Nanotechnology encompasses structures comparable in size with biomolecules and enables new ways to measure and detect biology, perturb cells, and treat patients. Here, Wong et al. review nanotechnology-based approaches for precisely measuring and perturbing living systems that may yield unexpected insights into systems biology as well as new therapeutic strategies for personalized medicine.
Keywords: Nanoparticle, microfluidics, single cell, biomarker, microneedle, targeted delivery
Abstract
Historically, biomedical research has been based on two paradigms. First, measurements of biological behaviors have been based on bulk assays that average over large populations. Second, these behaviors have then been crudely perturbed by systemic administration of therapeutic treatments. Nanotechnology has the potential to transform these paradigms by enabling exquisite structures comparable in size with biomolecules as well as unprecedented chemical and physical functionality at small length scales. Here, we review nanotechnology-based approaches for precisely measuring and perturbing living systems. Remarkably, nanotechnology can be used to characterize single molecules or cells at extraordinarily high throughput and deliver therapeutic payloads to specific locations as well as exhibit dynamic biomimetic behavior. These advances enable multimodal interfaces that may yield unexpected insights into systems biology as well as new therapeutic strategies for personalized medicine.
In the context of biology and medicine, nanotechnology encompasses the materials, devices, and systems whose structure and function are relevant for small length scales, from nanometers (10−9 m) through microns (10−6 m) (Whitesides 2003). This size regime is associated with intriguing phenomena in both living systems and artificial devices. In particular, the fundamental building blocks of life fall within this range, including biological macromolecules and cells. For example, dsDNA is a chain-like macromolecule with a diameter of 2 nm, cell membranes are sheet-like structures with a thickness of ∼10 nm, and (suspended) eukaryotic cells are approximately spherical with a diameter of ∼10 um. Remarkably, artificial nanostructures can also be constructed with comparable dimensions, including nanopores with ∼2-nm openings, inorganic nanowires of ∼10-nm diameter, and spherical nanoparticles of 10- to 100-nm diameter (Box 1). Moreover, this size regime is associated with unexpected chemistry and physics where molecular effects can play a significant role. This can be understood from a purely geometric argument—a width of ∼10 nm in a silicon nanowire corresponds to only ∼40 atoms. A consequence of this is that the surface area of these nanostructures is extremely large relative to the volume. If the same silicon nanowire has a square cross-section, then approximately one out of every 10 atoms is associated with the surface. In contrast, if we have a macroscopic wire with a width of 10 mm, only one in 10,000 atoms is associated with the surface. Finally, as a consequence of these small dimensions, electrons are spatially confined, resulting in exceptional electrical, magnetic, optical, and thermal properties (Alivisatos 2004). An exciting prospect is to bridge across these biomolecular and physical realms, leveraging unique capabilities from one realm for application in the other.
Box 1. Some assembly required: making nanostructures from molecular building blocks.
The production of nanostructures occurs through two general approaches (Madou 2012): (1) Top-down approaches impose a structure or pattern on a flat surface by successively adding or removing thin layers of materials. For example, photolithography uses UV light to pattern light-sensitive polymer into nanoscale features. Increased resolution can be achieved using electron or focused ion beams to pattern surfaces, particularly to mill or sculpt away material. (2) Bottom-up approaches are based on organizing atomic or molecular components into hierarchical assemblies. For instance, cytoskeletal polymers form through the sequential addition of monomeric protein units. Instead, carbon nanotubes or silicon nanowires are grown from chemical precursors in a gas or liquid phase. A key conceptual difference is that top-down fabrication is deterministic and well controlled, whereas bottom-up assembly is stochastic and may result in defects. Nevertheless, top-down fabrication is optimized for highly specific processing conditions that are incompatible with biologics, necessitating the use of bottom-up assembly.
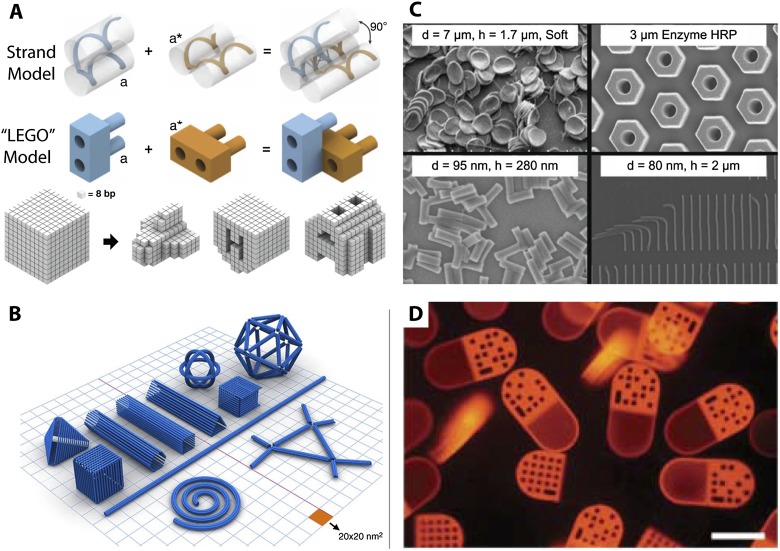
A particularly exciting example of bottom-up self-assembly has been the use of one-dimensional (1D) nucleic acid strands that organize into designed two-dimensional (2D) and three-dimensional (3D) architectures based on molecular recognition (Aldaye et al. 2008). Initial work based on the formation of branches and junctions inspired the design of 2D tiles that organized into hierarchical lattices based on logical rules (Seeman 2003). Subsequently, in “DNA origami,” a single long DNA strand could be guided with shorter “staple” strands to fold into more complex 2D shapes (Rothemund 2006). Most recently, these approaches have been extended to 3D structures. For instance, cuboidal “Lego-like” bricks have been produced, where the free ends are recognition sites that can bind uniquely to particular bricks in solution with a defined orientation (Fig. 2A; Ke et al. 2012) (). Nearly arbitrary “pixelated” objects can be designed by removing brick-like elements from a solid cube of ∼10 pixels per side (∼25 nm), with 100 different geometries demonstrated. Alternatively, at DNA strands can be woven into sheet-like structures that can be rolled, curved, and twisted (Fig. 2B; Dietz et al. 2009). One promising application for these techniques is to construct scaffolds where DNA-linked proteins, peptides, and inorganic nanostructures can be precisely positioned and dynamically actuated. Building on these concepts, a drug delivery “robot” has been demonstrated where logical aptamer recognition of cancer-associated antigens led to unveiling of antibody fragments that could molecularly interact with cell surface receptors (Douglas et al. 2012). Overall, DNA nanotechnology is extremely powerful for rationally designing 2D and 3D nanostructures with precise control that cannot be achieved using any other techniques. Nevertheless, a continuing challenge is to reduce the formation of defective and misfolded structures as well as establish the biocompatibility of these structures in vivo (Shih and Lin 2010).
