Abstract
A major challenge in cancer epidemiologic studies, especially those of rare cancers, is observing enough cases. To address this, researchers often join forces by bringing multiple studies together to achieve large sample sizes, allowing for increased power in hypothesis testing, and improved efficiency in effect estimation. Combining studies, however, renders the analysis difficult owing to the presence of heterogeneity in the pooled data. In this article, motivated by a collaborative nested case–control (NCC) study of ovarian cancer in three cohorts from United States, Sweden, and Italy, we investigate the use of penalty regularized partial likelihood estimation in the context of pooled NCC studies to achieve two goals. First, we propose an adaptive group lasso (gLASSO) penalized approach to simultaneously identify important variables and estimate their effects. Second, we propose a composite agLASSO penalized approach to identify variables with heterogeneous effects. Both methods are readily implemented with the group coordinate gradient decent algorithm and shown to enjoy the oracle property. We conduct simulation studies to evaluate the performance of our proposed approaches in finite samples under various heterogeneity settings, and apply them to the pooled ovarian cancer study.
Keywords: Cox's proportional hazards model, Group penalty, Heterogeneity, Nested case–control sampling, Ovarian cancer, Pooled studies, Shrinkage estimation
1. Introduction
Cancer is both a rare and a complex disease and thus a large number of subjects are often needed to elucidate the relationship between the disease and risk factors. As a single study is unlikely to be sufficient or definitive, researchers have devoted increasing efforts to build large pooled datasets by bringing multiple studies together, such as the projects in the NIH/NCI Cohort Consortium (http://epi.grants.cancer.gov/Consortia/cohort.html). The pooling strategy has allowed researchers to examine rare cancers, rare exposures, risk factors with small effects, and the interplay among multiple risk factors.
The motivating study for this paper is a collaboration between the New York University Women's Health Study (NYUWHS), the Northern Sweden Health and Disease Study (NSHDS), and the Italian Hormones and Diet in the Etiology of Cancer Study (ORDET) to assess the effects of circulating levels of inflammation markers on the risk of invasive epithelial ovarian cancer (Clendenen and others, 2011). This joint effort identified 230 cases from the three cohorts and, for each case, 2 controls were selected from the same cohort using the nested case–control (NCC) sampling design (Thomas, 1979). The inflammatory markers were measured from stored blood samples collected at enrollment, and other risk factors were assembled from the questionnaires. We know that heterogeneity is often present in pooled observational epidemiological studies (Friedenreich, 1993; Ioannidis and others, 2002). The potential source of heterogeneity for our study included recruitment, disease ascertainment methods, and sample types (serum/plasma). Implementing the NCC design that selected cases and controls nested within the same cohort facilitated the pooling and reduced the heterogeneity in our study. But we still found that some markers exhibited heterogeneous effects across cohorts when examining the heterogeneity using a likelihood ratio test comparing models with and without biomarker × cohort-membership interaction terms (Clendenen and others, 2011).
Statistical methods for pooled studies need to recognize heterogeneity. A commonly used method for pooled analysis is the two-stage method to combine study-specific results using either a fixed-effects model (Hedges and Olkin, 1985) or a random-effects model (DerSimonian and Laird, 1986). The method originates from the meta-analysis of randomized clinical trials and has been extended to meta-regression to adjust heterogeneous study characteristics (Greenland, 1994; Thompson and Higgins, 2002). Bayesian approaches to random-effects meta-analysis (Smith and others, 1995) and a Bayesian hierarchical model (Liu and others, 2011) have also been proposed to integrate multiple studies and accommodate heterogeneity. In recent papers focusing on variable selection in pooled genetic studies, group penalty regularized regressions that consider the effects of a genetic variant over multiple studies in a group manner have been adopted, e.g. the group lasso (gLASSO) penalized regression for genome-wide mapping in ancestry admixed population (Puniyani and others, 2010); gene selection in pooled microarray studies using the group minimax concave penalty (Ma and others, 2011) or the group bridge penalty (Ma and others, 2011).
In this article, we propose two approaches based on penalized partial likelihood with group selection feature to integrate multiple NCC studies with potential heterogeneity. In the first approach, we adopt the adaptive gLASSO (agLASSO) penalty technique proposed by Wang and Leng (2008) to incorporate heterogeneity to the analysis of pooled NCC studies and show that the agLASSO penalty regularized maximum partial likelihood estimators have the oracle property (Fan and Li, 2001) for selection and estimation. Furthermore, identifying covariates with heterogeneous effects has great implications for building accurate cancer risk prediction models. Statistical tests for heterogeneity, such as Cochran Q-test (Cochran, 1954) and 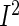 -test (Higgins and Thompson, 2002), often have low power. In our second approach, we introduce a hierarchical structure over a variable's effects by modeling heterogeneous effects through interactions of the variable with cohort-membership indicators, and propose a composite agLASSO (cagLASSO) method to identify this hierarchical structure for variables with heterogeneous effects. Our cagLASSO method generalizes Zhao and others (2009) by applying data-adaptive weights to different components in the cagLASSO penalty and can achieve a consistent selection for heterogeneous effects.
-test (Higgins and Thompson, 2002), often have low power. In our second approach, we introduce a hierarchical structure over a variable's effects by modeling heterogeneous effects through interactions of the variable with cohort-membership indicators, and propose a composite agLASSO (cagLASSO) method to identify this hierarchical structure for variables with heterogeneous effects. Our cagLASSO method generalizes Zhao and others (2009) by applying data-adaptive weights to different components in the cagLASSO penalty and can achieve a consistent selection for heterogeneous effects.
The rest of the article is organized as follows. In Section 2, we propose the agLASSO and cagLASSO approaches for pooled NCC studies with heterogeneity, and establish asymptotic properties for our proposed estimators. Numerical simulations and the analysis of the ovarian cancer study are presented in Section 3. A discussion with concluding remarks is presented in Section 4.
2. Penalized partial likelihood approaches for pooled NCC studies
Consider a pooled NCC study from K parent cohorts with each size of 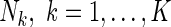 . Let
. Let 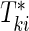 be the failure time and
be the failure time and 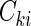 be the censoring time for the ith subject in cohort k. Denote the observed time-to-event by
be the censoring time for the ith subject in cohort k. Denote the observed time-to-event by 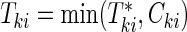 , failure status by
, failure status by 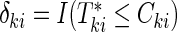 , and the counting process by Nki(t)=δkiI(T*ki≤t)
, and the counting process by Nki(t)=δkiI(T*ki≤t)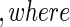 I(⋅) denotes the indicator function throughout. Within cohort k, cases are identified as subjects with
I(⋅) denotes the indicator function throughout. Within cohort k, cases are identified as subjects with 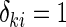 , and for a given case i, the NCC design randomly samples m controls without replacement from the risk set at
, and for a given case i, the NCC design randomly samples m controls without replacement from the risk set at 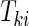 excluding the case itself. Let
excluding the case itself. Let 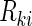 denote the indices of the case and its m selected controls. Covariates of interest
denote the indices of the case and its m selected controls. Covariates of interest  are ascertained for each case–control set at the case failure time.
are ascertained for each case–control set at the case failure time.
