Summary
Modification of specific Gram-negative bacterial cell envelope components, such as capsule, O-antigen and lipid A, are often essential for the successful establishment of infection. Francisella species express lipid A molecules with unique characteristics involved in circumventing host defences, which significantly contribute to their virulence. In this study, we show that NaxD, a member of the highly conserved YdjC superfamily, is a deacetylase required for an important modification of the outer membrane component lipid A in Francisella. Mass spectrometry analysis revealed that NaxD is essential for the modification of a lipid A phosphate with galactosamine in Francisella novicida, a model organism for the study of highly virulent Francisella tularensis. Significantly, enzymatic assays confirmed that this protein is necessary for deacetylation of its substrate. In addition, NaxD was involved in resistance to the antimicrobial peptide polymyxin B and critical for replication in macrophages and in vivo virulence. Importantly, this protein is also required for lipid A modification in F. tularensis as well as Bordetella bronchiseptica. Since NaxD homologues are conserved among many Gram-negative pathogens, this work has broad implications for our understanding of host subversion mechanisms of other virulent bacteria.
Introduction
Mammalian host defences include multiple pathways for recognition of, and action against, Gram-negative bacterial cell wall components including capsule, O-antigen and lipid A. Accordingly, many such pathogens have evolved modifications of these structural elements in order to evade host responses. The lipid A molecules of Francisella species have multiple unique modifications, although the details of the pathways involved in generating these alterations are still being elucidated.
Francisella tularensis is a Gram-negative intracellular pathogen and the causative agent of tularaemia. Due to its extreme infectivity, high morbidity and mortality rates, history of weaponization, and ease of aerosolization and dissemination, it is considered a category A select agent (potential bioweapon) by the Centers for Disease Control and Prevention (CDC) (Darling et al., 2002). Francisella novicida is a less virulent species that rarely causes disease in humans but is frequently used as a laboratory model as it causes a tularaemia-like disease in mice, is easily genetically manipulated, and is known to use many of the same virulence determinants as F. tularensis (Titball and Petrosino, 2007). These include the Francisella pathogenicity island (FPI), which is thought to encode a putative type VI secretion system, oxidative stress resistance proteins, siderophores, and outer membrane lipid A modifications that enable the bacteria to evade recognition and damage by host phagocytes (Bakshi et al., 2006; Gunn and Ernst, 2007; Nano and Schmerk, 2007; Ramakrishnan et al., 2008; Honn et al., 2012).
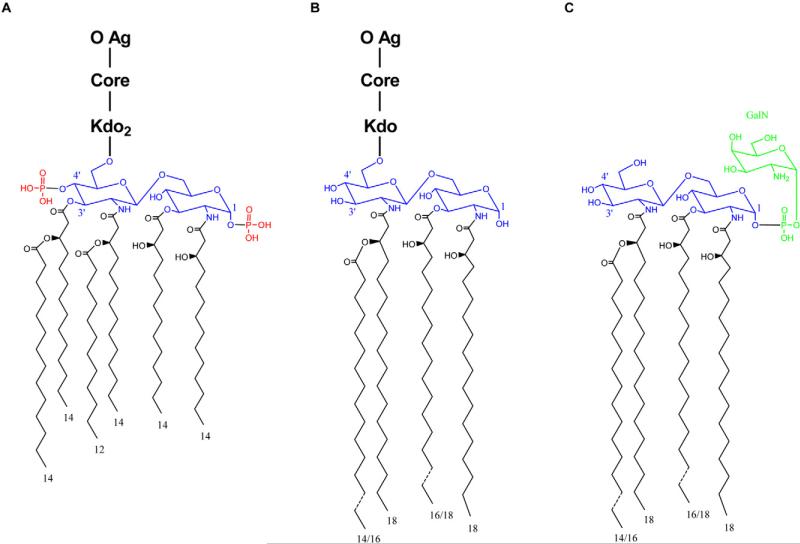
Francisella LPS has a unique lipid A moiety that is distinct from canonical lipid A structures of other Gram-negative pathogens. For example, compared with the hexa-acylated lipid A expressed by Escherichia coli, Francisella lipid A features only four acyl chains that are longer than those of E. coli by as many as six carbons (Raetz and Whitfield, 2002; Trent, 2004; Raetz et al., 2009) (Fig. 1A and B). In addition, Francisella LPS lacks both the 4′ and 1 position distal phosphates (Raetz et al., 2009). Also unique to Francisella species, 70% of the total lipid A in the outer membrane exists in a ‘free’ form that lacks the traditional Kdo, core and O-antigen polysaccha-rides of complete LPS (Wang et al., 2006; Zhao and Raetz, 2010) (Fig. 1C). Unlike the lipid A of complete LPS, free lipid A retains the 1 position phosphate that is further modified with a galactosamine residue.
Fig. 1.
E. coli and Francisella LPS and lipid A structures. Structures of (A) E. coli LPS, (B) complete LPS from Francisella species and (C) ‘free’ lipid A of Francisella species are compared. (A, B) O-antigen (O Ag), core sugars (Core) and the specific core sugar Kdo (Kdo) are indicated. For all structures, lipid A backbone disaccharides are highlighted in blue and acyl chains are represented in black with numbers denoting length. E. coli lipid A 4′ and 1 position phosphate groups (missing from the lipid A of complete Francisella LPS) are highlighted in red. Unlike the lipid A component of complete Francisella LPS, Francisella free lipid A includes a phosphate modified with galactosamine at the 1 position (highlighted in green).
As highly successful intracellular pathogens, Francisella species are able to utilize multiple phagocytic and non-phagocytic cell types for replication (Fujita et al., 1993; Qin and Mann, 2006; Hall et al., 2007; 2008; Schulert et al., 2009). Entry into host macrophages often occurs by a novel process involving the formation of unusually large and asymmetrical pseudopod loops (Clemens and Horwitz, 2007). One to 3 h after uptake by phagocytes, Francisella species escape the phagosome before replicating within the host cytosol. However, many of the details of Francisella's intracellular life cycle are still unknown (Clemens and Horwitz, 2007).
Although much progress has been made in understanding Francisella virulence mechanisms, there are still many questions regarding how this pathogen is able to so effectively replicate within host cells and cause disease. To begin to answer these questions, we performed a genome-wide in vivo negative selection screen to identify genes required for pathogenesis (Weiss et al., 2007). Next, we conducted an intracellular replication screen to determine which of those genes were important specifically for replication in macrophages (Llewellyn et al., 2011). FTN_0544 was identified in both of these screens. Although annotated as a hypothetical protein of unknown function in the NCBI database, FTN_0544 belongs to the YdjC superfamily of proteins. Interestingly, proteins belonging to this family are encoded by multiple Gram-negative pathogens including Bordetella bronchiseptica, Brucella abortus, Coxiella burnetii and Legionella pneumophila.
In this study, we show that FTN_0544 is a deacetylase involved in the galactosamine modification of Francisella's unique free lipid A molecules. We have thus renamed this protein NaxD (N-acetylhexosamine deacetylase). Furthermore, we show that the action of NaxD is required for resistance to the cationic antimicrobial peptide polymyxin B, intracellular replication and virulence in vivo. Importantly, we have shown that the role of this protein is conserved in human pathogenic F. tularensis, as well as B. bronchiseptica. Since NaxD is highly conserved in numerous Gram-negative pathogens, this work has broad implications for the elucidation of mechanisms of pathogenesis in other virulent bacteria.
Results
NaxD is a member of the YdjC superfamily of proteins
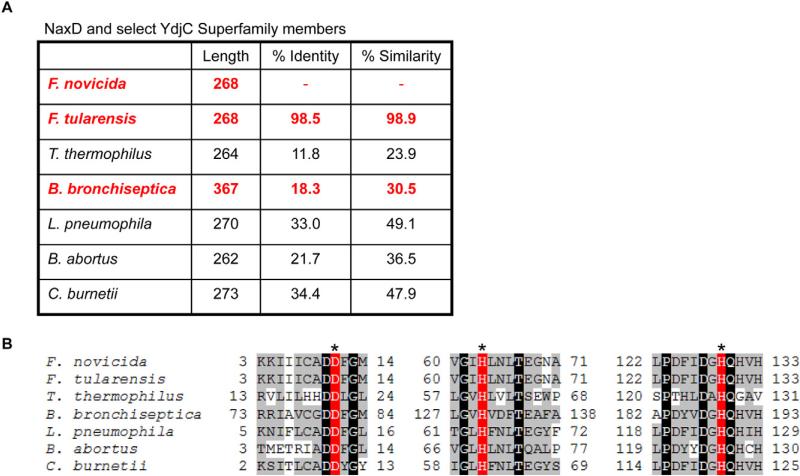
Although naxD is annotated as encoding a hypothetical protein in the NCBI database, protein sequence analysis revealed that NaxD belongs to the YdjC superfamily. This family is highly conserved, with over 3000 entries in the NCBI database. Homologues of NaxD are encoded by numerous pathogens including B. bronchiseptica, B. abortus, L. pneumophila and C. burnetii (Fig. 2A). While a member of this family from Bacillus stearothermophilus had been putatively identified as a part of a cryptic cellobiose metabolism operon (Lai and Ingram, 1993), another member from Thermus thermophilus, TTHB029, has been shown to have structural similarity to a deacetylase from Streptococcus pneumoniae (Imagawa et al., 2008). Structural analysis revealed a putative active site containing three potential catalytic residues (Imagawa et al., 2008). Importantly, these residues are conserved among YdjC superfamily proteins (Fig. 2B), suggesting that NaxD and other YdjC proteins may function as deacetylases.