A number of hybrid schemes have been developed that combine top-down and bottom-up approaches. For instance, in soft lithography, top-down features are replicated into plastic “stamps,” which can be coated with proteins or inorganic nanomaterials to be transferred to specific locations on a surface (Whitesides et al. 2001). A variation of this approach (PRINT) has been used to make polymeric particles, which are cast and removed using roll-to-roll manufacturing (Wang et al. 2011a). This approach can independently control particle size, shape, stiffness, material, and surface chemistry (Fig. 2C). This was used to systematically explore design rules for red blood cell-shaped particles, showing that eightfold softer particles displayed 30-fold enhanced circulation times (Merkel et al. 2011). Another example is the use of photolithography to cross-link hydrogels in microfluidic channels, allowing for layered composite architectures based on interfacial flows (Helgeson et al. 2011). This approach is particularly powerful for constructing complex geometries with different functionalities (Fig. 2D); for instance, multicompartment particles with barcode identifiers and regions of distinct nucleic acid probes for multiplexed analyte detection (Pregibon et al. 2007). This functionality could be further applied to generate heterogeneous textures, chemical patchiness, and continuous molecular gradients. Overall, these hybrid schemes take advantage of both bottom-up and top-down approaches for preparing nanostructures with greater consistency under more biocompatible conditions. However, a continuing challenge for widespread adoption will be to scale production for high-volume manufacturing.
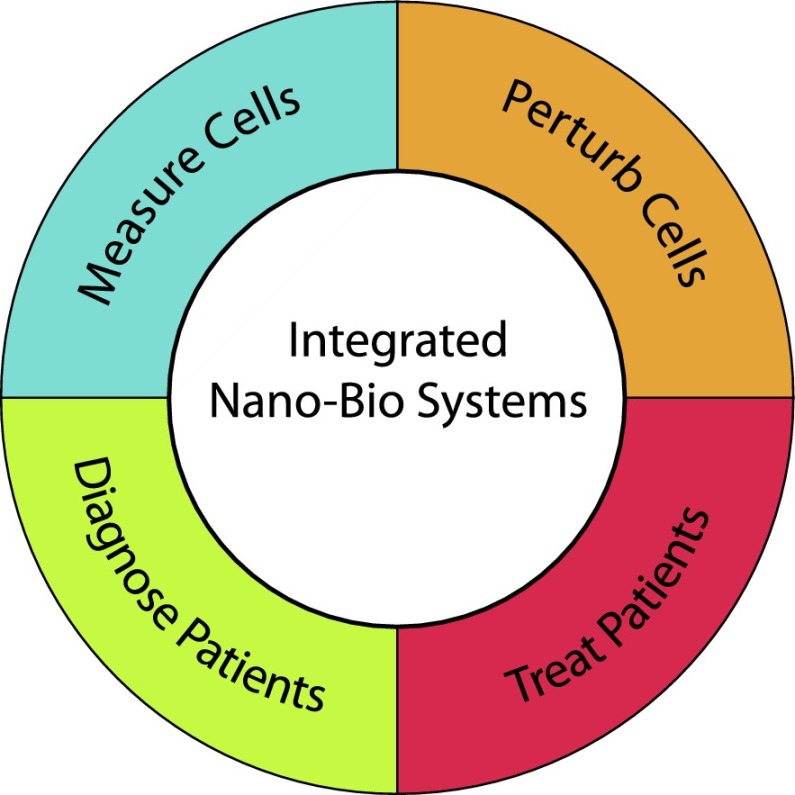
A conceptual framework for nano–bio interfaces can be structured based on two overarching themes (Fig. 1). First, nanotechnology enables new ways to measure and detect biology both in vitro and in vivo. For instance, nanoscale devices can sense minute differences at the level of single molecules and single cells. This exquisite sensitivity can be used, for instance, to characterize single-cell heterogeneity at extremely high throughput, revealing distinct hierarchies and subpopulations. This level of detail is often lost in traditional bulk assays, which measure “average” information about populations pooled from large numbers. Second, nanotechnology enables new ways to perturb cells and treat patients. For example, nanomaterials can be designed to precisely deliver therapeutics to targeted locations while overcoming or evading biological barriers, thus altering the inherent pharmacokinetics and biodistribution of the cargo (typically a drug). This avoids the complications associated with conventional delivery using soluble drugs, which are associated with low and inconsistent uptake. An exciting prospect is the synergistic integration of these two themes: nanotechnologies that can simultaneously detect and perturb biology. The design and construction of such biomimetic nanosystems, driven by feedback between nanomaterials and their environment, will give rise to emergent behaviors such as adaptation, self-organization, and amplification. These capabilities are potentially transformative for confronting the spatial and temporal complexity of living systems.
Figure 1.
Key themes underlying nanotechnology in biology and medicine, including new capabilities to measure or perturb cells in vitro as well as diagnose or treat patients. An exciting prospect is the future integration of these different capabilities for biomimetic behaviors such as adaption, self-organization, and amplification.
In this review, we review some illustrative examples of nanotechnology applied to biology and medicine. This is intended to make the new capabilities of nanotechnology accessible for nonexperts (Box1, Fig. 2), with particular emphasis on our respective areas of expertise in nanotechnology and microfluidics for mammalian cell biology and medicine. In particular, we address recent advances for characterizing biological systems: precision measurement of biomolecules and cells in vitro, manipulation of picoliter droplets, and the clinical evaluation of complex samples. We also examine innovative approaches for perturbing biological systems, including minimally invasive nanoneedles as well as rationally designed multifunctional nanoparticles. Due to space constraints, we only included a limited sampling of recent developments; we refer readers to more comprehensive reviews elsewhere for each topic. Finally, we consider the future prospects and challenges for these technologies.
Figure 2.