2.1. Variable selection and effect estimation
In each cohort k, the failure time follows a Cox proportional hazards (PHs) model (Cox, 1972):
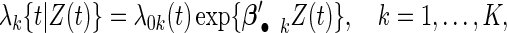 |
(2.1) |
where 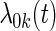 is the cohort-specific baseline hazard function, and
is the cohort-specific baseline hazard function, and 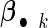 is a
is a 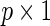 vector of coefficients characterizing the effects of covariates in cohort k. Parameters of interest are
vector of coefficients characterizing the effects of covariates in cohort k. Parameters of interest are
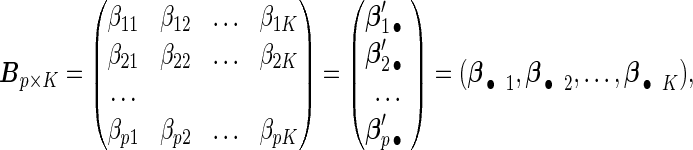 |
where each row βj •′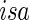 K×1 vector denoting the effects of the jth covariate across K studies. The Cox PH model assumes that covariates have multiplicative effects on the hazard function of failure time and yields interpretation of the coefficients as hazard ratios. Furthermore, the Cox PH model is commonly used to analyze NCC data because of its easy implementation using the partial likelihood technique (Thomas, 1979; Oakes, 1981). Under the Cox PH model and time-invariant covariates, the expression of Thomas’ partial likelihood function is equivalent to the conditional logistic likelihood. The study-specific log partial likelihood takes the form of
K×1 vector denoting the effects of the jth covariate across K studies. The Cox PH model assumes that covariates have multiplicative effects on the hazard function of failure time and yields interpretation of the coefficients as hazard ratios. Furthermore, the Cox PH model is commonly used to analyze NCC data because of its easy implementation using the partial likelihood technique (Thomas, 1979; Oakes, 1981). Under the Cox PH model and time-invariant covariates, the expression of Thomas’ partial likelihood function is equivalent to the conditional logistic likelihood. The study-specific log partial likelihood takes the form of
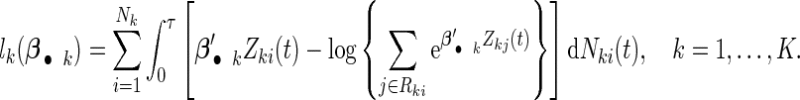 |
(2.2) |
Researchers often think that covariates have similar effects across pooled studies while acknowledging the existence of heterogeneity. This is essentially the idea behind the meta-analysis random-effects method in which study-specific effects are assumed to distribute around a central effect. Thus, it is natural to impose a group structure to each covariate's effects over K studies and select the covariate in the group manner. Denote the 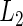 -norm of
-norm of 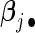 by
by 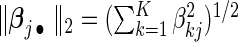 . The proposed agLASSO penalized partial likelihood estimator is defined as
. The proposed agLASSO penalized partial likelihood estimator is defined as
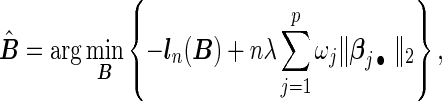 |
(2.3) |
where 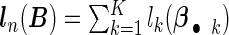 , n is the sample size of the pooled NCC studies, λ is a tuning parameter controlling the sparseness of the model, and
, n is the sample size of the pooled NCC studies, λ is a tuning parameter controlling the sparseness of the model, and 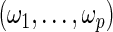 are data-dependent weights reflecting the relative importance of covariates. We choose
are data-dependent weights reflecting the relative importance of covariates. We choose 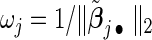 where
where 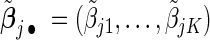 and
and 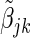 is the jth element of the maximum partial likelihood estimator for (2.2) within cohort k.
is the jth element of the maximum partial likelihood estimator for (2.2) within cohort k.
The convexity of the negative of log partial likelihood functions (2.2) (Goldstein and Langholz, 1992) and of the agLASSO penalty facilitates the optimization in (2.3). Meier and others (2008) propose a group coordinate gradient descent algorithm for the logistic regression with gLASSO penalty, and have implemented it in R-package grplasso. Using a quadratic approximation for the log-likelihood function, the algorithm iterates through each covariate group by first examining the penalized approximation function via the Karush–Kuhn–Tucker (KKT) condition to either set the coefficients to be exact zeros or estimate them to be non-zero, and then supplements the non-zero estimates with an inexact line search until convergence. We adapt the algorithm to our context with the partial likelihood function for NCC data and with adaptive weights.
To select the tuning parameter, we use a BIC-type criterion because of its consistence property for the adaptive LASSO estimation with least square approximation (Wang and Leng, 2007). Our numerical experiences also suggest the superior performance of the BIC-type criterion. Specifically, 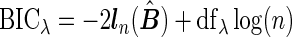 , where the degree of freedom
, where the degree of freedom 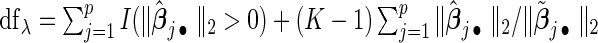 following Yuan and Lin (2006) for the gLASSO estimatorsQ6.
following Yuan and Lin (2006) for the gLASSO estimatorsQ6.
The proposed agLASSO method selects important variables that have large group norms of their effects over K studies and accommodates heterogeneity by allowing the variable to have different magnitudes or directions for its effects across studies. Sometimes it is also important to know which covariates have heterogeneous effects for model building using the pooling strategy, because heterogeneous effects need to be modeled by distinct parameters to avoid misrepresenting effects; the homogenous effect can be represented by a common coefficient across studies to reduce the model's complexity and improve efficiency. Therefore, we next develop a method to identify variables with heterogeneous effects.
2.2. Identification of heterogeneous effects
We reparameterize the cohort-specific Cox model (2.1) into
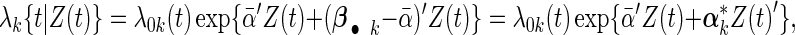 |
(2.4) |
where the 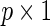 vector
vector 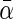 denotes the average covariate effects and
denotes the average covariate effects and 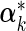 denotes the deviation of covariate effects in cohort k from
denotes the deviation of covariate effects in cohort k from 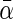 . To incorporate the constraint of
. To incorporate the constraint of 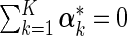 , we use the “sum to zero contrast” matrix
, we use the “sum to zero contrast” matrix 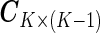 (e.g. contr.sum in R) and transform the original parameter matrix
(e.g. contr.sum in R) and transform the original parameter matrix 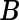 into
into
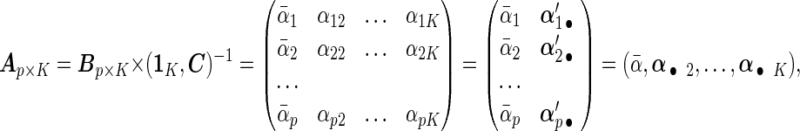 |
where 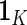 is a vector of
is a vector of 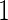 's. Now heterogeneous effects of each variable are represented by the coefficients of interaction terms of the variable with the contrast created from cohort membership, motivating us to consider the hierarchical selection method with composite absolute penalty (CAP) for identifying interactions (Zhao and others, 2009). We propose to estimate
's. Now heterogeneous effects of each variable are represented by the coefficients of interaction terms of the variable with the contrast created from cohort membership, motivating us to consider the hierarchical selection method with composite absolute penalty (CAP) for identifying interactions (Zhao and others, 2009). We propose to estimate 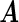 by minimizing the following cagLASSO penalized partial likelihood function, i.e.
by minimizing the following cagLASSO penalized partial likelihood function, i.e.