Fig. 2.
NaxD is a member of the YdjC superfamily. The amino acid sequences of F. novicida and F. tularensis NaxD (FTN_0544 and FTT_0453 respectively) were aligned with YdjC superfamily proteins from Thermus thermophilus (TTHB029), Bordetella bronchiseptica (BB4267), Legionella pneumophila (lp12_2472), Brucella abortus (BAbS19_II01260) and Coxiella burnetii (CBU_0580) using clustalo (http://www.ebi.ac.uk/Tools/msa/clustalo/).
A. The per cent amino acid identity and similarity to F. novicida NaxD are shown. Proteins in bold and highlighted in red are described in this manuscript.
B. Amino acids surrounding putative active-site residues are shown and numbers indicate their position in the sequence. Highlighting indicates conserved putative active-site residues (red, asterisk), identical (black) and similar (grey) residues.
NaxD is required for F. novicida replication in macrophages and virulence in vivo
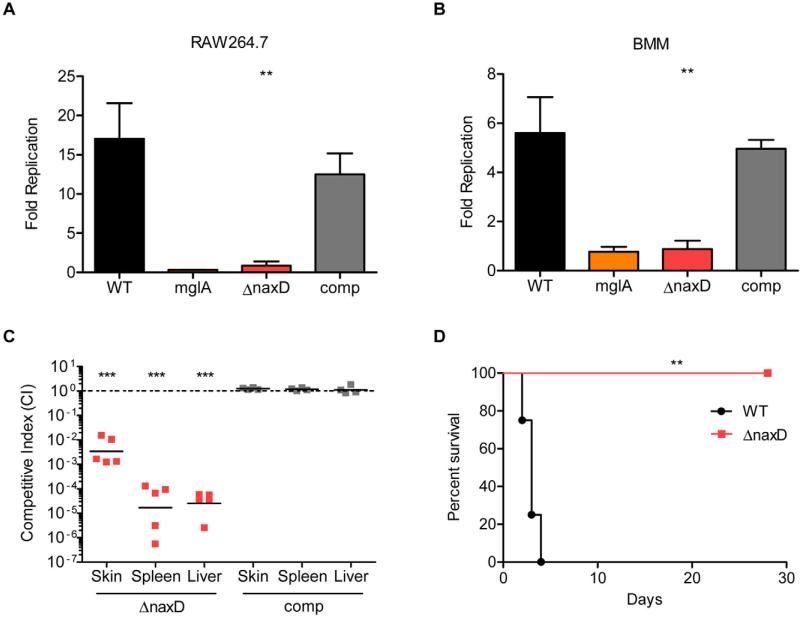
We originally identified naxD as being required for virulence in an in vivo genome-wide negative selection screen (Weiss et al., 2007). In addition, we showed that this gene was required for intracellular proliferation in a macrophage replication screen (Llewellyn et al., 2011). Since both of these screens utilized transposon mutants, we wanted to ensure that the observed phenotypes resulted from disruption of naxD and not unintended secondary mutations. To do this, we generated an F. novicida naxD deletion mutant and a complemented strain. The naxD mutant exhibited wild-type growth kinetics in both rich and minimal media (Fig. S1). Macrophage replication experiments revealed that the naxD mutant was unable to replicate in either RAW264.7 macrophages or primary murine bone marrow-derived macrophages (BMM) (Fig. 3A and B). In fact, the level of attenuation of the naxD mutant was similar to that of a previously characterized strain lacking a functional copy of the gene encoding the virulence factor MglA, which is known to persist but not replicate in macrophages (Baron and Nano, 1998). In addition, the naxD complemented strain replicated to levels similar to wild-type. Given that Francisella must escape the phagosome in order to replicate, we used fluorescence microscopy to measure escape kinetics via colocalization of intracellular bacteria with the phagosomal marker LAMP-1 (Fig. S2). These experiments demonstrated that wild-type and naxD mutant F. novicida escaped the phagosomes of BMM with similar kinetics, indicating that the mutant's attenuation in macrophages is not due to a deficiency in phagosomal escape (Fig. S2). Overall, these results show that NaxD is required for intracellular proliferation but not for phagosomal escape.
Fig. 3.
NaxD is required for replication in murine macrophages and mice.
A and B. (A) RAW264.7 or (B) primary murine bone marrow-derived macrophages (BMM) were infected with a 20:1 moi of wild-type F. novicida (WT), the mglA mutant (mglA), the naxD deletion strain (ΔnaxD) or the complemented strain (comp). Colony-forming units from lysates 30 min post infection were compared with those from (A) 24 or (B) 6 h post infection to determine fold intracellular replication (n = 3 biological replicates).
C. Mice were subcutaneously infected with a 1:1 mixture of 105 cfu each of wild-type and ΔnaxD (red) or wild-type and the complemented strain (grey). Forty-eight hours after infection, organs were harvested, cfu enumerated and the competitive index (CI) calculated for the skin at the site of infection, spleen and liver. CI = (cfu mutant output/cfu WT output)/(cfu mutant input/cfu WT input). Bars represent the geometric mean CI values from each group of mice (n = 5 mice). CI values below 1 (dashed line) indicate attenuation of the mutant strain.
D. Mice were subcutaneously infected with 2 × 107 cfu of either wild-type or ΔnaxD and sacrificed if they appeared moribund (n = 4 mice).
In (A) and (B), bars represent the average and error bars represent the standard deviation of three biological replicates from one experiment. Data shown in all panels are representative of at least three independent experiments. Asterisks indicate significance as compared with wild-type (A, B, D) or compared with 1 (C). **P < 0.005, ***P < 0.0005.
While our in vivo negative selection screen identified naxD as being important for virulence, it did not provide quantitative data regarding the degree of attenuation of a naxD mutant. To determine this, we performed competition experiments. Briefly, mice were infected with a 1:1 ratio of wild-type F. novicida and either the naxD deletion mutant or complemented strain. Forty-eight hours post infection, the naxD mutant displayed an approximate 2.5 log attenuation in the skin and a nearly 5 log attenuation in both the liver and spleen compared with wild-type (Fig. 3C). All mutant phenotypes were restored to wild- type levels in the complemented strain (Fig. 3C). To determine the consequence of the naxD mutant's virulence defect, we infected mice with either the wild-type or mutant strain and monitored survival. While mice infected with wild-type bacteria were moribund by 4 days after infection, the mutant did not kill mice up to 28 days post infection (Fig. 3D). Taken together, these data show that naxD is required for both replication in host macrophages and virulence in vivo.
NaxD is involved in altering surface charge and resistance to polymyxin B
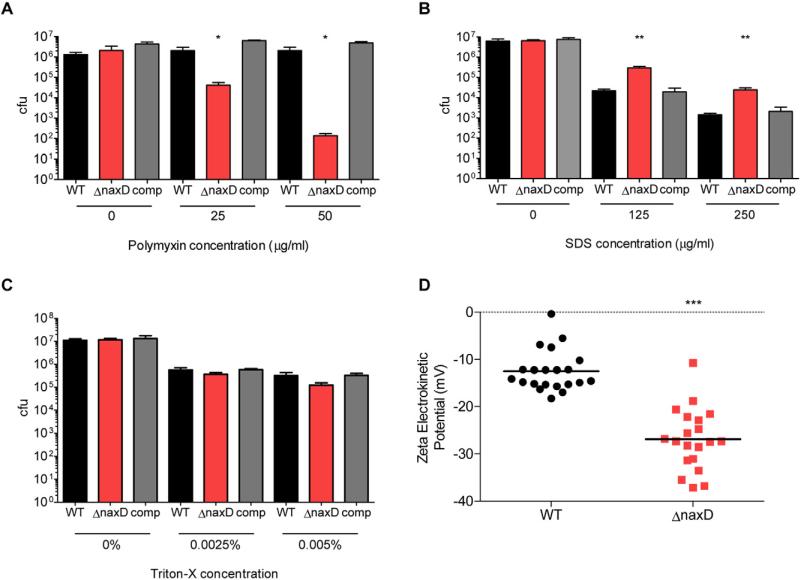
After validating the importance of NaxD in F. novicida infection of macrophages and mice, our next aim was to determine the role of this protein in pathogenesis. To further characterize the phenotypes of the naxD mutant, we subjected both wild-type and the deletion mutant to different antimicrobials and compared the survival of each strain. While the wild-type was unaffected at the concentrations tested, the mutant displayed dose-dependent sensitivity to the cationic antimicrobial peptide polymyxin B (Fig. 4A), which acts on Gram-negative bacteria by binding to the negatively charged lipid A component of LPS (Morrison and Jacobs, 1976). Conversely, the mutant showed increased resistance to the anionic detergent SDS (Fig. 4B) and displayed wild-type levels of sensitivity to the non-ionic detergent Triton X-100 (Fig. 4C). The altered response of the mutant to charged antimicrobials that act on the cell membrane suggested that NaxD might be involved in altering the net charge of the bacterial surface. To test this hypothesis, we measured the zeta electrokinetic potential of each strain, which gives an indirect reading of the bacterial surface charge. We determined that the mutant exhibited approximately a twofold decrease in zeta potential compared with wild-type bacteria (Fig. 4D), indicating that NaxD is involved in increasing the charge of the bacterial surface. Taken together, the naxD mutant's decreased surface charge and increased sensitivity to cationic polymyxin B, which targets negatively charged lipid A, suggested that NaxD could be required for a modification to lipid A that alters its charge.