(A) DNA “Lego-like” bricks for three-dimensional cuboid structures. (B) DNA sheets assembled into curved and anisotropic geometries. (C) Nanoparticles of different materials, shapes, and sizes fabricated using PRINT. (D) Multicomponent barcoded particles fabricated using flow lithography. From Pregibon et al. (2007) and Ke et al. (2012). Reprinted with permission from AAAS. Reprinted from Shih and Lin (2010), with permission from Elsevier. Reprinted from Wang et al. (2011a); © 2011 Wiley-VCH Verlag GmbH & Co., KGaA, Weinheim.
Counting one by one: precision measurements of biomolecules and cells
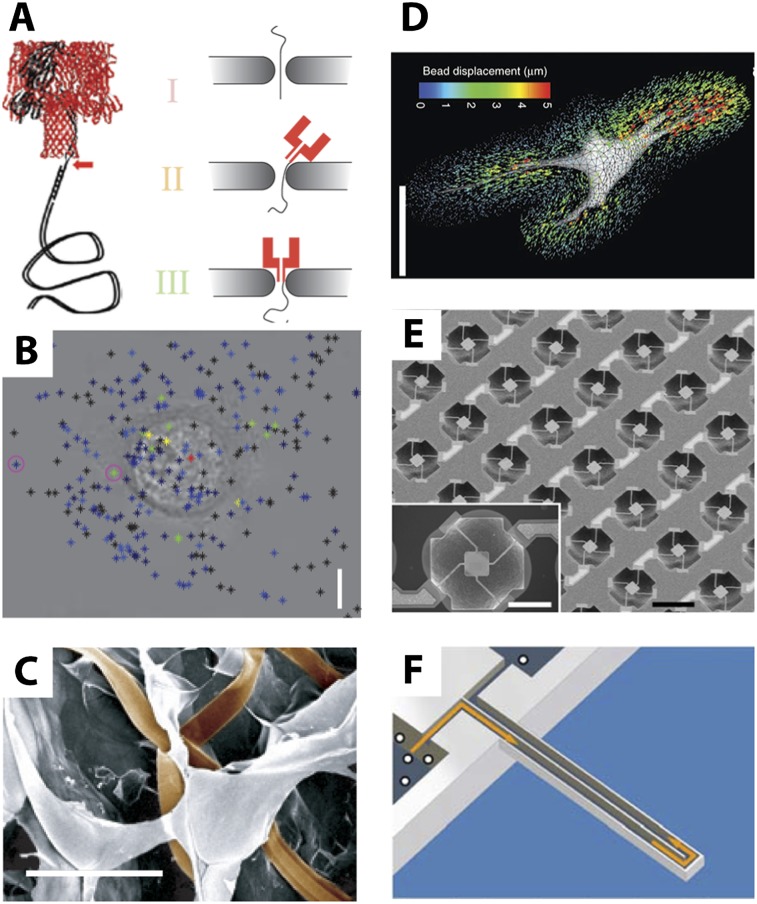
Historically, measurements of biomolecules and cells have occurred using bulk assays, such as lysates pooled from populations of thousands or millions of cells at the completion of the experiment. Instead, the small sizes associated with nanostructures enable them to probe and manipulate the dynamics of single cells and molecules with unprecedented resolution. One example of this is the use of nanoscale pores that can discriminate between molecules on the basis of size and biochemical characteristics (Branton et al. 2008). In particular, biological channels and pores are capable of regulating ion transport with a selectivity on the order of angstroms (0.1 nm or 10−10 m) (Bayley and Cremer 2001). Inspired by this biological functionality, it has been hypothesized that nanopores could unwind and unzip DNA so that individual nucleotides translocate sequentially in single file. Nevertheless, building precisely controlled pores with ∼1-nm diameter has been challenging (Venkatesan and Bashir 2011). Biological protein pores such as α-haemolysin (αHL) present well-defined apertures but generally require incorporation into mechanically delicate lipid bilayers to maintain stability. In contrast, artificial nanopores lack the chemical complexity of proteins and exhibit reduced selectivity. A hybrid scheme was recently implemented by directly inserting an αHL protein into a slightly larger inorganic nanopore (Fig. 3A; Hall et al. 2010). This scheme displays the benefits of a biological pore with increased selectivity and sensitivity but also the mechanical stability of an inorganic scaffold. This device could potentially be scaled so that large numbers of nanopores could operate in parallel. In principle, this approach could enable long reads of single molecules at high translocation velocities. An ongoing challenge for this technology is to achieve sensitivity with single-base-pair resolution due to the stochastic motion of DNA as well as the measurement sensitivity at fast translocation speeds.
Figure 3.
(A) Sequencing DNA with hybrid biological–artificial nanopores. (B) Measuring cell-secreted ROS using carbon nanotubes. (C) Electrophysiology in three-dimensonal (3D) scaffolds using nanowire arrays. (D) Mechanical deformation of 3D scaffolds using traction force microscopy. (E) Growth of adherent cells measured using microresonators. (F) Growth of cells in solution measured using suspended mass resonators. Reprinted by permission from Macmillan Publishers Ltd. from Hall et al. (2010), Jin et al. (2010), Legant et al. (2010), Son et al. (2012), and Tian et al. (2012). Reproduced with permission from Park et al. (2010).
The small sizes of nanostructure-based biosensors can be harnessed most effectively when they are patterned at high densities around single cells, enabling highly localized measurements at submicron and subcellular length scales. For example, existing technologies cannot measure the cellular secretion of growth factors, cytokines, and other signaling molecules into extracellular space (Love 2010). Nanostructures are advantageous not only because they are comparable in size with biomolecules but also because their size enables enhanced optical characteristics (Brolo 2012). One recent study used carbon nanotube sensors to detect reactive oxygen species (ROS) with single-molecule resolution based on optical fluorescence quenching (Jin et al. 2010). In response to epidermal growth factor (EGF) stimulation of A431 carcinoma cells, transient “hot spots” of high ROS concentration were observed in the proximity of the cell membrane (Fig. 3B) even when cells were fixed. An analogous approach was used to measure intracellular cytochrome C dynamics using plasmonic spectroscopy of gold nanoparticles (Choi et al. 2009). Remarkably, in response to proapoptotic stimuli, they were able to measure the release of cytochrome C from mitochrondria into the cytoplasm. Finally, an array of nanoplasmonic resonators patterned with submicron spacing was used to characterize the autocrine signaling of IL-2 in T lymphocytes, revealing a highly localized secretion profile (Wang et al. 2011c). Overall, these optical nanostructures enable exquisite molecular sensitivity with high spatial and temporal resolution, capable of distinguishing between molecules secreted by a particular cell and those in the background. Nevertheless, producing these nanostructures requires specialized fabrication techniques that are challenging to scale up, which will need to be addressed before they can be widely used.