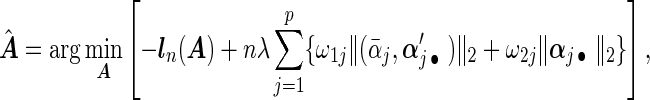 |
(2.5) |
where 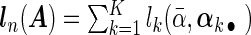 ,
, 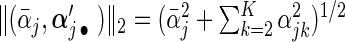 ,
, 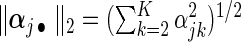 , and weights
, and weights 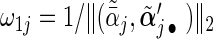 and
and 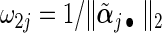 with
with 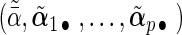 being the maximizer of
being the maximizer of 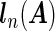 . The two penalty terms in (2.5) have overlap on αj • and thus can yield a hierarchical selection that once heterogeneous effects αj • deviate from zero, both
. The two penalty terms in (2.5) have overlap on αj • and thus can yield a hierarchical selection that once heterogeneous effects αj • deviate from zero, both 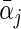 and αj • will be estimated to be non-zero together (Zhao and others, 2009). The extra penalty on αj • leads to the identification of only a non-zero average effect without heterogeneous effects.
and αj • will be estimated to be non-zero together (Zhao and others, 2009). The extra penalty on αj • leads to the identification of only a non-zero average effect without heterogeneous effects.
The calculation of (2.5) can be carried out by slightly modifying the algorithm described in Section 2.1. Specifically, in each iteration over the group of coefficients of the jth covariate, we examine the penalized approximation function via two KKT conditions to either set both 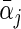 and αj • to be zero, or only set αj •=0 and obtain a non-zero estimate for
and αj • to be zero, or only set αj •=0 and obtain a non-zero estimate for 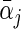 , or estimate both to be non-zero. We apply the inexact line search to the non-zero estimates and iterate the algorithm until convergence. The tuning parameter is selected using
, or estimate both to be non-zero. We apply the inexact line search to the non-zero estimates and iterate the algorithm until convergence. The tuning parameter is selected using 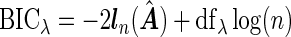 with the
with the 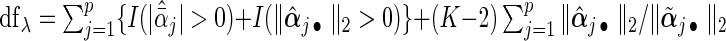 .
.
2.3. Theoretical properties
We study the asymptotic properties of the proposed agLASSO and cagLASSO estimators, with respect to estimation consistency, selection consistency, and oracle property. For the discussion of the agLASSO estimator, we assume that the first 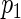 rows of the true parameter matrix
rows of the true parameter matrix 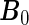 are the effects of important variables, i.e. ∥β0j •∥2>0
are the effects of important variables, i.e. ∥β0j •∥2>0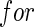 j≤p1 and
j≤p1 and 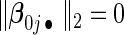 for
for 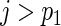 . We reorganize the parameters into a vector form as β=(β′1 •,…,β′p •)′ and decompose it into βa=(β′1 •,…,β′p1 •)′ and βb=(β′(p1+1) •,…,β′p •)′. Accordingly, we have β0=(β′0a,β′0b)′ and
. We reorganize the parameters into a vector form as β=(β′1 •,…,β′p •)′ and decompose it into βa=(β′1 •,…,β′p1 •)′ and βb=(β′(p1+1) •,…,β′p •)′. Accordingly, we have β0=(β′0a,β′0b)′ and 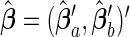 . We denote the Fisher information matrix of
. We denote the Fisher information matrix of 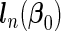 by
by 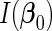 , which is positive-definite under the regularity conditions (1)–(6) given in Goldstein and Langholz (1992). Let
, which is positive-definite under the regularity conditions (1)–(6) given in Goldstein and Langholz (1992). Let 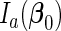 denote the upper left
denote the upper left 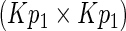 submatrix of
submatrix of 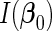 . Following the arguments in Wang and Leng (2008), we define
. Following the arguments in Wang and Leng (2008), we define 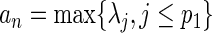 and
and 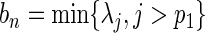 where
where 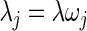 .
.
Theorem 2.1 —
Under the regularity conditions, the agLASSO estimator in (2.3) satisfies:
(a. Estimation consistency) if
, then
;
(b. Selection consistency) if
and
, then
;
(c. Oracle property) if
and
, then
.
For the ease of discussion of the cagLASSO estimator, we assume that the first 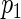 rows of true parameter matrix
rows of true parameter matrix 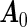 have ∥α′0j •∥2>0 the next p2 rows have
have ∥α′0j •∥2>0 the next p2 rows have 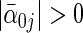 and ∥α′0j •∥2=0; the remaining rows have
and ∥α′0j •∥2=0; the remaining rows have 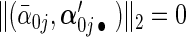 . Then we denote the vector form of
. Then we denote the vector form of 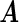 by
by 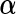 and decompose it into two parts:
and decompose it into two parts: 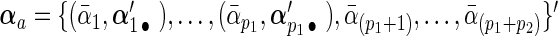 and
and 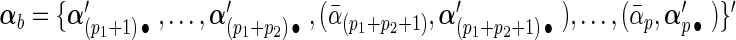 . Accordingly, let
. Accordingly, let 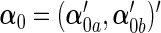 ,
, 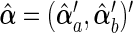 , and
, and 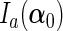 be the upper left
be the upper left 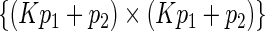 submatrix of the Fisher information matrix
submatrix of the Fisher information matrix 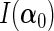 . We further define
. We further define 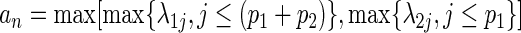 and
and 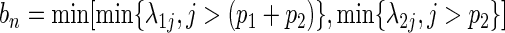 .
.
Theorem 2.2 —
Under the regularity conditions, the cagLASSO estimator in (2.5) satisfies:
(a. Estimation consistency) if
, then
;
(b. Selection consistency) if
and
, then
;
(c. Oracle property) if
and
, then
.
Proofs for the theorems are given in Appendix of supplementary material available at Biostatistics online. The theorems show that the agLASSO and cagLASSO estimators consistently select and estimate variables, and, furthermore, the cagLASSO estimator consistently identifies covariates with heterogeneous effects or with non-zero average effects. We estimate the covariance of the proposed estimators using the local quadratic approximation method proposed by Fan and Li (2001) to incorporate the penalty effect on finite sample estimation. We denote the non-zero components of the agLASSO estimator by 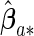 and estimate its covariance matrix by the following sandwich formula:
and estimate its covariance matrix by the following sandwich formula:
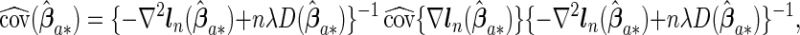 |
(2.6) |
where 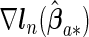 ,
, 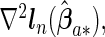 and
and 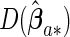 are the corresponding components of sample estimates for the gradient vector ∇ln(β)=∂ln(β)/∂β, hessian matrix ∇2ln(β)=∂2ln(β)/∂β∂β′, and D(β)=diag(ω11K/∥β1 •∥2,…,ωp1K/∥βp •∥2). For the cagLASSO estimator
are the corresponding components of sample estimates for the gradient vector ∇ln(β)=∂ln(β)/∂β, hessian matrix ∇2ln(β)=∂2ln(β)/∂β∂β′, and D(β)=diag(ω11K/∥β1 •∥2,…,ωp1K/∥βp •∥2). For the cagLASSO estimator 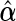 , denote its non-zero components by
, denote its non-zero components by 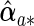 and estimate its covariance using (2.6) with D(β)
and estimate its covariance using (2.6) with D(β)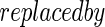 D(α)=diag{D1(α),…,Dp(α)} where
D(α)=diag{D1(α),…,Dp(α)} where 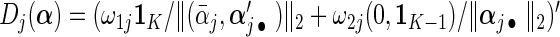 .