Fig. 4.
NaxD is involved in resistance to cationic antimicrobials and alteration of bacterial surface charge.
A–C. Wild-type F. novicida (WT), the naxD deletion mutant (ΔnaxD) or the complemented strain (comp) were incubated with the indicated concentrations of (A) polymyxin B, (B) SDS or (C) Triton X-100 for 6 h. Cultures were then serially diluted and plated for cfu (n = 3 biological replicates).
D. The zeta potential of wild-type and ΔnaxD was measured (n = 10 technical replicates) and the results of three independent experiments were combined for statistical analysis.
In (A)–(C), bars represent the average and error bars represent the standard deviation of three biological replicates from one experiment. Data shown are representative of at least three independent experiments. Asterisks indicate significance as compared with wild-type. *P < 0.05, **P < 0.005, ***P < 0.0005.
NaxD is required for lipid A modification with galactosamine
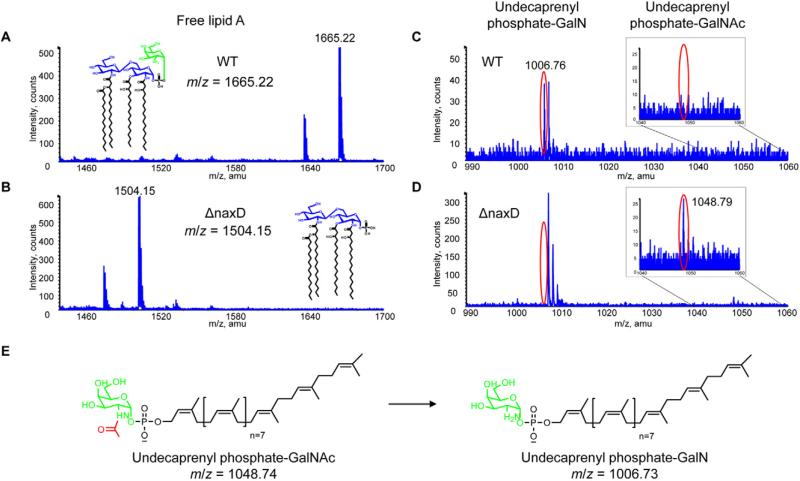
In order to determine if NaxD is involved in lipid A modification, we analysed the lipid fractions of the wild-type and mutant strains using liquid chromatography electro-spray ionization mass spectrometry (LC-ESI/MS). As mentioned previously, the majority of Francisella lipid A exists as free lipid A (Vinogradov et al., 2002; Wang et al., 2006). ESI/MS analysis via direct infusion of wild-type F. novicida free lipid A revealed an anticipated peak at m/z 1665.22 (Fig. 5A) (Phillips et al., 2004; Wang et al., 2006), while the naxD mutant lipid A exhibited a peak at m/z 1504.15 (Fig. 5B). Interestingly, this shift corresponds to the molecular weight of galactosamine, and wild-type Francisella free lipid A is modified with a galactosamine at the 1 position phosphate (Phillips et al., 2004; Wang et al., 2006; 2009; Schilling et al., 2007; Shaffer et al., 2007; Kanistanon et al., 2008; Kalhorn et al., 2009; Song et al., 2009; Soni et al., 2010; Beasley et al., 2012). The absence of the galactosamine moiety on the free lipid A of the mutant would result in an exposed, negatively charged phosphate group, which correlates with the decreased surface charge of the mutant strain (Fig. 4D) (Phillips et al., 2004; Wang et al., 2006).
Fig. 5.
NaxD is required for the galactosamine modification of F. novicida free lipid A.
A–D. Total lipids were extracted from (A, C) wild-type F. novicida (WT) and (B, D) naxD mutant (ΔnaxD) strains in mid-log phase and (A, B) free lipid A, (C, D) undecaprenyl phosphate-N-acetylgalactosamine (GalNAc), and undecaprenyl phosphate-galactosamine (GalN) were analysed by ESI/MS in negative ion mode via direct infusion.
E. A schematic for the deacetylation of undecaprenyl phosphate-GalNAc (expected m/z = 1048.74) to undecaprenyl phosphate-GalN (expected m/z = 1006.73) is shown.
(A, E) Galactosamine is highlighted in green and (E) the acetyl group is highlighted in red.
To determine why the naxD deletion mutant lacks galactosamine and where NaxD might act in the lipid A biosynthetic pathway, we measured the presence and quantities of precursor molecules required for the galactosamine modification. Galactosamine is added to Francisella free lipid A from undecaprenyl phosphate-galactosamine (undecaprenyl phosphate-GalN) (Song et al., 2009), a complex of the sugar with a lipid carrier molecule. Analysis of the wild-type lipid fraction revealed a singly charged peak corresponding to undecaprenyl phosphate-GalN at m/z 1006.76 (Fig. 5C). However, this glycolipid was not present in the mutant strain (Fig. 5D). Instead, the mutant exhibited a peak at m/z 1048.79, corresponding to undecaprenyl phosphate-N-acetylgalactosamine (undecaprenyl phosphate-GalNAc), the acetylated precursor of undecaprenyl phosphate-GalN (Fig. 5D and E). Conversely, this acetylated precursor was not detected in the wild-type lipid fraction (Fig. 5C). Taken together, these data show that NaxD is required for deacetylation of undecaprenyl phosphate-GalNAc and that the absence of this deacetylation event prevents the galactosamine modification to F. novicida free lipid A.
NaxD is necessary for deacetylation of undecaprenyl phosphate-GalNAc
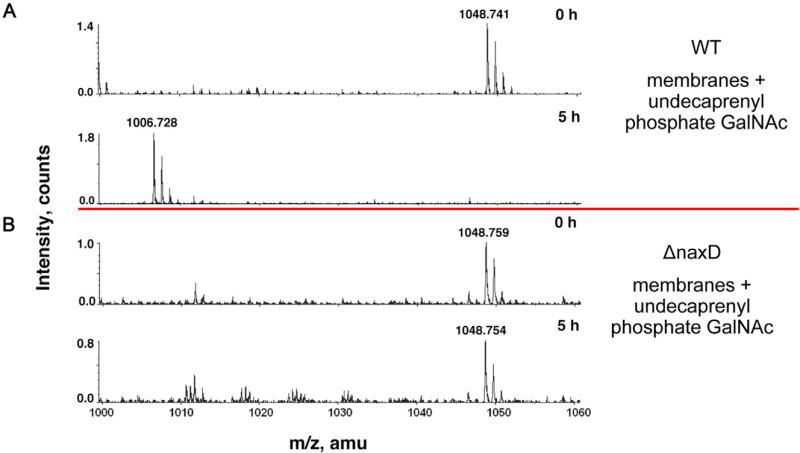
After MS analysis revealed NaxD was involved in deacetylation of undecaprenyl phosphate-GalNAc, we set out to determine whether NaxD was directly responsible for this reaction. First, using a strain in which NaxD was labelled with an 8× histidine tag, we found that NaxD localizes to the F. novicida membrane fraction (Fig. S4). To determine if NaxD could deacetylate undecaprenyl phosphate-GalNAc in the membrane, we harvested crude membrane fractions from either wild-type or mutant strains and incubated them with synthetic undecaprenyl phosphate-GalNAc. The lipids were extracted from each reaction and analysed using LC-ESI/MS. For both wild-type and mutant, the substrate peak (undecaprenyl phosphate-GalNAc, expected m/z 1048.74) was present at time zero (Fig. 6). After a 5 h incubation, deacetylation of undecaprenyl phosphate-GalNAc was observed in the wild-type reaction, since a peak consistent with undecaprenyl phosphate-GalN was detected (Fig. 6A). In contrast, no product peak was observed in the mutant reaction (Fig. 6B). These results showed that NaxD in the membrane fraction was necessary for undecaprenyl phosphate-GalNAc deacetylation.
Fig. 6.
NaxD is necessary for deacetylation of undecaprenyl phosphate-N-acetylgalactosamine. Deacetylase activity assays using synthesized undecaprenyl phosphate-N-acetylgalactosamine (GalNAc) and (A) 0.5 mg ml−1 F. novicida wild-type (WT) membrane fraction or (B) F. novicida naxD mutant (ΔnaxD) membrane fraction were incubated at 30°C for the indicated times and then analysed using LC-ESI/MS in negative ion mode.
