al., (2001)]. Finally, SPOCS identifies cliques in each graph by breaking it into subgraphs and using the branch and bound clique-finding algorithm (Konc and Janezic, 2007) to iteratively find all cliques in each subgraph.
We classify the graphs into four subjective categories: Complete, SemiComplete, Incomplete and Degenerate. The Complete graphs include no more than one protein per species and every protein (node in the graph) is connected to every other protein by a reciprocal best hit (edge), i.e., a maximum clique. SemiComplete graphs may be reported in which no more than one protein per species is present but with less than the maximal number of edges (this is a tunable parameter for which the default is 5%). By definition, these Complete and SemiComplete cliques are the result of single unique graphs that resolve to single unique cliques. The Incomplete graphs have no more than one protein per species, and less than the maximal number of edges (more missing edges than specified for SemiComplete). The Degenerate graphs have more than one protein (node) from one or more of the species. For the Incomplete and Degenerate graphs, the clique-finding algorithm will identify as many maximum cliques as possible.
2.3 Visualization
Using the –H flag with the stand-alone software, or the html radio button on the web application will generate self-contained html output, which allows the user to navigate through the graph space and view gene relationships. The graph structure is rendered using a force-directed layout engine, which uses charged particles to place related genes closer together in the graph; the html output provides this graph image as well as a link to the fasta file of those proteins and a table providing their functional annotation. Generation of the html output does not appreciably affect run-time, but will dramatically increase the number of files generated. Though no upper limit has been imposed on the number of genomes that SPOCS can process, the visualization functions were designed to facilitate the analysis of targeted datasets chosen to explore specific hypotheses, and can become difficult to interpret as the number of genomes increases.
3 CONCLUSION
SPOCS provides a flexible and extensible software tool for the prediction of orthologs and paralogs among closely related genomes. In addition to a standard table of orthologs, the html output presents a visualization of the predicted orthology/paralogy relationships, provides hooks back to the protein sequence data and allows the overlay of gene/protein expression metadata.
ACKNOWLEDGEMENTS
The authors thank Elena Peterson for assistance setting up the web application. Web application jobs are executed on the PNNL Institutional Computing system at PNNL.
Funding: This work was supported by U.S. Department of Energy, Office of Biological and Environmental Research (DOE-BER) Genome Science Program, and performed at the Pacific Northwest National Laboratory, a multiprogram national laboratory operated by Battelle for the DOE under Contract DE-AC06-76RL01830.
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister SJ, et al. Comparative bacterial proteomics: analysis of the core genome concept. PLoS One. 2008;3:e1542. doi: 10.1371/journal.pone.0001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konc J, Janezic D. An improved branch and bound algorithm for the maximum clique problem. Match-Commun. Math Co. 2007;58:569–590. [Google Scholar]
- Konstantinidis KT, et al. Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc. Natl Acad. Sci. USA. 2009;106:15909–15914. doi: 10.1073/pnas.0902000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen DM, et al. Computational methods for gene orthology inference. Brief. Bioinform. 2011;12:379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, et al. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Sjölander K, et al. Ortholog identification in the presence of domain architecture rearrangement. Brief. Bioinform. 2011;12:413–422. doi: 10.1093/bib/bbr036. [DOI] [PMC free article] [PubMed] [Google Scholar]