.
3. Numerical studies
3.1. Simulations to evaluate the agLASSO method
We simulated a pooled study consisting of NCC samples from three parent cohorts with sample sizes of 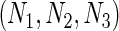 . For subjects in cohort k, we generated failure times from the Cox PH model
. For subjects in cohort k, we generated failure times from the Cox PH model 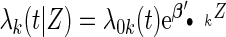 , where covariate vector Z was a 20D multivariate normal random vector with mean of 1, variance of 1, and pairwise correlation of
, where covariate vector Z was a 20D multivariate normal random vector with mean of 1, variance of 1, and pairwise correlation of 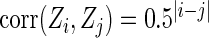 . Four covariates
. Four covariates 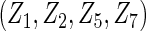 were associated with disease risk and the rest were assumed to be null covariates. Specifically, β1 •=(0.4,0.4,0.4) represented a homogeneous effect; β2 •=(0.4,0.6,0.3) was heterogeneous with small differences; β5 •=(−0.4,0,−0.3) was heterogeneous with moderate differences and had one zero effect; β7 •=(0,−0.7,0) was heterogeneous with big differences and had two zero effects. Four settings were used: (i) equal cohort size of
were associated with disease risk and the rest were assumed to be null covariates. Specifically, β1 •=(0.4,0.4,0.4) represented a homogeneous effect; β2 •=(0.4,0.6,0.3) was heterogeneous with small differences; β5 •=(−0.4,0,−0.3) was heterogeneous with moderate differences and had one zero effect; β7 •=(0,−0.7,0) was heterogeneous with big differences and had two zero effects. Four settings were used: (i) equal cohort size of 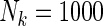 and an equal baseline rate
and an equal baseline rate 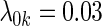 ; (ii)
; (ii) 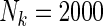 and equal baseline rate
and equal baseline rate 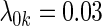 ; (iii) different a cohort sizes of
; (iii) different a cohort sizes of 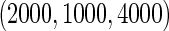 and equal baseline disease rate of
and equal baseline disease rate of 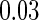 ; and (iv) different cohort sizes of
; and (iv) different cohort sizes of 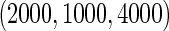 and different baseline disease rates of
and different baseline disease rates of 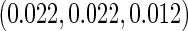 . Censoring times were generated from a uniform distribution
. Censoring times were generated from a uniform distribution 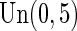 and yielded censoring rates ranging from
and yielded censoring rates ranging from 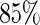 to
to 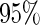 with different settings. We conducted 200 runs of simulations for each setting.
with different settings. We conducted 200 runs of simulations for each setting.
We compared the performance of our proposed agLASSO method with (i) cohort-specific method analyzing each cohort separately; (ii) pooled method ignoring any heterogeneity; (iii) meta-analyses using random- and fixed- effects models; and (iv) the gLASSO method. The cohort-specific method selects variables based on the 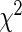 -test combining cohort-specific estimates, and the pooled and meta-analysis methods select variables based on the Wald test, all at the
-test combining cohort-specific estimates, and the pooled and meta-analysis methods select variables based on the Wald test, all at the 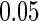 significance level. Table 1 summarizes the model selection results. The average mean squared error (MSE)
significance level. Table 1 summarizes the model selection results. The average mean squared error (MSE) 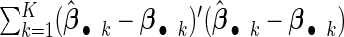 is used to measure the prediction accuracy, and the relative MSE (RMSE) with respect to the result of the cohort-specific method is also reported. Overall, our proposed method outperforms all other competitors in terms of being the closest to the true model size, the smallest number of false positives, the highest percentage of correctly fitted models, and having the smallest MSE. The proposed agLASSO method improves its performance as sample size increases, and handles different situations of heterogeneity reasonably well. Although the cohort-specific method is unbiased for estimation, it shows the largest MSEs in all settings due to large variances from only using the data of each sub-study. The meta-analysis methods generally improves the model selection, but the random-effects method can be very conservative, partially because when large heterogeneous effects are modeled by a random effect, the estimated variance for the random effect would mask the central effect. The gLASSO method tends to over-select variables with large numbers of false positives.
is used to measure the prediction accuracy, and the relative MSE (RMSE) with respect to the result of the cohort-specific method is also reported. Overall, our proposed method outperforms all other competitors in terms of being the closest to the true model size, the smallest number of false positives, the highest percentage of correctly fitted models, and having the smallest MSE. The proposed agLASSO method improves its performance as sample size increases, and handles different situations of heterogeneity reasonably well. Although the cohort-specific method is unbiased for estimation, it shows the largest MSEs in all settings due to large variances from only using the data of each sub-study. The meta-analysis methods generally improves the model selection, but the random-effects method can be very conservative, partially because when large heterogeneous effects are modeled by a random effect, the estimated variance for the random effect would mask the central effect. The gLASSO method tends to over-select variables with large numbers of false positives.
Table 1.
Simulation results on model selection and MSE
| Method | No. of identified variables | No. of false positives | No. of false negatives | Under-fitted% | Over-fitted% | Correct-fitted% | MSE | RMSE% |
|---|---|---|---|---|---|---|---|---|
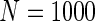 | ||||||||
| Cohort specific | 4.700 | 1.045 | 0.345 | 31.0 | 38.5 | 30.5 | 2.273 | REF |
| Pooled analysis | 4.190 | 0.835 | 0.645 | 55.0 | 25.0 | 20.0 | 1.024 | 45.1 |
| Meta-random | 2.915 | 0.645 | 1.730 | 98.0 | 1.5 | 0.5 | 1.203 | 52.9 |
| Meta-fixed | 4.050 | 0.860 | 0.810 | 64.5 | 18.0 | 17.5 | 1.170 | 51.5 |
| gLASSO | 4.350 | 0.550 | 0.200 | 18.5 | 37.0 | 44.5 | 0.532 | 23.4 |
| agLASSO | 3.635 | 0.140 | 0.505 | 38.0 | 9.5 | 52.5 | 0.489 | 21.5 |
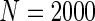 | ||||||||
| Cohort specific | 4.840 | 0.860 | 0.020 | 2.0 | 51.0 | 47.0 | 0.887 | REF |
| Pooled analysis | 4.625 | 0.700 | 0.075 | 7.5 | 46.5 | 46.0 | 0.717 | 80.8 |
| Meta-random | 3.065 | 0.600 | 1.535 | 99.5 | 0.5 | 0 | 0.758 | 85.5 |
| Meta-fixed | 4.700 | 0.805 | 0.105 | 10.5 | 49.5 | 40.0 | 0.751 | 84.7 |
| gLASSO | 4.850 | 0.855 | 0.005 | 0.5 | 61.5 | 38.0 | 0.295 | 33.3 |
| agLASSO | 3.945 | 0.045 | 0.100 | 10.0 | 4.5 | 85.5 | 0.218 | 24.6 |
| Different cohort sizes | ||||||||
| Cohort specific | 4.865 | 0.930 | 0.065 | 6.5 | 58.0 | 35.5 | 1.212 | REF |
| Pooled analysis | 4.280 | 0.900 | 0.620 | 62.0 | 21.5 | 16.5 | 0.750 | 61.9 |
| Meta-random | 3.240 | 0.645 | 1.405 | 99.5 | 0 | 0.5 | 0.759 | 62.6 |
| Meta-fixed | 4.185 | 0.905 | 0.750 | 72.0 | 18.0 | 10.0 | 0.793 | 65.4 |
| gLASSO | 4.840 | 0.840 | 0.001 | 0 | 58.5 | 41.5 | 0.347 | 28.6 |
| agLASSO | 4.105 | 0.150 | 0.045 | 4.0 | 14.0 | 82.0 | 0.262 | 21.6 |
| Different cohort sizes and disease rates | ||||||||
| Cohort specific | 4.600 | 0.860 | 0.260 | 24.5 | 41.5 | 34.0 | 1.926 | REF |
| Pooled analysis | 4.100 | 0.790 | 0.690 | 63.5 | 17.5 | 19.0 | 0.877 | 45.5 |
| Meta-random | 3.070 | 0.595 | 1.525 | 98.0 | 0.5 | 1.5 | 0.973 | 50.5 |
| Meta-fixed | 3.965 | 0.790 | 0.825 | 76.0 | 9.0 | 15.0 | 0.970 | 50.4 |
| gLASSO | 4.765 | 0.810 | 0.045 | 4.0 | 52.0 | 44.0 | 0.457 | 23.7 |
| agLASSO | 3.910 | 0.225 | 0.315 | 28.5 | 16.5 | 55.0 | 0.413 | 21.4 |
“Cohort specific” refers to the method treating each cohort separately and the significance test is based on the χ2-test combining cohort-specific results; “Pooled analysis” refers to the pooled analysis ignoring any heterogeneity; “Meta-random” refers to the meta-analysis with random-effects modeling; “Meta-fixed” refers to the meta-analysis with fixed-effects modeling; “gLASSO” refers to the group LASSO method; “agLASSO” refers to the adaptive group LASSO method; “No. of identified variables” refers to the average number of identified variables by each method in 200 runs of simulations; “No. of false positive” refers to the average number of selected variables out of those true values being 0 by each method in 200 runs of simulations; “No. of false negatives” refers to the average number of missed variables out of those true non-zero values by each method in 200 runs of simulations; “Under-fitted%” refers to the percentage of simulation runs that miss at least one true variable; “Over-fitted%” refers to the percentage of simulation runs that include all true variables and at least one null covariate; “Correct-fitted%” refers to the percentage of simulation runs that correctly identify all variables.