Another readout of interest for single cells is electrical and ion channel activity. Such elecrophysiological measurements are highly relevant for neuroscience (Alivisatos et al. 2013) as well as pharmacological testing (Dunlop et al. 2008). Nevertheless, existing patch clamps and electrodes are relatively bulky and cumbersome so that they integrate poorly with three-dimensional (3D) tissues and do not achieve single-cell resolution. Nanostructures add new capabilities here, since their high surface area to volume ratios make them extremely sensitive to electrical changes. There has been extensive work to develop integrated circuits that are highly flexible, stretchable, and biocompatible (Kim et al. 2012a). One recent study incorporated a flexible mesh-like array of electrical sensors that formed an interpenetrating network with a 3D scaffold (Fig. 3C). Subsequent culturing of neurons showed good viability over several weeks, and simultaneous recordings of electrical activity from different cells was possible. This degree of seamless integration may have occurred because the synthetic architectural elements were comparable in size with the neurons. Future work with increased densities of electrical sensors could potentially be used to map out neural circuits (Alivisatos et al. 2013). One difficulty with this approach is the positioning of each electrode to interface with specific cells of interest.
The mechanical forces exerted by cells to sense and remodel their surroundings are extremely important for understanding how they regulate cell and tissue function (Nelson and Bissell 2006). This has been previously explored in the context of cells adhering to flat two-dimensional (2D) substrates. Recently, there has been increasing interest in measuring how cells interact with 3D microenvironments, which may be more physiologically relevant (Baker and Chen 2012). The forces applied by cells to deform 3D scaffolds was quantified by tracking the motion of tens of thousands of embedded fluorescent nanoparticles (Fig. 3D; Legant et al. 2010). Remarkably, they found that fibroblasts in 3D exerted strong traction forces near the tips of extensions, analogous to forces measured in 2D near lamellipodia. An important next step will be to adapt this technique to systems more reminiscent of the extracellular matrix (ECM), which exhibit highly heterogeneous and dynamic architectures (Even-Ram and Yamada 2005).
Finally, nanostructures can be used to directly measure single-cell phenotypes. In particular, one source of cell-to-cell variability is the cell cycle, driving growth dynamics and division events (Snijder and Pelkmans 2011). Resonating sensors were used to measure the mass of adherent cells over 50 h (Fig. 3E; Park et al. 2010). Alternatively, a resonating sensor with suspended geometry was used to measure the mass of suspended cells over 35 h (Fig. 3F; Son et al. 2012). Remarkably, both groups observed that growth rate is not constant but increases with increasing cell size. These growth rates varied at certain checkpoints in the cell cycle and also varied at the single-cell level across the population. These techniques may help to reveal mechanically driven feedback mechanisms used to constrain variability and maintain homeostasis at the cell and tissue level (Kafri et al. 2013).
Overall, nanostructures enable sensitive measurements of single molecular and cellular behaviors, enabled by sizes comparable with these biological building blocks and enhanced optical, electronic, and mechanical characteristics. Nevertheless, the general schemes described here require close proximity to cells, which often raises questions of biocompatibility for extended long-term measurements. Moreover, these techniques require highly specialized fabrication techniques as well as measurement infrastructure. As such, they are currently much more effective for in vitro measurements compared with in vivo.
Divide and conquer: droplets for high-throughput biology
Another approach for accessing single-molecule dynamics and single-cell heterogeneity is based on using microfluidics to partition bulk solutions into smaller volumes that contain discrete numbers of molecules or cells. For example, droplets of aqueous solution can be formed with volumes of ∼1 pL (10−12 L) or ∼10-μm diameter. At these small sizes, the effective concentration of reagents is increased, and the diffusion distance is considerably decreased (Leamon et al. 2006; Vincent et al. 2010). This may be transformative for high-throughput assays (∼109 droplets per milliliter) where extremely large numbers of reactions must be performed in parallel while efficiently using expensive reagents (Guo et al. 2012; Schneider et al. 2013).
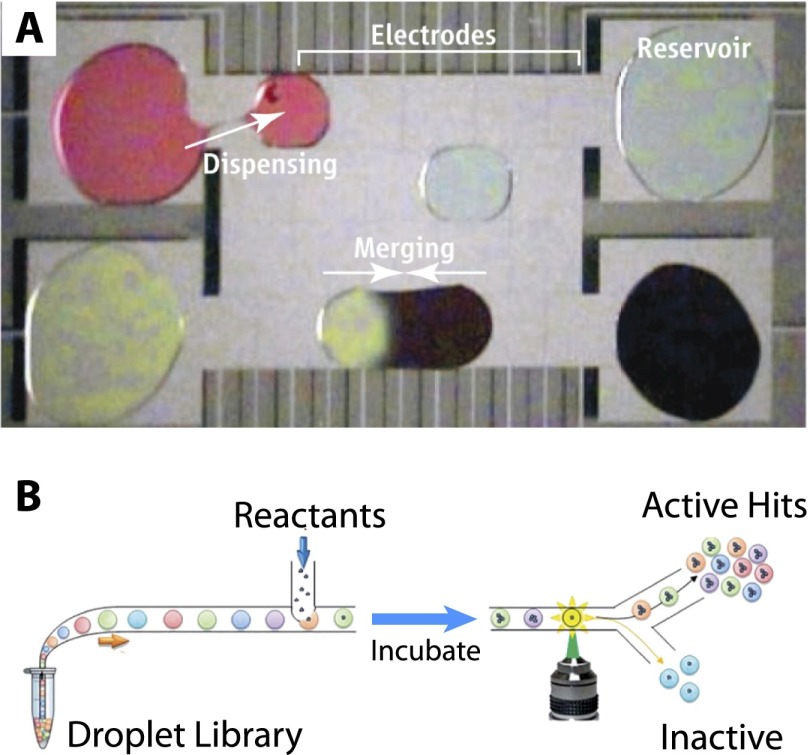
One device is based on the electrical manipulation of droplets on flat surfaces (digital microfluidics) (Wheeler 2008). A modular set of operations could be performed on droplets, including moving, merging, mixing, splitting, and dispensing from different reservoirs (Fig. 4A). This scheme is particularly advantageous for solid-phase or liquid–liquid extraction using nonaqueous solvents, which are generally incompatible with conventional plastic microfluidic devices. For example, estrogen could be directly extracted from 1 μL of tissue, blood, or serum in ∼10 min without user intervention, corresponding to a 1000-fold reduction in sample volume and a 20-fold increase in speed (Mousa et al. 2009). These analytes were then sensitively quantified using mass spectrometry. Dried blood spots could also be analyzed using a similar technique, avoiding the issues of fouling and clogging that challenge channel-based microfluidic devices (Jebrail et al. 2011).