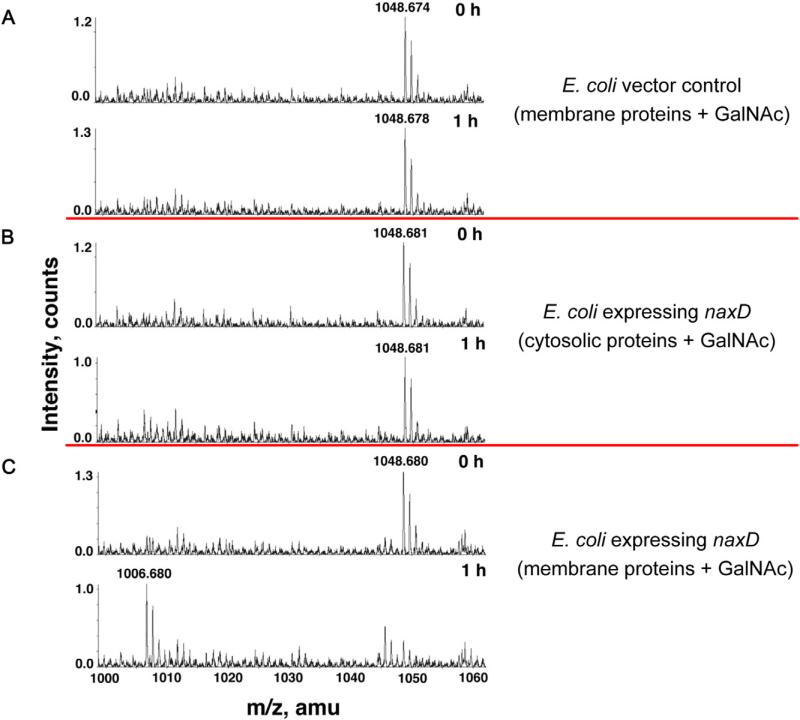
To determine if NaxD was responsible for this enzymatic activity, we overexpressed naxD in E. coli, which does not encode a NaxD homologue, does not synthesize undecaprenyl phosphate-GalNAc or undecaprenyl phosphate-GalN, and does not modify its lipid A with galactosamine. We demonstrated that NaxD localized to the E. coli membrane fraction (Fig. S5), isolated membranes from strains that were transformed with either an empty vector control or the naxD expression plasmid, and assayed for enzymatic activity as described for F. novicida above. LC-ESI/MS analysis revealed that there was no deacetylated product (expected m/z 1006.73) detected for the reactions using a whole-cell lysate from the E. coli empty vector control strain (Fig. 7A) or with the soluble fraction from E. coli expressing naxD (Fig. 7B). However, the deacetylated product, undecaprenyl phosphate-GalN, was detected in assays that contained the membrane fraction from E. coli expressing NaxD (Fig. 7C). Together, these results using membrane fractions from F. novicida and E. coli respectively demonstrate that NaxD is necessary and suggest that it is sufficient for deacetylation of undecaprenyl phosphate-GalNAc.
Fig. 7.
NaxD is required for deacetylation of undecaprenyl phosphate-N-acetylgalactosamine when exogenously expressed in E. coli. Deacetylase activity assays using synthesized undecaprenyl phosphate-N-acetylgalactosamine (GalNAc) and (A) whole-cell lysate from E. coli with an empty vector, (B) the soluble fraction from E. coli expressing naxD, or (C) the membrane fraction from E. coli expressing naxD were incubated at 30°C for the indicated times and then analysed using LC-ESI/MS in negative ion mode.
The NaxD orthologue in F. tularensis is required for lipid A modification and virulence
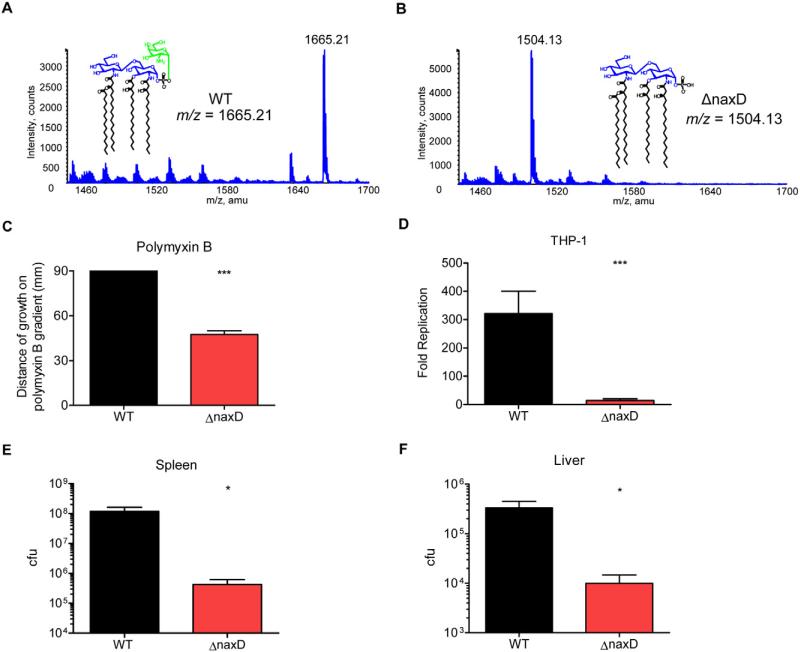
The NaxD orthologue from human pathogenic F. tularensis, FTT_0453, retains 99% amino acid identity with F. novicida NaxD (Fig. 2A). In order to ascertain if NaxD function is conserved in F. tularensis, we generated a FTT_0453 (naxD) deletion mutant in strain SchuS4. LC-ESI/MS analysis of wild-type F. tularensis free lipid A revealed the m/z 1665.21 peak that corresponds to Fran-cisella lipid A modified with galactosamine (Fig. 8A), although this species was not detected in the mutant. Instead, the naxD mutant displayed a peak at m/z 1504.13 that corresponds to lipid A without galactosamine (Fig. 8B). This demonstrates the conserved role of NaxD in lipid A modification in highly virulent F. tularensis.
Fig. 8.
NaxD function is conserved in human pathogenic F. tularensis.
A and B. Total lipids were extracted from (A) wild-type F. tularensis and (B) the ΔnaxD strain in mid-log phase and lipid A composition was analysed by LC-ESI/MS. (A) Galactosamine is highlighted in green.
C. The distance of growth of wild-type or ΔnaxD along a 0–2 mg ml−1 gradient of polymyxin B was measured (n = 3 biological replicates).
D. Human PMA-differentiated THP-1 macrophage-like cells were infected with either wild-type F. tularensis (WT) or the naxD deletion mutant (ΔnaxD) at a 50:1 moi. Colony-forming units recovered from macrophages lysed 24 h after infection were compared with cfu recovered at 2 h post infection to calculate fold replication (n = 3 biological replicates).
E and F. Mice were subcutaneously infected with 50 cfu of either wild-type or ΔnaxD and 48 h after infection, organs were harvested and plated and cfu were enumerated for the (E) spleen and (F) liver (n = 5 mice).
In (C) and (D), bars represent the average and error bars represent the standard deviation of three biological replicates from one experiment. Data shown in (C)–(F) are representative of at least three independent experiments. Asterisks indicate significance as compared with wild-type. *P < 0.05, ***P < 0.0005.
Next we tested the functional role of F. tularensis NaxD in polymyxin B resistance, intracellular replication and in vivo survival. The naxD deletion mutant displayed an increased susceptibility to polymyxin B as compared with wild-type (Fig. 8C). Given that F. tularensis is a virulent human pathogen, we wanted to determine the importance of NaxD function during infection of human cells. Indeed, we observed a severe defect in replication of the naxD deletion mutant compared with wild-type F. tularensis 24 h after infection of human THP-1 macrophage-like cells (Fig. 8D). In addition, similar to the F. novicida naxD mutant (Fig. 3B), the F. tularensis naxD deletion mutant was unable to proliferate in primary murine BMM (Fig. S3). Importantly, NaxD was also required for replication in mice, since 48 h after subcutaneous infection, wild-type F. tularensis was recovered at a level 2.5 logs higher in the spleen of mice (Fig. 8E) and nearly 1.5 logs higher in the liver than the mutant strain (Fig. 8F). These data show that NaxD function is conserved in human pathogenic F. tularensis, in which it is required for the addition of galactosamine to free lipid A and is required for resistance to the cationic antimicrobial peptide polymyxin B, replication within human cells and virulence in mice.
Conserved role of the NaxD homologue in Bordetella bronchiseptica
Given that the YdjC superfamily of proteins is conserved among many virulent bacteria, we wanted to determine if a NaxD homologue from a different pathogen shared a similar function in lipid A modification. To test this, we generated a B. bronchiseptica deletion mutant lacking the gene encoding the NaxD homologue BB4267 (Fig. 2). B. bronchiseptica is a Gram-negative bacterium that colonizes mammalian respiratory tracts and is considered a primary pathogen of domestic animals such as dogs, cats, rabbits and pigs, but can also establish chronic infections in immunocompromised humans (Egberink et al., 2009). Lipid A from this pathogen has two phosphate groups that are known to be modified with glucosamine, a stereoisomer of galactosamine (Tirsoaga et al., 2007; Marr et al., 2008; Basheer et al., 2011). Although BB4267 is nearly 100 amino acids larger than NaxD, the majority of the protein is comprised of the YdjC superfamily domain, including the conserved putative active-site residues, which suggests conserved function (Fig. 2B).