Table 2 presents the results on selecting individual variables. For covariates 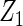 and
and 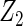 where effect signals are strong with no or minor heterogeneity, all methods can pick up the signal with good power. For null covariates
where effect signals are strong with no or minor heterogeneity, all methods can pick up the signal with good power. For null covariates 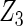 and
and 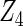 , the proposed agLASSO method shows the best performance excluding these variables. For covariates
, the proposed agLASSO method shows the best performance excluding these variables. For covariates 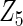 and
and 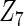 with moderate or large heterogeneity, the cohort-specific method, and the penalized methods of gLASSO and agLASSO perform well. The pooled analysis method deteriorates dramatically for
with moderate or large heterogeneity, the cohort-specific method, and the penalized methods of gLASSO and agLASSO perform well. The pooled analysis method deteriorates dramatically for 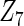 under the settings of different cohort sizes and/or different disease rates that aggravate the heterogeneity across cohorts. Meta-analysis methods yield unsatisfactory results as well.
under the settings of different cohort sizes and/or different disease rates that aggravate the heterogeneity across cohorts. Meta-analysis methods yield unsatisfactory results as well.
Table 2.
Simulation results on individual variable selection frequency 
| Method | 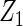 |
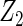 |
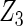 |
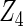 |
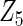 |
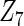 |
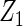 |
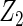 |
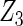 |
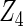 |
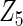 |
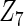 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
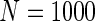 |
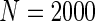 |
|||||||||||
| Cohort specific | 98.0 | 98.5 | 5.0 | 8.5 | 75.5 | 93.5 | 100 | 100 | 8.0 | 5.0 | 98.0 | 100 |
| Pooled analysis | 99.5 | 99.0 | 4.0 | 3.0 | 67.5 | 69.5 | 100 | 100 | 4.0 | 4.0 | 96.0 | 96.5 |
| Meta-random | 97.5 | 89.5 | 2.0 | 3.5 | 35.5 | 4.5 | 100 | 97.5 | 4.0 | 5.0 | 47.5 | 1.5 |
| Meta-fixed | 99.0 | 98.5 | 2.5 | 4.0 | 67.0 | 54.5 | 100 | 100 | 6.0 | 6.0 | 96.5 | 93.0 |
| gLASSO | 100 | 99.5 | 4.0 | 3.5 | 82.0 | 98.5 | 100 | 100 | 5.0 | 3.5 | 99.5 | 100 |
| agLASSO | 91.5 | 96.5 | 1.0 | 1.5 | 68.0 | 93.5 | 100 | 100 | 0.0 | 0.0 | 90.0 | 100 |
| Different cohort sizes | Different cohort sizes and disease rates | |||||||||||
| Cohort specific | 100 | 100 | 4.5 | 4.0 | 99.0 | 94.5 | 100 | 100 | 6.5 | 5.5 | 89.0 | 85.0 |
| Pooled analysis | 100 | 100 | 5.5 | 4.0 | 99.5 | 38.5 | 100 | 100 | 6.0 | 4.0 | 91.5 | 39.5 |
| Meta-random | 100 | 98.5 | 3.5 | 3.5 | 60.0 | 1.0 | 97.5 | 94.0 | 4.0 | 4.0 | 53.5 | 2.5 |
| Meta-fixed | 100 | 100 | 4.0 | 4.0 | 99.5 | 28.5 | 100 | 100 | 4.5 | 5.5 | 91.0 | 26.5 |
| gLASSO | 100 | 100 | 3.5 | 3.0 | 100 | 100 | 100 | 100 | 5.0 | 5.0 | 97.0 | 98.5 |
| agLASSO | 100 | 99.5 | 1.0 | 1.0 | 96.5 | 99.5 | 98.0 | 99.5 | 0.5 | 0.5 | 79.5 | 91.5 |
“Cohort specific” refers to the method treating each cohort separately and the significance test is based on the χ2-test combining cohort-specific results; “Pooled analysis” refers to the pooled analysis ignoring any heterogeneity; “Meta-random” refers to the metaanalysis with random-effects modeling; “Meta-fixed” refers to the meta-analysis with fixed-effects modeling; “gLASSO” refers to the group LASSO method; “agLASSO” refers to the adaptive group LASSO method.
To examine the accuracy of the proposed variance calculation in Section 2.3, we compare the sample standard deviation of estimates over simulations and the average formula-based estimates of standard errors in Table 3. First, the proposed agLASSO method shows smaller standard errors than the cohort-specific method across the board, indicating that our proposed method can improve efficiency by integrating information across multiple studies. Second, when heterogeneity is small in 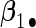 and
and 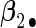 , the agLASSO asymptotic variance estimates are close to their sample counterparts, especially when the sample size is large. Third, for
, the agLASSO asymptotic variance estimates are close to their sample counterparts, especially when the sample size is large. Third, for 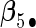 and
and 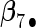 with moderate or large heterogeneity, the agLASSO asymptotic variance estimates display some discrepancy with the sample standard deviations. One explanation could be that applying the same group penalty to a heterogeneous vector tends to penalize the small coefficients more than the large ones, and thus the estimates for small coefficients demonstrate smaller variability.
with moderate or large heterogeneity, the agLASSO asymptotic variance estimates display some discrepancy with the sample standard deviations. One explanation could be that applying the same group penalty to a heterogeneous vector tends to penalize the small coefficients more than the large ones, and thus the estimates for small coefficients demonstrate smaller variability.
Table 3.