Figure 4.
(A) One-picoliter droplets can be dispensed, moved, merged, mixed, and split in an open geometry using electric fields. (B) Droplet libraries can be challenged against a reactant and then sorted at ultrahigh throughput in channels. From Wheeler (2008). Reprinted with permission from AAAS. Reproduced from Guo et al. (2012) with permission of The Royal Society of Chemistry.
Alternatively, a channel geometry was implemented where droplets of aqueous solution are compartmentalized by an oil phase and lined up single file (Fig. 4B). Libraries of droplets encoding different analytes such as drug compounds, viruses, antibodies, or enzymes could be screened against cells or other reactants (Guo et al. 2012). These droplets can be interrogated by fluorescence and sorted at high speeds on the order of 108 droplets per day. This technology was applied to the directed evolution of the enzyme horseradish peroxidase, which generated mutants with catalytic rates >10 times faster than the parent, with a 1000-fold increase in speed and a 1 million-fold reduction in cost (Agresti et al. 2010). Another application is digital PCR, where DNA can be compartmentalized in the single-molecule limit and amplified without bias (Baker 2012). One challenge for droplet microfluidics is to interface with the macroscopic world, whether to isolate individual droplets of interest or for highly combinatorial assays (Guo et al. 2012). One possibility to address these difficulties could be the use of on-chip logic and computation (Prakash and Gershenfeld 2007) for large-scale integration (Melin and Quake 2007).
Finding a needle in a haystack: extreme clinical diagnostics
Blood and other biofluids encode a tremendous amount of information about the healthy or diseased state of patients (Toner and Irimia 2005). Unlike in vitro measurements, deciphering clinically relevant information often requires isolating certain biomarkers from an extraordinarily complex and heterogeneous mixture of proteins (Anderson and Anderson 2002) or cells (Mach et al. 2013). These biomarkers of interest may be extremely rare and obscured by the complex background of the overall population. Moreover, effective point-of-care diagnostics must be rapid, accurate, and low cost to serve patients and inform clinical diagnostics (Chin et al. 2012). Nanostructures are advantageous, since their high surface areas can be used to capture clinically relevant biomarkers through molecular recognition processes. The enhanced chemical and physical properties can be then be used to detect or isolate these biomarkers.
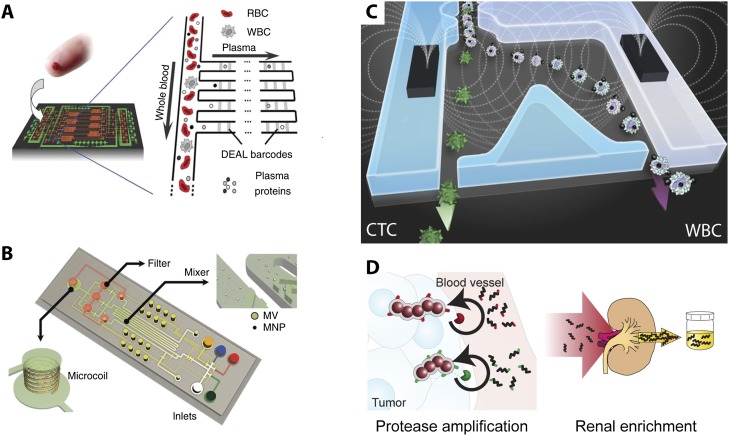
Rapid, multiplexed proteomic analysis has been demonstrated from microliter quantities of blood using integrated microfluidic chips (Fan et al. 2008). A robust and modular patterning scheme was developed where nonoverlapping sets of DNA linkers immobilize antibodies at discrete locations (Fig. 5A). The highly reproducible patterning of these biomolecules at high density was critical to achieve this multiplexed readout. These antibodies are then used in a sandwich assay with a secondary antibody, allowing measurement of twelve separate biomarkers with high dynamic range. The correlations in cytokine, growth factor, and antigen expression could be used to stratify patient disease state. This approach was also applied to measure secreted signals from immune cells, revealing high functional heterogeneity at the single-cell level (Ma et al. 2011).
Figure 5.
(A) Integrated barcode chip for proteomic biomarkers from blood. (B) Protein typing of circulating microvesicles using miniaturized NMR. (C) Antigen-dependent and -independent enrichment of rare tumor cells using magnetic nanoparticles. (D) Multiplexed synthetic biomarkers are amplified by enzymatic cleavage near tumors followed by renal concentration for noninvasive monitoring of disease progression. Reprinted by permission from Macmillan Publishers Ltd. from Fan et al. (2008) and Shao et al. (2012).
Microvesicles secreted from tumors have also been characterized as biomarkers (Shao et al. 2012). Microvesicles were labeled using clusters of target-specific magnetic nanoparticles, which could be sensitively detected using micro-NMR (nuclear magnetic resonance) technology based on changes in spin–spin relaxation time (T2) (Fig. 5B; Lee et al. 2008). This approach revealed striking heterogeneity in single microvesicles compared with the bulk analyses that have historically been performed. Remarkably, the proteomic signature of these microvesicles was consistent with the cells of origin. Furthermore, microvesicle expression profiles tracked efficacy of drug treatment and were predictive of differences in treatment mechanisms.