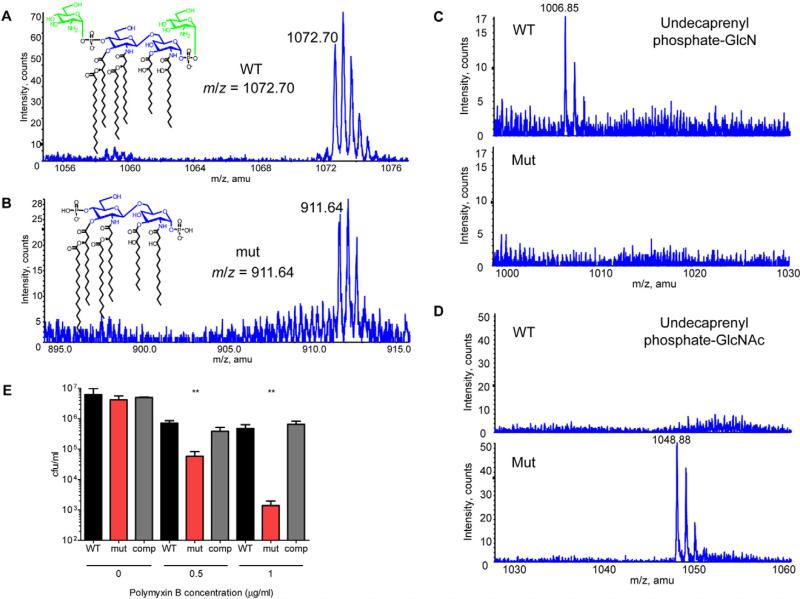
Indeed, LC-ESI/MS analysis of the wild-type lipid fractions revealed a doubly charged lipid A peak at m/z 1072.70 that corresponds to lipid A with glucosamine modifications at both the 4′ and 1 position phosphates (Fig. 9A). This peak was absent in the mutant fractions, which instead displayed a doubly charged peak at m/z 911.64, corresponding to the lipid A molecule missing both glucosamine modifications (Fig. 9B). These data show that the NaxD homologue BB4267, like NaxD in Francisella, is required for the modification of lipid A phosphates with hexosamine sugars.
Fig. 9.
Conserved role of the Bordetella bronchiseptica NaxD homologue in lipid A modification.
A–D. Total lipids were extracted from wild-type (WT) and naxD homologue mutant (mut) strains of B. bronchiseptica in mid-log phase and (A, B) lipid A, (C) undecaprenyl phosphate-glucosamine (GlcN) and (D) undecaprenyl phosphate-N-acetylglucosamine (GlcNAc) analysed by LC-ESI/MS. In (A), glucosamine groups are highlighted in green.
E. WT, mut or the complemented strain (comp) were incubated with the indicated concentrations of polymyxin B for 6 h and cfu were enumerated (n = 3 biological replicates). Bars represent the average and error bars represent the standard deviation of three biological replicates from one experiment. Data shown are representative of at least three independent experiments. Asterisks indicate significance as compared with wild-type. **P < 0.005.
Similar to the addition of galactosamine to Francisella lipid A, the modification of B. bronchiseptica lipid A with glucosamine requires the deacetylation of undecaprenyl phosphate-N-acetylglucosamine (undecaprenyl phosphate-GlcNAc) to form undecaprenyl phosphate-glucosamine (undecaprenyl phosphate-GlcN). LC-ESI/MS from the lipid fraction of the wild-type strain showed a singly charged peak at m/z 1006.85, corresponding to undecaprenyl phosphate-GlcN (Fig. 9C). In contrast, the bb4267 mutant displayed a peak at m/z 1048.88, corresponding to undecaprenyl phosphate-GlcNAc (Fig. 9D), which was undetectable in the wild-type. This demonstrates that, similar to NaxD function in Francisella, BB4267 is required for a deacetylation reaction in B. bronchiseptica. To test the functional relevance of the lipid A modification with glucosamine, we measured the polymyxin B sensitivity of the wild-type and mutant strains and found that the bb4267 mutant exhibited a dose-dependent increase in susceptibility to polymyxin B as compared with wild-type (Fig. 9E). These data confirm that the function of NaxD is conserved among multiple Gram-negative pathogens.
Discussion
Using in vivo negative selection (Weiss et al., 2007) and intramacrophage replication (Llewellyn et al., 2011) screens, we have recently identified NaxD as an important virulence factor of Francisella. Here we have extended those findings by showing that this member of the YdjC protein superfamily is a deacetylase that is required for lipid A modifications that render bacteria more resistant to killing by the cationic antimicrobial peptide polymyxin B. Given our findings that the B. bronchiseptica NaxD homologue is also required for lipid A modification, this report suggests that NaxD/YdjC proteins are likely to have an important role in the pathogenesis of other virulent Gram-negative bacteria.
This work contributes to a greater understanding of the mechanisms by which Francisella is able to so effectively evade killing by antimicrobial peptides compared with other Gram-negative pathogens (Ishimoto et al., 2006; Mohapatra et al., 2007). For example, Francisella is nearly 1000× more resistant to polymyxin B than E. coli (Mohapatra et al., 2007). Because polymyxin B is known to bind Gram-negative lipid A, it is interesting that the majority of the exposed surface of the Francisella outer membrane consists of free lipid A (Zhao and Raetz, 2010). To our knowledge, Francisella is the only Gram-negative pathogen shown to exhibit this sort of unique outer membrane composition. Mutants that lack the galactosamine modification on free lipid A have an exposed phosphate at the 1 position (Phillips et al., 2004; Bina et al., 2006; Schilling et al., 2007; Shaffer et al., 2007; Kanistanon et al., 2008; Kalhorn et al., 2009; Song et al., 2009; Wang et al., 2009; Soni et al., 2010; Beasley et al., 2012), significantly altering the charge and likely the topography of the majority of the outer leaflet of the outer membrane. Interestingly, the small percentage of lipid A that is part of complete LPS lacks the 1 position phosphate and, therefore, the galactosamine modification as well (Zhao and Raetz, 2010). It is not clear why Francisella produces such a large amount of lipid A without O-antigen, given that O-antigen is critical for virulence (Sandstrom et al., 1988; Sorokin et al., 1996; Clay et al., 2008). However, since the majority of Francisella's outer membrane is composed of free lipid A, it is intuitive that the galactosamine modification to this moiety would be critical in resistance to host stresses, similar to the importance of modifications to complete LPS in other bacteria (Wang and Quinn, 2010). Indeed, given that this modification is important for resistance to polymyxin B, replication in macrophages, and during in vivo infection, it likely contributes to Francisella resistance to host cationic antimicrobial peptides such as cathelicidins, defensins and ubiquicidin (Weiss et al., 2007; Kanistanon et al., 2008; Flannagan et al., 2009; Llewellyn et al., 2011). Future work will aim to elucidate whether there are advantages conferred by the novel cell surface component free lipid A, e.g. whether free lipid A promotes enhanced resistance to host antimicrobials compared with full LPS.
The Gram-negative pathogen B. bronchiseptica has been shown to express lipid A species that have one or both phosphates modified with glucosamine, a stereoisomer of galactosamine (Tirsoaga et al., 2007; Marr et al., 2008; Basheer et al., 2011). Like galactosamine, addition of glucosamine neutralizes the negative charge of lipid A phosphates (Marr et al., 2008). In this study we show that the B. bronchiseptica NaxD homologue BB4267 is required for the glucosamine modification of lipid A phosphates and specifically is necessary for the deacetylation of undecaprenyl phosphate-N-acetylglucosamine. Similar to the Francisella galactosamine modification, we show that this glucosamine modification is important for resistance to the lipid A-binding cationic antimicrobial peptide polymyxin B. While no bacteria other than Francisella species have been reported to utilize free lipid A, our B. bronchiseptica data indicate that NaxD homologues could be involved in modifying the lipid A component of complete LPS of other Gram-negative pathogens.
This study has generated new insight into Francisella pathogenesis and lipid A biosynthesis as well as the function of YdjC superfamily proteins. Significantly, this study has broad implications for host–pathogen interactions of other highly virulent NaxD homologue-encoding Gram-negative bacteria, particularly intracellular pathogens such as B. abortus, L. pneumophila and C. burnetii. Future studies on the role of this family of proteins will likely further illuminate the virulence mechanisms of other NaxD homologue-encoding pathogenic bacteria.