Simulation results on the standard errors for the estimates
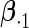 |
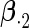 |
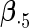 |
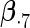 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | 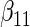 |
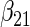 |
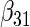 |
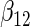 |
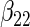 |
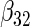 |
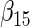 |
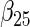 |
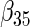 |
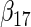 |
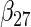 |
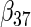 |
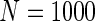 | ||||||||||||
| Cohort specific | 0.184 | 0.193 | 0.184 | 0.211 | 0.205 | 0.191 | 0.188 | 0.201 | 0.193 | 0.174 | 0.229 | 0.195 |
| 0.166 | 0.173 | 0.165 | 0.186 | 0.199 | 0.183 | 0.185 | 0.188 | 0.182 | 0.180 | 0.202 | 0.180 | |
| agLASSO | 0.134 | 0.139 | 0.132 | 0.127 | 0.160 | 0.121 | 0.101 | 0.071 | 0.093 | 0.085 | 0.153 | 0.085 |
| 0.124 | 0.124 | 0.125 | 0.124 | 0.126 | 0.125 | 0.119 | 0.119 | 0.121 | 0.115 | 0.118 | 0.117 | |
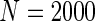 | ||||||||||||
| Cohort specific | 0.108 | 0.101 | 0.124 | 0.119 | 0.119 | 0.124 | 0.121 | 0.116 | 0.119 | 0.116 | 0.125 | 0.118 |
| 0.109 | 0.113 | 0.110 | 0.121 | 0.129 | 0.120 | 0.121 | 0.123 | 0.121 | 0.119 | 0.131 | 0.119 | |
| agLASSO | 0.094 | 0.090 | 0.103 | 0.098 | 0.111 | 0.090 | 0.095 | 0.047 | 0.082 | 0.068 | 0.104 | 0.067 |
| 0.092 | 0.093 | 0.093 | 0.092 | 0.094 | 0.092 | 0.086 | 0.086 | 0.086 | 0.084 | 0.087 | 0.085 | |
| Different cohort sizes | ||||||||||||
| Cohort specific | 0.108 | 0.189 | 0.078 | 0.119 | 0.209 | 0.086 | 0.121 | 0.185 | 0.086 | 0.116 | 0.195 | 0.088 |
| 0.109 | 0.172 | 0.075 | 0.121 | 0.196 | 0.083 | 0.121 | 0.187 | 0.083 | 0.119 | 0.200 | 0.082 | |
| agLASSO | 0.095 | 0.127 | 0.079 | 0.102 | 0.145 | 0.079 | 0.094 | 0.080 | 0.067 | 0.062 | 0.152 | 0.039 |
| 0.091 | 0.120 | 0.068 | 0.092 | 0.122 | 0.068 | 0.085 | 0.116 | 0.062 | 0.084 | 0.117 | 0.062 | |
| Different cohort sizes and disease rates | ||||||||||||
| Cohort specific | 0.135 | 0.229 | 0.126 | 0.139 | 0.238 | 0.139 | 0.157 | 0.255 | 0.147 | 0.136 | 0.289 | 0.121 |
| 0.128 | 0.203 | 0.120 | 0.141 | 0.233 | 0.131 | 0.142 | 0.222 | 0.131 | 0.139 | 0.238 | 0.130 | |
| agLASSO | 0.115 | 0.142 | 0.110 | 0.114 | 0.162 | 0.106 | 0.105 | 0.079 | 0.080 | 0.067 | 0.179 | 0.060 |
| 0.104 | 0.134 | 0.099 | 0.104 | 0.135 | 0.099 | 0.098 | 0.132 | 0.093 | 0.096 | 0.131 | 0.092 | |
“Cohort specific” refers to the method treating each cohort separately; “agLASSO” refers to the adaptive group LASSO method. For each parameter's estimate, sample standard deviation of estimates is on the top and the average of estimated standard errors is at the bottom.
Additional simulations with a cohort size of 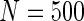 were also conducted and showed that all methods had low power to select important variables when the sample size is small. The penalized methods of gLASSO and agLASSO generally exhibited better performances than others, but the superiority of the agLASSO over gLASSO was not obvious under situations with low sample size.
were also conducted and showed that all methods had low power to select important variables when the sample size is small. The penalized methods of gLASSO and agLASSO generally exhibited better performances than others, but the superiority of the agLASSO over gLASSO was not obvious under situations with low sample size.
3.2. Simulations to evaluate the cagLASSO method
We compared the proposed cagLASSO method, the interaction method including marker×cohort-membership terms, and the CAP method with group 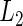 -penalties (Zhao and others, 2009), regarding their performance for identifying important variables and variables with heterogeneous effects. Data generation remained the same as in Section 3.1. Tables 4 and 5 summarize the results on overall model selection and individual variable selection, respectively. The proposed cagLASSO approach accurately selects the four important variables and yields the smallest MSE under all settings (Table 4). Note that three out of the four important variables have heterogeneous effects and demonstrate different magnitudes of heterogeneity. Although the cagLASSO method seems to be inferior to the other two methods in selecting variables with heterogeneous effects, it has comparable performance for selecting the correct heterogeneous variables. From Table 5, we note that the cagLASSO method has the lowest false-positive rate to correctly recognize the absence of heterogeneity in
-penalties (Zhao and others, 2009), regarding their performance for identifying important variables and variables with heterogeneous effects. Data generation remained the same as in Section 3.1. Tables 4 and 5 summarize the results on overall model selection and individual variable selection, respectively. The proposed cagLASSO approach accurately selects the four important variables and yields the smallest MSE under all settings (Table 4). Note that three out of the four important variables have heterogeneous effects and demonstrate different magnitudes of heterogeneity. Although the cagLASSO method seems to be inferior to the other two methods in selecting variables with heterogeneous effects, it has comparable performance for selecting the correct heterogeneous variables. From Table 5, we note that the cagLASSO method has the lowest false-positive rate to correctly recognize the absence of heterogeneity in 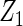 . For
. For 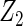 with small heterogeneous effects, none of the methods detects the heterogeneity signal with good power. For variables of
with small heterogeneous effects, none of the methods detects the heterogeneity signal with good power. For variables of 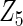 and
and 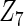 with moderate or large heterogeneity, the cohort-specific method works generally well, but performances by the CAP estimator and the cagLASSO estimator fluctuate. In general, we find that the cagLASSO estimator shows better performance than the CAP method for
with moderate or large heterogeneity, the cohort-specific method works generally well, but performances by the CAP estimator and the cagLASSO estimator fluctuate. In general, we find that the cagLASSO estimator shows better performance than the CAP method for 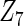 with large heterogeneity but lower selection frequency for
with large heterogeneity but lower selection frequency for 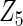 with moderate heterogeneity. This observation can be due to the fact that, in finite samples, the data-adaptive weight
with moderate heterogeneity. This observation can be due to the fact that, in finite samples, the data-adaptive weight 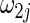 in (2.5) may over-penalize the heterogeneous effects when the group norm of heterogeneous effect is small.
in (2.5) may over-penalize the heterogeneous effects when the group norm of heterogeneous effect is small.
Table 4.