Circulating tumor cells (CTCs) shed by metastatic tumors into the blood have generated much recent interest. However, CTCs are extremely rare, intermixed with blood cells at concentrations of approximately one per billion in clinical samples (Yu et al. 2011). An established biomarker for CTCs is EpCAM, which is expressed by cells of epithelial origin but lacking in blood cells. One approach for “CTC-Chips” was based on the enhanced surface area associated with micro/nanostructures for adhesion-based capture using arrays of EpCAM-functionalized micropillars (Nagrath et al. 2007). This technology was clinically validated for tracking disease progression and informing therapeutic treatments (Maheswaran et al. 2008). Nevertheless, one difficulty posed by laminar fluid flow at these length scales is relatively slow mixing (Stone et al. 2004), which limited the transport of CTCs to the capture surface. The second generation “HB-Chip” addressed this issue by using topographically patterned “herringbones” to promote chaotic mixing (Stott et al. 2010). Remarkably, this technology also captured multicellular CTC “clusters,” raising the possibility that collective or cooperative behaviors may be advantageous for metastatic dissemination. Additional texturing of the capture surface using silicon nanowires has also demonstrated excellent capture efficiencies (Wang et al. 2011b).
The third-generation CTC-iChip was based on the separation of cells bound with ferromagnetic nanoparticles (Ozkumur et al. 2013). This approach was highly controlled using deterministic particle ordering through inertial focusing (Di Carlo et al. 2007), allowing precise deflection of selected cells into collection channels (Fig. 5C) at extremely high throughput (8 mL/h ∼ 107 cells/sec). The small size of these nanoparticles is advantageous for binding efficiently to surface receptors on CTCs—even those with relatively low expression that may not be captured using existing immunoaffinity techniques. Furthermore, a negative depletion approach enriches for cancer cells that do not express established epithelial cell surface markers, including triple-negative breast cancers, pancreatic cancers, and melanoma. These isolated CTCs displayed morphological similarities to tumors of origin but considerable variability in size. This single-cell heterogeneity was further interrogated by multigene microfluidic quantitative RT–PCR of 15 different prostate cancer cells, revealing distinct subsets with variable expression of epithelial, mesenchymal, and stem cell biomarkers as well as androgen receptors. This approach is highly promising for monitoring and guiding personalized therapies.
One unresolved question for biomedical diagnostics is how early biomarkers can be detected during disease progression. Recently, a quantitative framework was proposed for estimating measurable levels of biomarker based on their secretion from tumors (relative to healthy tissues), transport into vasculature, and subsequent dilution, degradation, and elimination (Hori and Gambhir 2011). A troubling conclusion was a sizable mismatch between tumor size and detection limits, translating to a period of years before a tumor could be diagnosed.
A new approach was recently demonstrated for amplifying biomarkers so that they can be more easily detected (Kwong et al. 2013). A library of synthetic, mass-encoded peptides was linked to nanoparticles using protease-cleavable linkers (Fig. 5D). During liver fibrosis or colorectal cancer, increased expression of proteases such as matrix metalloproteins (MMP) led to repeated catalytic cleavage of synthetic linkers, leading to liberation and concentration of peptides in the urine. Samples were then tested using mass spectroscopy for highly sensitive measurements of the mass-encoded “synthetic biomarkers.” This scheme overcomes the limitations of conventional biomarker detection by amplifying the initial signal, increasing sensitivity using stable synthetic peptides that do not occur biologically (low background), and multiplexing. This initial demonstration in small animals shows great promise for further clinical applications with humans.
The integration of microfluidics and nanotechnology has enabled new modalities for point-of-care diagnostics as well as implantable devices for monitoring patients, particularly for early detection and to measure drug efficacy. In particular, microfluidic technologies with integrated sensors for processing bodily fluids such as blood or saliva can be manufactured very reliably in a high-throughput format and also are very robust. Nanostructures, by virtue of their small dimensions, can interact precisely with molecular and cellular components as well as exhibit exceptional electronic, magnetic, and optical properties. As a consequence, nanostructures can considerably enhance the sensitivity of these assays. However, these nanostructures may require highly specialized production techniques that are difficult to scale up (Box 1). A crucial challenge for the future will be to translate these technologies to the bedside or point of care with sufficiently high accuracy and low cost.
Lying on a bed of nails: minimally invasive delivery using nanoneedles
A continuing challenge for therapeutic applications is targeted delivery of agents to defined locations in cells and tissues. Biological barriers such as the cell membrane, mucosal layers, or skin have evolved to be highly efficient at excluding unrecognized materials. For instance, eukaryotic cell membranes partition the interior of a cell from the surrounding extracellular environment and are comprised of amphiphilic lipids with a hydrophilic head and a hydrophobic tail (De Weer 2000). These lipids are arranged to form a hydrophobic core region that restricts the transport of hydrophilic and polar molecules across the bilayer. As a consequence, using such molecules to perturb cellular function often requires special techniques to bypass the cell membrane, such as electroporation or delivery agents, which may have a detrimental effect on cell viability. As a consequence, molecular agents such as plasmid DNA, siRNA, peptides, proteins, and small molecule drugs may be ineffective when delivered systemically. Nanostructures with small diameters but a high aspect ratio are a highly promising alternative for bypassing these barriers in order to deliver agents to defined locations.