Experimental procedures
Bacterial strains and growth conditions
Wild-type F. novicida strain U112 and a previously described mglA point mutant, GB2 (Baron and Nano, 1998), were a generous gift from Dr Denise Monack (Stanford University, Stanford, CA). These strains, the naxD deletion mutant and the naxD complemented strain were grown at 37°C on a rolling drum in tryptic soy broth (TSB; Difco/BD, Sparks, MD) supplemented with 0.02% L-cysteine (Sigma-Aldrich, St. Louis, MO). F. novicida was plated for colony-forming units (cfu) on tryptic soy agar (TSA; Difco/BD) and supplemented with 0.01% L-cysteine, with the exception of bacteria from mouse experiments, which were plated on modified Mueller Hinton (mMH) agar plates (Difco/BD) supplemented with 0.025% ferric pyrophosphate (Sigma-Aldrich), 0.1% glucose (Sigma-Aldrich) and 0.01% L-cysteine. When appropriate, kanamycin (Fisher Scientific, Fair Lawn, NJ) was added to media at a concentration of 30 μg ml-1. F. tularensis strains were grown in mMH broth or on Brain Heart Infusion (BHI) agar (BHI supplemented with 50 μg ml−1 haemin, 1.4% agar (w/v) and 1% (v/v) IsoVitalex (BBL, Cockeysville, MD). Counter selection for resolution of F. tularensis FTT0453 deletion plasmid co-integrants was performed on cysteine heart agar containing 5% sucrose. Kanamycin was added to the plates when necessary at 10 μg ml−1 for F. tularensis. B. bronchiseptica wild-type strain RB50, the bb4267 deletion mutant and the bb4267 complemented strain were grown under similar conditions as F. novicida, except using Stainer-Scholte broth supplemented with nicotinic acid, glutathione and ascorbic acid as previously described (Hulbert and Cotter, 2009), or Bordet-Gengou blood agar plates (Remel, Lenexa, KS). When appropriate, streptomycin (Fisher Scientific) and kanamycin were added at concentrations of 25 μg ml−1 and 50 μg ml−1 respectively.
Mutagenesis and complementation
To generate the naxD deletion mutant in F. novicida, PCR was used to amplify flanking DNA regions upstream and downstream of the gene of interest. A kanamycin resistance cassette was sewn in between these flanking regions using overlapping PCR reactions. The final linear PCR product was then gel purified and transformed into chemically competent wild-type strain U112 as previously described (Anthony et al., 1991). The primers used to create the kanamycin-resistant deletion mutant contained FRT sites flanking the kanamycin resistance cassette, which allowed a clean deletion of each mutant to be made using the plasmid pFFlp encoding the Flp-recombinase as previously described (Gallagher et al., 2008). A construct for the complementation of the mutant was generated by overlapping PCR using PCR-amplified fragments of the wild-type gene of interest, upstream and downstream flanking regions, and a kanamycin resistance cassette. These constructs were then transformed into the chemically competent naxD clean deletion mutant. Verification of allelic replacement in the mutant and complemented strains was performed using check primers in PCR reactions on purified genomic DNA from each strain. PCR products of the correct size were subsequently sequenced (MWG Operon, Huntsville, AL) for final verification of allelic replacement. PCR constructs for F. tularensis mutagenesis were amplified as for F. novicida and then cloned into plasmid pXB186 containing a kanamycin resistance cassette and the sacB counter-selectable marker (making plasmid pΔFTT0453). To generate the FTT0453 deletion mutant, the deletion plasmid was introduced by electroporation into electrocompetent F. tularensis SchuS4. Electrocompetent cells were prepared on the day of the transformation as described in the supplemental experimental procedures. The resulting clones were screened by PCR for the FTT0453 deletion. To generate the B. bronchiseptica bb4267 deletion mutant, linear PCR deletion constructs were amplified as described above for F. novicida. This bb4267-deleting fragment was cloned into the Bordetella allelic exchange plasmid pSS4245. The resulting plasmid was then transformed into wild-type B. bronchiseptica strain RB50, following procedures described previously (Inatsuka et al., 2010). The loss of bb4267 was confirmed by PCR and subsequent enzymatic digestions. To generate the complementation plasmid, the two external primers used to produce the bb4267-deleting fragment were employed and this complementing fragment was cloned into pUC18-Mini-TN7 plasmid that has a Tn7 integration sequence, which, along with a helper plasmid pTNS3, was mated into the deletion strain via tri-parental mating (Choi et al., 2005) to generate the complemented strain. All primers and plasmids used in this study are listed in Table S1.
Antimicrobial assays
The antimicrobial peptide polymyxin B (USB, Cleveland, OH) was dissolved in peptide buffer (0.01% acetic acid, 0.2% BSA) and then serially diluted in the same buffer to desired concentrations. The detergents sodium-dodecyl-sulphate (SDS; Fisher Scientific) and Triton X-100 (Fisher Scientific) were serially diluted in 25% TSB. Overnight cultures of bacteria were diluted to 1 × 107 cfu ml−1 in 25% TSB. Ninety microlitres of diluted cultures were then added to 96-well plates containing 10 ml of the appropriate antimicrobial. Plates were incubated at 37°C on shaking platforms for 6 h. Cultures were then serially diluted and plated to enumerate cfu. F. tularensis antimicrobial susceptibility was determined by the gradient agar plate method as previously described (Szybalski and Bryson, 1952; Bina et al., 2006; 2008). Briefly, 35 ml of BHI-chocolate agar (without polymyxin B) was poured into a square Petri dish and allowed to solidify as a wedge by elevating one side of the plate. After the agar solidified, 35 ml of BHI-chocolate agar containing polymyxin B at 2 mg ml−1 was added to the levelled plate and allowed to solidify. Theses plates were inoculated with overnight mMH broth cultures of each respective strain and incubated at 37°C for 2 days when the length of growth along the polymyxin B gradient was recorded. The gradient agar plate tests were performed a minimum of three times and representative results are presented.
Zeta electrokinetic potential
Overnight cultures of bacteria were subcultured and grown to OD600 = 1.0. The bacteria were then pelleted (10 000 g, 3 min) and resuspended at a 5× concentration in 20 mM potassium chloride. Twenty microlitres of the concentrated bacteria were added to 3.2 ml of 20 mM potassium chloride in the zeta potential electrokinetic cuvette from Brookhaven Instruments Corporation (BIC, Holtsville, NY). The bacterial sizes and zeta electrokinetic potentials were measured using the 90Plus size and zeta potential analyser (BIC). Data were analysed using BIC Zeta Potential Analyser Software Version 5.20, which takes into account the size of the bacteria when calculating the zeta potential.
Macrophages
RAW264.7 murine macrophages (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (high glucose, L-glutamine; DMEM; Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal calf serum (FCS; HyClone, Logan, UT). Bone marrow-derived macrophages (BMM) were isolated from wild-type C57BL/6 mice and cultured as described previously (Schaible and Kaufmann, 2002) in DMEM supplemented with 10% heat-inactivated FCS and 10% macrophage colony-stimulating factor (M-CSF)-conditioned medium (collected from M-CSF-producing L929 cells). THP-1 monocyte-like cells (ATCC) were cultured in RPMI (Lonza) with 10% heat-inactivated fetal calf serum (HyClone). Macrophages were incubated before and during infection at 37°C with 5% CO2.
Macrophage infections
RAW264.7 macrophages were seeded in 24-well plates at 5 × 105 cells per well and incubated overnight. The following day, overnight cultures of the indicated strains were pelleted (10 000 g, 3 min) and resuspended in DMEM/10% FCS. After removal of the overnight media, the macrophages were infected with bacteria at an moi of 20:1 (bacteria to macrophage), centrifuged for 15 min at 900 g, and then incubated for 30 min. Next, the macrophages were washed twice with warm DMEM and then incubated in DMEM/10% FCS with 10 μg ml−1 gentamicin. At 30 min and 24 h post infection, the macrophages were washed twice and then lysed with 1% saponin in phosphate-buffered saline (PBS). Macrophage lysates were serially diluted and plated on mMH agar, the resulting cfu were enumerated and the fold replication of each strain was determined. The same protocol as above was followed for the BMM infections with the following alterations: 3 × 105 BMM were plated per well, DMEM/10% FCS/10% M-CSF was used throughout, and the final time point was 6 h instead of 24 h. The difference in time point was due to the fact that F. novicida triggers inflammatory mediated cell death in BMM (Mariathasan et al., 2005). Therefore, bacterial replication was measured at 6 h post infection to minimize loss of bacterial counts as a consequence of the host cell death response. It is likely that we did not observe this early cell death in RAW264.7 macrophages because this cell line is known to be deficient in ASC/caspase-1 inflammasome-mediated cell death (Pelegrin et al., 2008). For F. tularensis experiments, THP-1 cells or BMM were seeded into 24-well tissue culture plates (3 × 105 cells per well) in a total volume of 1 ml of culture medium. THP-1 cells were treated with 200 nM phorbol 12-myristate 13-acetate (PMA) immediately after cells were plated. The cells were infected 24 h later with the indicated strains at an moi of 50:1 bacteria to macrophage. Fifty micrograms per millilitre of gentamicin was added 2 h later to kill any remaining extracellular bacteria. At 2 or 24 h after infection, wells were washed twice with PBS, lysed, and then bacteria were enumerated by dilution plating in duplicate using an IUL Eddy Jet Spiral plater and a Flash and Go automated colony counter (Neutec Group, Farmingdale, NY).