Simulation results on model selection and identification of heterogeneous effects
| Method | No. of identified variable (4) | No. of correct variable | No. of identified hetero. eff. (3) | No. of correct hetero. eff. | MSE |
|---|---|---|---|---|---|
 | |||||
| Cohort interaction | 4.350 | 3.295 | 2.500 | 1.425 | 1.242 |
| CAP | 4.255 | 3.690 | 1.565 | 1.480 | 0.333 |
| cagLASSO | 3.910 | 3.670 | 1.325 | 1.280 | 0.280 |
 | |||||
| Cohort interaction | 4.720 | 3.910 | 2.840 | 1.930 | 0.497 |
| CAP | 4.575 | 3.985 | 2.220 | 2.080 | 0.234 |
| cagLASSO | 4.105 | 3.960 | 1.740 | 1.730 | 0.164 |
| Different cohort sizes | |||||
| Cohort interaction | 4.715 | 3.805 | 2.640 | 1.650 | 0.708 |
| CAP | 4.265 | 3.690 | 1.580 | 1.400 | 0.327 |
| cagLASSO | 3.950 | 3.760 | 1.240 | 1.180 | 0.259 |
| Different cohort sizes and disease rates | |||||
| Cohort interaction | 4.312 | 3.497 | 2.382 | 1.372 | 1.147 |
| CAP | 4.106 | 3.613 | 1.327 | 1.201 | 0.362 |
| cagLASSO | 3.900 | 3.678 | 1.186 | 1.106 | 0.293 |
“Cohort interaction” refers to the pooled analysis using the interaction terms of covariates with cohort-specific membership indicators; “CAP” refers to the composite absolute penalty method; “cagLASSO” refers to the composite adaptive group lasso penalty method; “No. of identified variable” refers to the average number of identified important variables with non-zero effect by each method in 200 runs of simulations; “No. of correct variable” refers to the average number of identified variables with non-zero effect out of those with true non-zero effects; “No. of identified hetero. eff.” refers to the average number of identified variables with non-zero heterogeneous effect by each method in 200 runs of simulations; “No. of correct hetero. eff.” refers to the average number of identified variables with non-zero heterogeneous effect out of those with true non-zero heterogeneous effects.
Table 5.
Simulation results on the variable selection frequency 
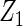 |
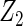 |
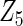 |
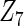 |
|||||
|---|---|---|---|---|---|---|---|---|
| Method | Ave. (0.4) | Hetero. (0, 0) | Ave. (0.43) | Hetero. (0.17, −0.13) | Ave. (−0.23) | Hetero. (0.23, −0.06) | Ave. (−0.23) | Hetero. (−0.47, 0.23) |
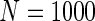 | ||||||||
| Cohort interaction | 99.0 | 8.5 | 98.5 | 19.0 | 67.0 | 36.5 | 65.0 | 87.0 |
| CAP | 100 | 2.0 | 99.0 | 19.0 | 82.0 | 42.5 | 88.0 | 86.5 |
| cagLASSO | 99.0 | 1.5 | 98.5 | 10.5 | 77.5 | 33.0 | 92.0 | 84.5 |
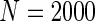 | ||||||||
| Cohort interaction | 100 | 5.0 | 100 | 35.0 | 95.5 | 58.0 | 95.5 | 100 |
| CAP | 100 | 5.5 | 100 | 32.5 | 98.5 | 75.5 | 100 | 100 |
| cagLASSO | 100 | 0.0 | 100 | 19.0 | 96.0 | 54.5 | 100 | 99.5 |
| Different cohort sizes | ||||||||
| Cohort interaction | 100 | 6.5 | 100 | 29.0 | 88.0 | 40.0 | 92.5 | 96.0 |
| CAP | 100 | 4.0 | 100 | 26.5 | 100 | 44.5 | 69.0 | 69.0 |
| cagLASSO | 100 | 1.5 | 100 | 19.0 | 99.5 | 23.5 | 76.5 | 75.5 |
| Different cohort sizes and different disease rates | ||||||||
| Cohort interaction | 100 | 6.5 | 100 | 23.0 | 71.5 | 31.5 | 78.5 | 82.5 |
| CAP | 100 | 4.0 | 100 | 18.0 | 97.5 | 38.0 | 64.0 | 64.0 |
| cagLASSO | 100 | 1.5 | 100 | 15.0 | 93.0 | 23.0 | 75.0 | 72.5 |
“Cohort interaction” refers to the pooled analysis using the interaction terms of covariates with cohort-specific membership indicators; “CAP” refers to the composite absolute penalty method; “cagLASSO” refers to the composite adaptive group LASSO penalty method; “Ave.” refers to the average effect of the covariate and its true value is listed below in the parenthesis; “Hetero.” refers to the heterogeneous effects of the covariate and the true values are listed below in the parenthesis.
3.3. The pooled ovarian cancer study
We applied the proposed agLASSO method to the pooled NCC study on ovarian cancer to identify important risk factors and compared its results with those from the cohort-specific analysis, pooled analysis, meta-analysis with random-effects and fixed-effects models, and gLASSO method. Specifically, we considered all 17 inflammation markers and 6 potential confounders including pregnancy history, use of oral contraceptive, use of hormone replacement therapy, age at menarche, body mass index, and current smoking status. The analysis was based on 229 ovarian cancer cases and 429 matched controls (NYUWHS: 81 cases and 160 controls; ORDET: 41 cases and 82 controls; NSHDS: 107 cases and 187 controls). Four subjects were removed due to missing data. All biomarker data were log-transformed and standardized within each cohort.
Both the gLASSO and agLASSO methods select interleukin-4 (IL4) as an important risk factor for ovarian cancer and thus we present the results from different methods focused on IL4 in Table 6. The cohort-specific method indicates that IL4 may have heterogeneous effects across three cohorts: it shows positive association with disease risk in NYUWHS 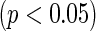 and NSHDS but negative association in ORDET although the results from NSHDS and ORDET are not statistically significant. The results are consistent with the findings in Clendenen and others (2011). The proposed agLASSO method reaches a similar conclusion as the cohort-specific analysis but with more regularized estimates and tighter confidence intervals by integrating three studies together. Under this pattern of heterogeneity, however, the pooled analysis or meta-analysis does not show any significant result. Furthermore, our proposed cagLASSO method also demonstrates the existence of heterogeneity in IL4 effects across three cohorts and estimates (in the log of hazards ratio scale) the average effect as
and NSHDS but negative association in ORDET although the results from NSHDS and ORDET are not statistically significant. The results are consistent with the findings in Clendenen and others (2011). The proposed agLASSO method reaches a similar conclusion as the cohort-specific analysis but with more regularized estimates and tighter confidence intervals by integrating three studies together. Under this pattern of heterogeneity, however, the pooled analysis or meta-analysis does not show any significant result. Furthermore, our proposed cagLASSO method also demonstrates the existence of heterogeneity in IL4 effects across three cohorts and estimates (in the log of hazards ratio scale) the average effect as 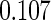 and the heterogeneous effects as
and the heterogeneous effects as 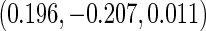 for NYUWHS, ORDET, and NSHDS, respectively.
for NYUWHS, ORDET, and NSHDS, respectively.
Table 6.
Results on estimation of the effects of IL4 in the ovarian cancer study
| Method | HR | 95% CI |
|---|---|---|
| Cohort specific | ||
| NYU | 2.596 | (1.211, 5.565)* |
| ORDET | 0.568 | (0.278, 1.161) |
| NSHDS | 1.833 | (0.822, 4.086) |
| Pooled analysis | 1.260 | (0.905, 1.755) |
| Meta-random | 1.377 | (0.551, 3.439) |
| Meta-fixed | 1.326 | (0.856, 2.053) |
| agLASSO | ||
| NYU | 1.343 | (1.033, 1.747)* |
| ORDET | 0.847 | (0.614, 1.168) |
| NSHDS | 1.088 | (0.872, 1.357) |
“HR” refers to the hazards ratio estimate corresponding to 1 standard deviation increase of the IL4 level (in the logscale); “95% CI” refers to the 95% confidence interval. *The statistical significance at 0.05 level.