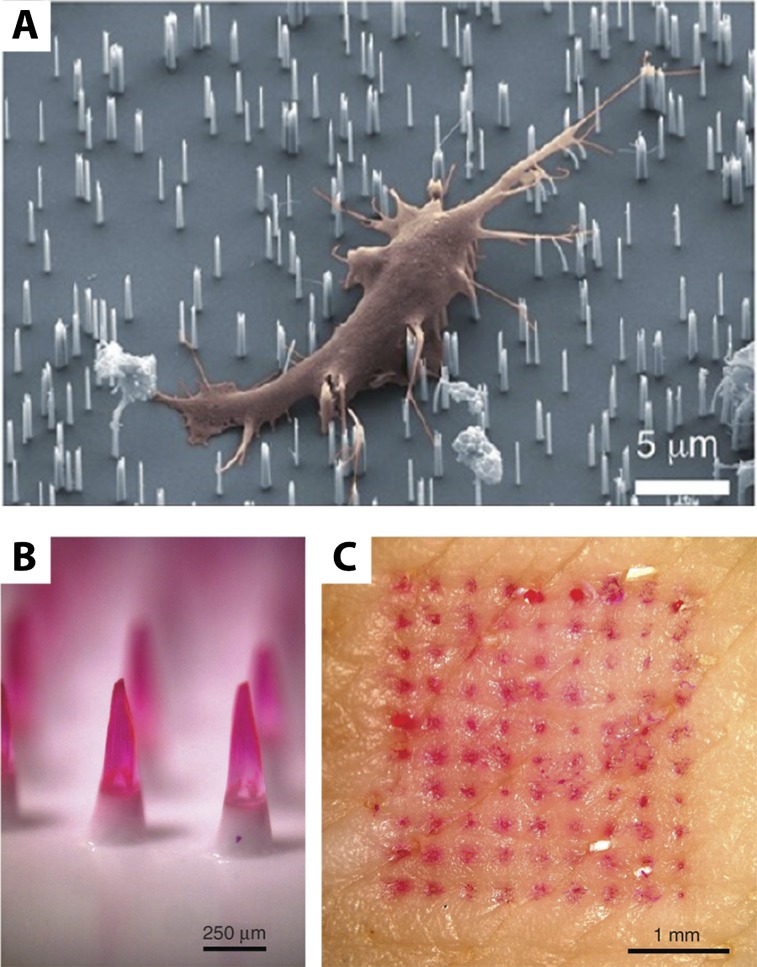
A nanopatterned surface consisting of an array of silicon nanowires with an ∼50-nm diameter and ∼1-μm height was functionalized with a variety of molecular agents in a microarray format (Shalek et al. 2010, 2012). Remarkably, these nanowires spontaneously inserted into cells without inducing apoptosis (Fig. 6A). This led to robust delivery of molecular agents (>95%) into the cellular interior to guide neuronal growth, siRNA knockdown, and inhibition of apoptosis as well as target proteins to organelles. These nanopatterned surfaces are fully compatible with existing printing technologies, enabling “living-cell microarrays” for high-throughput screens of cell stimuli with greatly increased efficiency. Nevertheless, one limitation of this technique is that the concentration and release kinetics of drugs are not well controlled. An alternate approach was based on an array of hollow alumina “nanostraws” that served as fluid conduits from an adjacent reservoir (VanDersarl et al. 2012). The composition of molecular species in the reservoir could be defined using microfluidics, allowing spatial gradients or transient pulses of molecular species. Further control over molecular delivery and intracellular transport was achieved by applying spatially localized electrical pulses, which enabled >70% transfection efficiency (Xie et al. 2013). Future work will be necessary to explore the role of physical nanotopography on cell behavior (Kim et al. 2012b) as well as confirm long-term cell viability.
Figure 6.
(A) Arrays of silicon nanowires can deliver siRNA into the interior of dendritic cells. Dissolving polymer microneedles (B) can deliver influenza vaccine to defined locations beneath the skin (C). Reprinted by permission from Macmillan Publishers Ltd. from Sullivan et al. (2010). Reprinted with permission from Shalek et al. (2012). © 2012 American Chemical Society.
Human skin also displays barrier properties that limit molecular transport (Prausnitz et al. 2004). The outermost layer of the epidermis, known as the stratum corneum, displays a high degree of structural heterogeneity. As a consequence, molecular transport across this layer must occur through highly tortuous pathways and can be strongly impeded. One approach to deliver molecular agents is to use hypodermic needles that inject past the stratum corneum. However, this is a painful, highly invasive process that cannot be self-administered. Instead, an array of polymeric microneedles with a smaller diameter can painlessly insert into skin, delivering drugs to a controlled depth (Fig. 6B,C; Sullivan et al. 2010). This is particularly useful for vaccination, since immunogenic Langerhans cells are often localized ∼100 μm from the surface. These microneedles degrade within minutes, allowing the skin to reseal and recover barrier function to prevent infection. Moreover, these microneedles were found to improve on antibody and cellular immune response in mice compared with conventional needles. Variations on this concept include multilayer-coated microneedles, which can be used for programmed and sustained delivery profiles (DeMuth et al. 2013).
Deliver to zip code: multifunctional nanoparticles for therapy and diagnostics
Drug-loaded nanoparticles can also be advantageous for delivering concentrated doses to target sites through prolonged circulation times and increased uptake combined with reduced toxicity elsewhere (Shi et al. 2010). Size appears to play an important role in spherical particle transport, since micrometer-scale particles are often cleared by macrophage phagocytosis or filtration in the liver or spleen (Mitragotri and Lahann 2009). Nevertheless, these behaviors are also affected by physical characteristics such as shape, mechanics, and surface chemistry (Nel et al. 2009). For example, pathogens such as bacteria and viruses display distinctive asymmetric and anisotropic shapes, which enable specific interactions with target cells and evasion of the immune response (Yoo et al. 2011). In contrast, red blood cells exhibit a characteristic flexible, discoidal shape that avoids filtration in the spleen, despite their relatively large size (Skalak and Branemark 1969). Inspired by this physical phenomenon, long, flexible polymeric micelles were designed that persist in circulation for extended periods with minimal phagocytosis (Geng et al. 2007). Recent advances in synthesis and fabrication have enabled the design of biomimetic nanosystems that can cooperate to synergistically improve their functionality (Box 1).
Nanoparticles can also incorporate advanced functionality such as targeting to specific organs and cells, responsiveness to external stimuli, imaging capabilities, and drug delivery (Bao et al. 2013). For example, passive targeting of tumors based on enhanced permeability and retention (EPR) in “leaky vasculature” (Chauhan et al. 2011) can be complimented by “active” targeting of cancer cells. For instance, tumor cells and vasculature often display distinct molecular “zip codes” that can be targeted by short peptide sequences (Ruoslahti 2012). Remarkably, certain peptides can actually penetrate deep into tissues, thereby overcoming elevated interstitial pressures (Sugahara et al. 2009). These findings, which originated in phage display and further developed through the imaging of targeted nanomaterials, have led to new fundamental insights on the trafficking of particulate materials in vivo, termed the CendR pathway (Teesalu et al. 2009).