Mice
For F. novicida experiments, female C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) between 7 and 10 weeks of age were kept under specific pathogen-free conditions in filter-top cages at Emory University and provided with sterile food and water ad libitum. Experimental studies were performed in accordance with the Emory University Institutional Animal Care and Use Committee (IACUC) guidelines. For F. tularensis experiments, C57BL/6 mice were purchased from Charles River Laboratories. Mice were age-matched and used between 7 and 10 weeks of age. Mice were housed in sealed Allentown caging and HEPA-filtered cage racks with food and water ad libitum. All experimental protocols were reviewed and approved by the University of Tennessee Health Science Center IACUC.
Mouse experiments
For competition experiments, mice were inoculated subcutaneously with a 1:1 ratio of kanamycin-resistant deletion mutant and kanamycin-sensitive wild-type F. novicida for a total of 2 × 105 cfu in 50 μl of sterile PBS. After 48 h, the mice were sacrificed and the spleen, liver and skin at the site of infection were harvested, homogenized, plated for cfu on MH plates with and without kanamycin, and then incubated overnight at 37°C. Competitive index (CI) values were determined using the formula: (cfu mutant output/cfu WT output)/(cfu mutant input/cfu WT input). For survival experiments, mice were infected subcutaneously with 2 × 105 cfu of either the deletion mutant or wild-type strain in 50 μl of sterile PBS and then monitored for signs of illness and sacrificed if they appeared moribund.
For F. tularensis infection experiments, mice were challenged subcutaneously with 50 cfu in 100 μl of sterile PBS. All procedures were performed under BSL3 containment according to standard operating procedures that have been fully vetted by the UTHSC Committee On Biocontainment and Restricted Entities (COBRE). Spleens, livers and lungs of challenged mice were homogenized with a disposable tissue homogenizer in 1 ml of sterile PBS and then 0.25 ml disruption buffer (2.5% saponin, 15% BSA, in PBS) was added with light vortexing. Appropriate dilutions of each sample were then plated in duplicate using an Eddy Jet spiral plater on mMH agar plates supplemented with 5% calf serum and incubated at 37°C for 48–72 h. Colonies were counted using a Flash & Go automated colony counter.
Preparation of total lipids
Overnight cultures of F. novicida U112 wild-type or naxD mutant strains were subcultured to OD600 = 0.02 and grown at 37°C in TSB supplemented with L-cysteine until the OD600 = 1.0. The cells were collected by centrifugation (5000 g, 20 min) and washed with PBS. The cell pellets were resuspended in a single-phase Bligh-Dyer mixture (Bligh and Dyer, 1959) consisting of chloroform, methanol and water (1:2:0.8, v/v), incubated at room temperature for 60 min, and centrifuged (10 000 g, 20 min) to remove insoluble debris. The supernatant was converted to a two-phase Bligh-Dyer system by adding chloroform and water to generate a mixture consisting of chloroform, methanol and water (2:2:1.8, v/v). The two phases of Bligh-Dyer system were separated under centrifugation and the lower phase was dried by rotary evaporation and under a stream of nitrogen. The total lipids were analysed using thin-layer chromatography (TLC) and LC-ESI/MS. The TLC plate was developed using the solvent chloroform, methanol, pyridine, acetic acid and water (25:10:5:4:3, v/v). Lipids were detected by spraying 10% of sulphuric acid in ethanol and charring at 300°C.
Negative ion mode electrospray ionization (ESI) mass spectrometry (MS) and MS/MS analysis
All ESI/MS and MS/MS spectra were acquired on a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA) equipped with an ESI source. Lipid A samples were dissolved in chloroform and methanol (2:1, v/v) containing 1% piperidine and subjected to ESI/MS in the negative ion mode via direct infusion (Garrett and Yost, 2006; Guan et al., 2007; Wang et al., 2009). Nitrogen was used as the collision gas for MS/MS experiments (Garrett and Yost, 2006; Guan et al., 2007; Wang et al., 2009). Data acquisition and analysis were performed using the instrument's Analyst QS software.
Liquid chromatography/mass spectrometry (LC/MS)
LC/MS of lipids was performed using a Shimadzu LC system (comprising a solvent degasser, two LC-10A pumps and an SCL-10A system controller) coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (as above). LC was performed at a flow rate of 200 μl min−1 with a linear gradient as follows: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 4 min. Mobile phase A consisted of methanol/acetonitrile/aqueous 1 mM ammonium acetate (60:20:20, v/v/v). Mobile phase B consisted of 100% ethanol containing 1 mM ammonium acetate. A Zorbax SB-C8 reversed-phase column (5 m, 2.1 × 50 mm) was obtained from Agilent (Palo Alto, CA). The postcolumn splitter diverted ~ 10% of the LC flow to the ESI source of the mass spectrometer.
Membrane fractionation
Fifty millilitres of F. novicida strains were harvested at OD600 = 1.0 by centrifugation for 20 min at 5000 g at 4°C. Cell pellets were washed with 50 mM K + HEPES, pH 7.5, resus-pended in 5 ml of the same buffer and passed through a French pressure cell at 18 000 p.s.i. Unbroken cells were then removed by centrifugation at 10 000 g for 20 min at 4°C. Membrane fractions were pelleted from whole-cell lysates by ultracentrifugation at 200 000 g for 2 h at 4°C. F. novicida fractionation and protein localization were verified using Western blotting (see supplemental experimental procedures). For E. coli, fractions were prepared similarly with the following exceptions: E. coli C41 (DE3) strains transformed with the empty vector or vector encoding naxD were grown in LB broth (1% tryptone, 0.5% yeast extract and 1% NaCl) with 100 μg ml−1 ampicillin and were induced using 1 mM IPTG when cell density reached OD600 = 0.8, then harvested when the OD600 = 2.0. NaxD protein expression was analysed using 12% SDS-PAGE gel and Coomassie staining.
Undecaprenyl phosphate-GalNAc deacetylase assay
These assays measured the deacetylase activity of proteins from the F. novicida wild-type or naxD mutant membrane fractions, whole-cell lysate of E. coli transformed with the empty vector, and the membrane and cytosolic fractions of E. coli transformed with vector encoding naxD (grown under inducing conditions). The 100 μl reaction mixture included 50 μg ml−1 protein from the bacterial fractions, 4.0 μM synthesized undecaprenyl phosphate-GalNAc (Song et al., 2009), 1 mM MnCl2, 150 mM KCl, 1.0 mg ml−1 BSA, 0.1% Triton X-100 and 50 mM HEPES (pH 7.5) and was incubated at 30°C. A 20 μl sample was removed at 0 and 5 h for F. novicida and 0 and 1 h for E. coli. Samples were converted to a two-phase Bligh-Dyer system by the addition of chloroform and methanol. After centrifugation, the lower phase was dried under nitrogen and analysed using LC-ESI/MS.
Statistical analysis
All macrophage replication, single infection, killing assay and zeta potential data were analysed for significance using the unpaired Student's t-test. For zeta potential, values beyond three standard deviations of the mean were excluded as outliers. The CI values from the mouse competition experiments were analysed with the one-sample Student's t-test and compared with 1. The mouse survival infection data were analysed for significance using the Gehan–Breslow–Wilcoxon test.