4. Discussion
In this article, we develop the penalized partial likelihood methods for variable selection and estimation of the Cox PH model in pooled NCC studies. The proposed methods can be easily extended to pooled analysis of other types of data, or even for combining studies with different designs, such as matched case–control and unmatched case–control studies. Other choices of penalty function can also be used to incorporate investigators’ prior knowledge on the magnitude or structure of heterogeneity. For example, we can use the group Lq penalty with 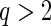 to shrink coefficients toward the diagonal and thus can encourage similarity of effects across multiple studies; the proposed cagLASSO method can adopt other penalties with hierarchical structure induced from a directed graph. Also from our numerical experience, when the sample size is sufficiently large, the simple definition for the degree of freedom that counts the number of non-zero coefficient estimates (Wang and others, 2007) can be used for calculating the BIC to select the final model. After significant heterogeneity is detected across studies, the sparse gLASSO penalty also can be used to select cohort-specific important variables.
to shrink coefficients toward the diagonal and thus can encourage similarity of effects across multiple studies; the proposed cagLASSO method can adopt other penalties with hierarchical structure induced from a directed graph. Also from our numerical experience, when the sample size is sufficiently large, the simple definition for the degree of freedom that counts the number of non-zero coefficient estimates (Wang and others, 2007) can be used for calculating the BIC to select the final model. After significant heterogeneity is detected across studies, the sparse gLASSO penalty also can be used to select cohort-specific important variables.
Using the adaptive weights in our penalized methods is important for achieving the oracle property and good practical performance. When the number of covariates is large, the data-dependent weights from the cohort-specific estimates may not always be estimable. We can use some initial estimates that are zero-consistent as the weights (Huang and others, 2006). Some other non-convex penalty regularized methods also have the oracle property, such as using the group bridge penalty (Ma and others, 2011), however, are more complex in terms of both numerical implementation and theory. In addition, when small heterogeneous effects are present, a very large sample would be necessary for the proposed methods to achieve a perfect fit on all variables.
Lastly, it is of great interest to study other survival models that can relax the PHs assumption for the analysis of pooled NCC studies with heterogeneity. The inverse selection probability weighted technique (Samuelsen, 1997) potentially can be used for this purpose, but the construction of an effective loss function that can simultaneously accommodate the selection weights and couple with penalties needs further investigation.
Supplementary material
Supplementary material is available at http://biostatistics.oxfordjournals.org.
Funding
This work was supported by the National Cancer Institute (R01 CA140632, R03 CA153083, R21 CA116585, R21 CA169739).
Supplementary Material
Acknowledgements
The authors would like thank the Editor, an Associate Editor, and two referees for their valuable suggestions. Conflict of Interest: None declared.
References
- Clendenen T. V., Lundin E., Zeleniuch-Jacquotte A., Koenig K. L., Berrino F., Lukanova A., Lokshin A. E., Idahl A., Ohlson N., Hallmans G. Circulating inflammation markers and risk of epithelial ovarian cancer. Cancer Epidemiology, Biomarkers & Prevention. 2011;20:799–810. doi: 10.1158/1055-9965.EPI-10-1180. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran W. G. The combination of estimates from different experiments. Biometrics. 1954;10:101. [Google Scholar]
- Cox D. R. Regression models and life-tables. Journal of the Royal Statistical Society Series B. 1972;34:187–220. [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Fan J., Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. Journal of the American Statistical Association. 2001;96:1348–1360. [Google Scholar]
- Friedenreich C. M. Methods for pooled analyses of epidemiologic studies. Epidemiology. 1993;4:295–302. doi: 10.1097/00001648-199307000-00004. [DOI] [PubMed] [Google Scholar]
- Goldstein L., Langholz B. Asymptotic theory for nested case-control sampling in the cox regression model. Annals of Statistics. 1992;20:1903–1928. [Google Scholar]
- Greenland S. Invited commentary: a critical look at some popular meta-analytic methods. American Journal of Epidemiology. 1994;140:290–296. doi: 10.1093/oxfordjournals.aje.a117248. [DOI] [PubMed] [Google Scholar]
- Hedges L. V., Olkin I. Statistical Methods for Meta-Analysis. Orlando: Academic Press; 1985. [Google Scholar]
- Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Huang J., Ma S., Zhang C.-H. Adaptive lasso for sparse high-dimensional regression models. Statistica Sinica. 2006;18:1603–1618. [Google Scholar]
- Ioannidis J. P. A., Rosenberg P. S., Goedert J. J., O’Brien T. R. Commentary: meta-analysis of individual participants’ data in genetic epidemiology. American Journal of Epidemiology. 2002;156:204–210. doi: 10.1093/aje/kwf031. [DOI] [PubMed] [Google Scholar]
- Liu F., Dunson D., Zou F. High-dimensional variable selection in meta-analysis for censored data. Biometrics. 2011;67:504–512. doi: 10.1111/j.1541-0420.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Huang J., Song X. Integrative analysis and variable selection with multiple high-dimensional data sets. Biostatistics. 2011a;12:763–775. doi: 10.1093/biostatistics/kxr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Huang J., Wei F., Xie Y., Fang K. Integrative analysis of multiple cancer prognosis studies with gene expression measurements. Statistics in Medicine. 2011b;30:3361–3371. doi: 10.1002/sim.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier L., Van De Geer S., Bühlmann P. The group lasso for logistic regression. Journal of the Royal Statistical Society Series B. 2008;70:53–71. [Google Scholar]
- Oakes D. Survival times: aspects of partial likelihood. International Statistical Review. 1981;49:235–252. [Google Scholar]
- Puniyani K., Kim S., Xing E. P. Multi-population GWA mapping via multi-task regularized regression. Bioinformatics. 2010;26:208–216. doi: 10.1093/bioinformatics/btq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsen S. O. A pseudolikelihood approach to analysis of nested case-control studies. Biometrika. 1997;84:379–394. [Google Scholar]
- Smith T. C., Spiegelhalter D. J., Thomas A. Bayesian approaches to random-effects meta-analysis: a comparative study. Statistics in Medicine. 1995;14:2685–2699. doi: 10.1002/sim.4780142408. [DOI] [PubMed] [Google Scholar]
- Thomas D. C. Addendum to methods of cohort analysis—appraisal by application to asbestos mining by Liddell F. D. K. and McDonald J. C. and and Thomas D. C. Journal of the Royal Statistical Society A. 1979;140:469–491. [Google Scholar]
- Thompson S. G., Higgins J. P. T. How should meta-regression analyses be undertaken and interpreted? Statistics in Medicine. 2002;21:1559–1573. doi: 10.1002/sim.1187. [DOI] [PubMed] [Google Scholar]
- Wang H., Leng C. Unified LASSO estimation by least squares approximation. Journal of the American Statistical Association. 2007;102:1039–1048. [Google Scholar]
- Wang H., Leng C. A note on adaptive group lasso. Computational Statistics & Data Analysis. 2008;52:5277–5286. [Google Scholar]
- Wang H., Li R., Tsai C.-L. Tuning parameter selectors for the smoothly clipped absolute deviation method. Biometrika. 2007;94:553–568. doi: 10.1093/biomet/asm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society Series B. 2006;68:49–67. [Google Scholar]
- Zhao P., Rocha G., Yu B. The composite absolute penalties family for grouped and hierarchical variable selection. The Annals of Statistics. 2009;37:3468–3497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


