Abstract
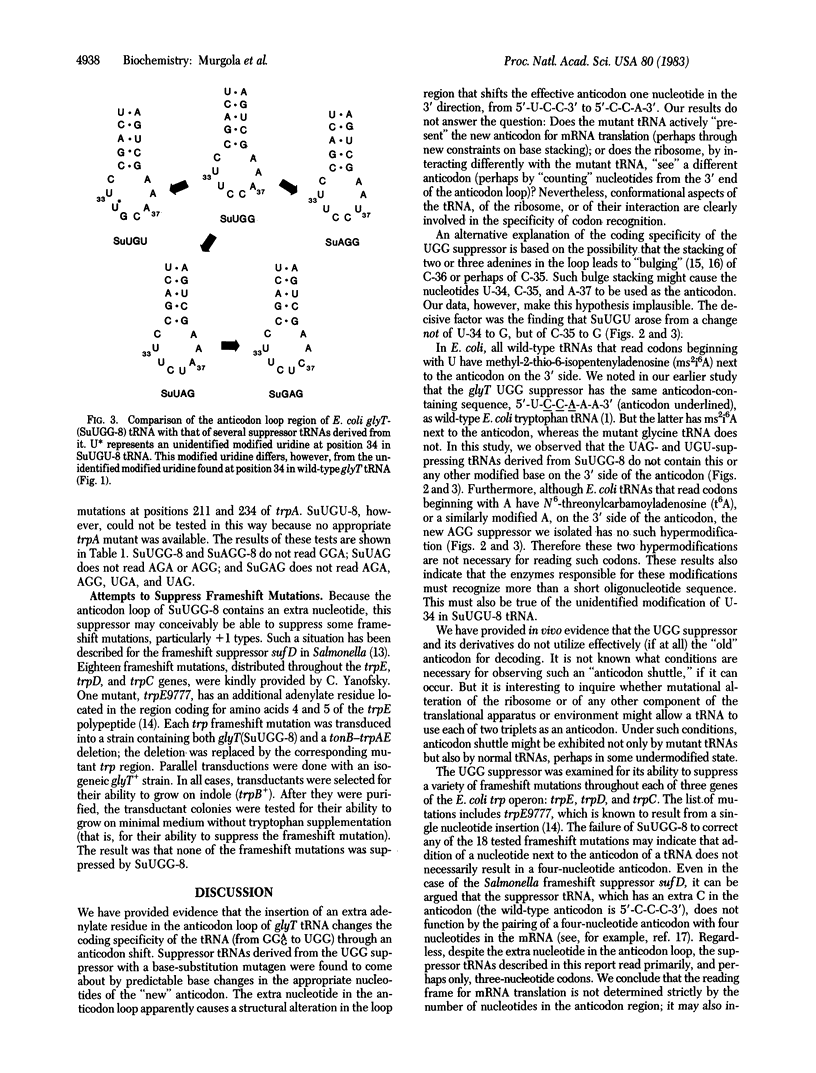
In a previous publication, an unusual UGG-reading missense suppressor caused by insertion of an extra adenylate residue in the anticodon loop of an Escherichia coli glycine tRNA was described. In this study, we provide in vivo evidence that the additional nucleotide causes an "anticodon shift" by one nucleotide in the 3' direction and that the "new" anticodon can explain the unanticipated coding properties of the suppressor. We converted the UGG suppressor with ethyl methanesulfonate, a base-substitution mutagen, to suppressors that read codons related to UGG by a single base change. Sequence analysis of each mutant tRNA revealed that its mutational alteration was an anticipated base change in one of the three nucleotides of the "new" anticodon. Although the new suppressors read codons beginning with A or U, the mutant tRNAs lack the customary hypermodified nucleosides on the 3' side of the anticodon. As determined on the basis of their in vivo coding specificities, the new mutant tRNAs do not continue to utilize the original anticodon triplet for decoding. Furthermore, the failure of the UGG suppressor to correct frameshift mutations throughout each of three genes of the trp operon suggests that the addition of a nucleotide to the anticodon loop of a tRNA does not necessarily result in out-of-frame decoding by the tRNA. Therefore, a "frameshift" mutation in a tRNA has principally changed the triplet codon recognition properties of the molecule.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Bronson M. J., Yanofsky C. Letter to the editor: Characterization of mutations in the tryptophan operon of Escherichia coli by RNA nucleotide sequencing. J Mol Biol. 1974 Oct 5;88(4):913–915. doi: 10.1016/0022-2836(74)90407-0. [DOI] [PubMed] [Google Scholar]
- Gupta R. C., Randerath K. Rapid print-readout technique for sequencing of RNA's containing modified nucleotides. Nucleic Acids Res. 1979 Aug 10;6(11):3443–3458. doi: 10.1093/nar/6.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H., Tinoco I., Jr Mutagen--oligonucleotide complexes with a bulged base as models for frameshift mutation. Nature. 1978 Aug 10;274(5671):609–610. doi: 10.1038/274609a0. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Adelberg E. A. Streptomycin-suppressible lethal mutations in Escherichia coli. J Bacteriol. 1970 Jul;103(1):20–26. doi: 10.1128/jb.103.1.20-26.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Childress J. R. Suppressors of a UGG missense mutation in Escherichia coli. J Bacteriol. 1980 Jul;143(1):285–292. doi: 10.1128/jb.143.1.285-292.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Pagel F. T. Codon recognition by glycine transfer RNAs of Escherichia coli in vivo. J Mol Biol. 1980 Apr 25;138(4):833–844. doi: 10.1016/0022-2836(80)90067-4. [DOI] [PubMed] [Google Scholar]
- Murgola E. J. Restricted wobble in UGA codon recognition by glycine tRNA suppressors of UGG. J Mol Biol. 1981 Jun 15;149(1):1–13. doi: 10.1016/0022-2836(81)90257-6. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Yanofsky C. Selection for new amino acids at position 211 of the tryptophan synthetase alpha chain of Escherichia coli. J Mol Biol. 1974 Jul 15;86(4):775–784. doi: 10.1016/0022-2836(74)90353-2. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Yanofsky C. Suppression of glutamic acid codons by mutant glycine transfer ribonucleic acid. J Bacteriol. 1974 Feb;117(2):439–443. doi: 10.1128/jb.117.2.439-443.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather N. E., Murgola E. J., Mims B. H. Nucleotide insertion in the anticodon loop of a glycine transfer RNA causes missense suppression. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7408–7411. doi: 10.1073/pnas.78.12.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather N. E., Murgola E. J., Mims B. H. Primary structure of an unusual glycine tRNA UGA suppressor. Nucleic Acids Res. 1981 Dec 11;9(23):6421–6428. doi: 10.1093/nar/9.23.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol. 1973 Apr 25;242(121):230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Ito J., Horn V. Amino acid replacements and the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:151–162. doi: 10.1101/sqb.1966.031.01.023. [DOI] [PubMed] [Google Scholar]