Supplementary Material
Acknowledgements
We dedicate this manuscript in memoriam of our friend, mentor, collaborator and renowned LPS biosynthesis expert Christian R. H. Raetz. We thank Larry Gallagher and Colin Manoil (University of Washington) for generously providing the pFFlp plasmid. In addition, we thank William Shafer, Thomas Henry, Brooke Napier and Tim Sampson for critical reading of this manuscript. The project described was supported by NIH Grant U54-AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Anthony LS, Gu MZ, Cowley SC, Leung WW, Nano FE. Transformation and allelic replacement in Francisella spp. J Gen Microbiol. 1991;137:2697–2703. doi: 10.1099/00221287-137-12-2697. [DOI] [PubMed] [Google Scholar]
- Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, Sellati TJ. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J Bacteriol. 2006;188:6443–6448. doi: 10.1128/JB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998;29:247–259. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- Basheer SM, Guiso N, Tirsoaga A, Caroff M, Novikov A. Structural modifications occurring in lipid A of Bordetella bronchiseptica clinical isolates as demonstrated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:1075–1081. doi: 10.1002/rcm.4960. [DOI] [PubMed] [Google Scholar]
- Beasley AS, Cotter RJ, Vogel SN, Inzana TJ, Qureshi AA, Qureshi N. A variety of novel lipid A structures obtained from Francisella tularensis live vaccine strain. Innate Immun. 2012;18:268–278. doi: 10.1177/1753425911401054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Wang C, Miller MA, Bina JE. The Bla2 beta-lactamase from the live-vaccine strain of Francisella tularensis encodes a functional protein that is only active against penicillin-class beta-lactam antibiotics. Arch Microbiol. 2006;186:219–228. doi: 10.1007/s00203-006-0140-6. [DOI] [PubMed] [Google Scholar]
- Bina XR, Lavine CL, Miller MA, Bina JE. The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol Lett. 2008;279:226–233. doi: 10.1111/j.1574-6968.2007.01033.x. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Clay CD, Soni S, Gunn JS, Schlesinger LS. Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J Immunol. 2008;181:5568–5578. doi: 10.4049/jimmunol.181.8.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Horwitz MA. Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann N Y Acad Sci. 2007;1105:160–186. doi: 10.1196/annals.1409.001. [DOI] [PubMed] [Google Scholar]
- Darling RG, Catlett CL, Huebner KD, Jarrett DG. Threats in bioterrorism. I: CDC category A agents. Emerg Med Clin North Am. 2002;20:273–309. doi: 10.1016/s0733-8627(02)00005-6. [DOI] [PubMed] [Google Scholar]
- Egberink H, Addie D, Belak S, Boucraut-Baralon C, Frymus T, Gruffydd-Jones T, et al. Bordetella bronchiseptica infection in cats. ABCD guidelines on prevention and management. J Feline Med Surg. 2009;11:610–614. doi: 10.1016/j.jfms.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Fujita H, Watanabe Y, Sato T, Ohara Y, Homma M. The entry and intracellular multiplication of Francisella tularensis in cultured cells: its correlation with virulence in experimental mice. Microbiol Immunol. 1993;37:837–842. doi: 10.1111/j.1348-0421.1993.tb01713.x. [DOI] [PubMed] [Google Scholar]
- Gallagher LA, McKevitt M, Ramage ER, Manoil C. Genetic dissection of the Francisella novicida restriction barrier. J Bacteriol. 2008;190:7830–7837. doi: 10.1128/JB.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TJ, Yost RA. Analysis of intact tissue by intermediate-pressure MALDI on a linear ion trap mass spectrometer. Anal Chem. 2006;78:2465–2469. doi: 10.1021/ac0522761. [DOI] [PubMed] [Google Scholar]
- Guan F, Uboh CE, Soma LR, Birks E, Chen J, Mitchell J, et al. LC-MS/MS method for confirmation of recombinant human erythropoietin and darbepoetin alpha in equine plasma. Anal Chem. 2007;79:4627–4635. doi: 10.1021/ac070135o. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Ernst RK. The structure and function of Francisella lipopolysaccharide. Ann N Y Acad Sci. 2007;1105:202–218. doi: 10.1196/annals.1409.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Craven RR, Fuller JR, Pickles RJ, Kawula TH. Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect Immun. 2007;75:1034–1039. doi: 10.1128/IAI.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect Immun. 2008;76:5843–5852. doi: 10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honn M, Lindgren H, Sjostedt A. The role of MglA for adaptation to oxidative stress of Francisella tularensis LVS. BMC Microbiol. 2012;12:14. doi: 10.1186/1471-2180-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert RR, Cotter PA. Laboratory Maintenance of Bordetella pertussis. Curr Protoc Microbiol. 2009 doi: 10.1002/9780471729259.mc04b01s15. Chapter 4: Unit 4B 1. [DOI] [PubMed] [Google Scholar]
- Imagawa T, Iino H, Kanagawa M, Ebihara A, Kuramitsu S, Tsuge H. Crystal structure of the YdjC-family protein TTHB029 from Thermus thermophilus HB8: structural relationship with peptidoglycan N-acetylglucosamine deacetylase. Biochem Biophys Res Commun. 2008;367:535–541. doi: 10.1016/j.bbrc.2007.12.144. [DOI] [PubMed] [Google Scholar]
- Inatsuka CS, Xu Q, Vujkovic-Cvijin I, Wong S, Stibitz S, Miller JF, Cotter PA. Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect Immun. 2010;78:2901–2909. doi: 10.1128/IAI.00188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto H, Mukae H, Date Y, Shimbara T, Mondal MS, Ashitani J, et al. Identification of hBD-3 in respiratory tract and serum: the increase in pneumonia. Eur Respir J. 2006;27:253–260. doi: 10.1183/09031936.06.00105904. [DOI] [PubMed] [Google Scholar]
- Kalhorn TF, Kiavand A, Cohen IE, Nelson AK, Ernst RK. A sensitive liquid chromatography/mass spectrometry-based assay for quantitation of aminocontaining moieties in lipid A. Rapid Commun Mass Spectrom. 2009;23:433–442. doi: 10.1002/rcm.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanistanon D, Hajjar AM, Pelletier MR, Gallagher LA, Kalhorn T, Shaffer SA, et al. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 2008;4:e24. doi: 10.1371/journal.ppat.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Ingram LO. Cloning and sequencing of a cellobiose phosphotransferase system operon from Bacillus stearothermophilus XL-65-6 and functional expression in Escherichia coli. J Bacteriol. 1993;175:6441–6450. doi: 10.1128/jb.175.20.6441-6450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE. 2011;6:e24201. doi: 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr N, Tirsoaga A, Blanot D, Fernandez R, Caroff M. Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgAS-activated ArnT ortholog. J Bacteriol. 2008;190:4281–4290. doi: 10.1128/JB.01875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, et al. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun. 2007;75:3305–3314. doi: 10.1128/IAI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DC, Jacobs DM. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry. 1976;13:813–818. doi: 10.1016/0019-2791(76)90181-6. [DOI] [PubMed] [Google Scholar]
- Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. Novel modification of lipid A of Francisella tularensis. Infect Immun. 2004;72:5340–5348. doi: 10.1128/IAI.72.9.5340-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 2006;6:69. doi: 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Guan Z, Ingram BO, Six DA, Song F, Wang X, Zhao J. Discovery of new biosynthetic pathways: the lipid A story. J Lipid Res. 2009;50(Suppl.):S103–S108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G, Meeker A, Dragulev B. fslE is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J Bacteriol. 2008;190:5353–5361. doi: 10.1128/JB.00181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom G, Lofgren S, Tarnvik A. A capsule-deficient mutant of Francisella tularensis LVS exhibits enhanced sensitivity to killing by serum but diminished sensitivity to killing by polymorphonuclear leukocytes. Infect Immun. 1988;56:1194–1202. doi: 10.1128/iai.56.5.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SHE. Studying trafficking of intracellular pathogens in antigen-presenting cells. Methods Microbiol. 2002;31:343–360. [Google Scholar]
- Schilling B, McLendon MK, Phillips NJ, Apicella MA, Gibson BW. Characterization of lipid A acylation patterns in Francisella tularensis, Francisella novicida, and Francisella philomiragia using multiple-stage mass spectrometry and matrix-assisted laser desorption/ ionization on an intermediate vacuum source linear ion trap. Anal Chem. 2007;79:1034–1042. doi: 10.1021/ac061654e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, Allen LA. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect Immun. 2009;77:1324–1336. doi: 10.1128/IAI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer SA, Harvey MD, Goodlett DR, Ernst RK. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1080–1092. doi: 10.1016/j.jasms.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Guan Z, Raetz CR. Biosynthesis of undecaprenyl phosphate-galactosamine and undecaprenyl phosphate-glucose in Francisella novicida. Biochemistry. 2009;48:1173–1182. doi: 10.1021/bi802212t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni S, Ernst RK, Muszynski A, Mohapatra NP, Perry MB, Vinogradov E, et al. Francisella tularensis blue-gray phase variation involves structural modifications of lipopolysaccharide o-antigen, core and lipid a and affects intramacrophage survival and vaccine efficacy. Front Microbiol. 2010;1:129. doi: 10.3389/fmicb.2010.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin VM, Pavlovich NV, Prozorova LA. Francisella tularensis resistance to bactericidal action of normal human serum. FEMS Immunol Med Microbiol. 1996;13:249–252. doi: 10.1111/j.1574-695X.1996.tb00246.x. [DOI] [PubMed] [Google Scholar]
- Szybalski W, Bryson V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J Bacteriol. 1952;64:489–499. doi: 10.1128/jb.64.4.489-499.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirsoaga A, El Hamidi A, Perry MB, Caroff M, Novikov A. A rapid, small-scale procedure for the structural characterization of lipid A applied to Citrobacter and Bordetella strains: discovery of a new structural element. J Lipid Res. 2007;48:2419–2427. doi: 10.1194/jlr.M700193-JLR200. [DOI] [PubMed] [Google Scholar]
- Titball RW, Petrosino JF. Francisella tularensis genomics and proteomics. Ann N Y Acad Sci. 2007;1105:98–121. doi: 10.1196/annals.1409.015. [DOI] [PubMed] [Google Scholar]
- Trent MS. Biosynthesis, transport, and modification of lipid A. Biochem Cell Biol. 2004;82:71–86. doi: 10.1139/o03-070. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Eur J Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Quinn PJ. Lipopolysaccharide: biosynthetic pathway and structure modification. Prog Lipid Res. 2010;49:97–107. doi: 10.1016/j.plipres.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, McGrath SC, Cotter RJ, Raetz CR. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, Raetz CR. Identification of undecaprenyl phosphate-beta-D-galactosamine in Francisella novicida and its function in lipid A modification. Biochemistry. 2009;48:1162–1172. doi: 10.1021/bi802211k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci USA. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Raetz CR. A two-component Kdo hydrolase in the inner membrane of Francisella novicida. Mol Microbiol. 2010;78:820–836. doi: 10.1111/j.1365-2958.2010.07305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.