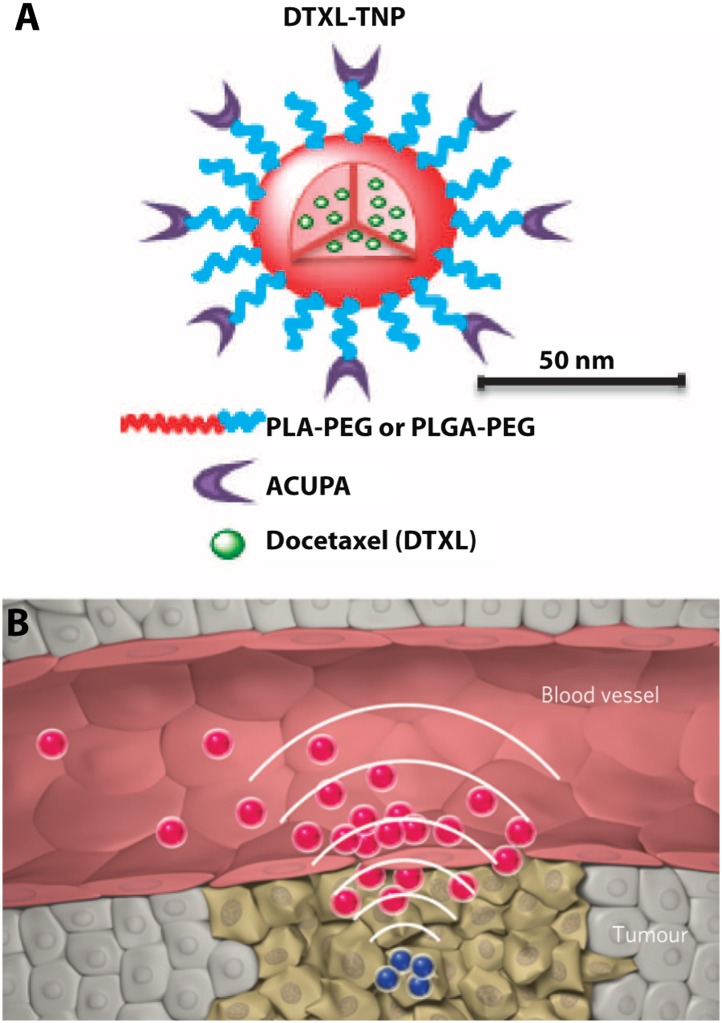
This approach was clinically translated as a nanoparticle that uses a targeting ligand for serum membrane antigen (PSMA) (Fig. 7A; Hrkach et al. 2012). This nanoparticle incorporates a “stealth” polymer brush shell that limits immunosurveilance as well as a degradable hydrophobic core that could be loaded with drugs. These optimized characteristics promoted circulation times while reducing liver accumulation. Moreover, these nanoparticles exhibited increased tumor accumulation relative to solvent-based drugs and prolonged tumor suppression. In preliminary patient trials, tumor shrinkage was observed even at diminished dosages of nanoparticles relative to solvent-based drugs.
Figure 7.
(A) Nanoparticle consisting of a degradable polymeric core loaded with drugs protected by a “stealth” polymer brush shell with targeting ligands. (B) Amplified targeting of nanoparticles by locally induced coagulation. From Hrkach et al. (2012). Reprinted with permission from AAAS. Reprinted by permission from Macmillan Publishers Ltd. from von Maltzahn et al. (2011).
A particularly challenging but attractive therapeutic cargo is siRNA. In principle, it can silence so-called “undruggable” targets but must be delivered repeatedly to the cytosol of the target cell to engage the silencing machinery in the RNA-induced silencing complex (RISC) (Whitehead et al. 2009). Moreover, siRNA is vulnerable to degradation by serum nucleases and renal filtration. Nanomaterials have the potential to address these issues by enabling targeted delivery of enhanced payloads while protecting the siRNA until needed (Davis et al. 2008). Exciting early clinical results have been demonstrated in cancer patients using a synthetic delivery based on a transferrin-targeted cyclodextran particle with a “stealth” polymer brush shell (Davis et al. 2010). Recently, tumor-penetrating nanocomplexes exploiting the CendR pathway have been developed to target essential oncogenes such as ID4 (Ren et al. 2012). An important next step will be to enhance the potency of siRNA, which has been demonstrated through the addition of lipid-like nanomaterials, enabling the silencing of multiple genes simultaneously (Love et al. 2010). It should be noted that nanomaterials have been used extensively to silence targets related to liver function, including for liver cancer, hypercholesterolemia, diabetes II, HBV, etc. (Whitehead et al. 2009) Nevertheless, additional work will be needed to extend delivery platforms beyond the liver and tumors to other normal and diseased tissues.
One unresolved issue is whether the additional functionality of these nanoparticles has trade-offs by increasing manufacturing complexity and the likelihood of unforeseen effects in vivo (Cheng et al. 2012). For instance, loading multiple drugs may result in cross-reactivity and difficulties in coordinating release profiles. Alternatively, given the finite volume available in a nanoparticle, loading both imaging and therapeutic agents may mean diminished effectiveness due to insufficient quantities of both. Rather that increase the complexity of individual nanoparticles, researchers have designed cooperative nanosystems where the action of simple, specialized particles are spatially and temporally coordinated. One approach relies on nanoparticles that can communicate through the environment to amplify the recruitment of therapeutics to a tumor site (von Maltzahn et al. 2011). In this system, gold nanorods passively accumulate in tumors, where they activate a coagulation response upon heating (Fig. 7B). This coagulation serves as a “signal” for “receiver” nanoparticles that target the transglutaminase or polymerized fibrin present in the coagulated environment. This communication enhances accumulation of receiving nanoparticles by 40-fold relative to noncommunicating controls, corresponding to an amplification of 35,000 molecules per signaling entity. In a similar manner, a two-particle system was designed with one nanoparticle increasing tumor permeability upon heating and inducing stress-related up-regulation of p32 surface receptors (Park et al. 2010). These receptors were then targeted by a second doxorubicin-loaded liposome, thereby amplifying therapeutic delivery to the permeated tumor. Such cooperative, synergistic behaviors offer an alternative route for the design of new therapeutic strategies.
Discussion
Nanotechnology offers exciting new approaches for both measuring living systems, including characterizing distinctive phenotypes and enriching clinically relevant biomarkers, and perturbing living systems, such as targeted drug delivery modalities that can overcome biological barriers. These various examples can be generalized as structural or functional interfaces for communicating information between biotic and abiotic systems. In this context, several key themes emerge that motivate the ongoing development of nanotechnology for biology and medicine. First, nanotechnology enables interfaces between the macroscale of humans down to the nanoscale of molecules and cells. These tools can be used to precisely examine and perturb individuals within a population, rather than the relatively crude bulk methods that have been used historically. Second, nanotechnology is based on translation and conversion between different types of signals. For example, cells and tissues often signal using biochemical messengers in ionic solution, which is very different from the conventional technologies used by humans. Nevertheless, the exceptional chemical and physical properties of nanomaterials, including enhanced electronic, optical, and thermal functionality, can be used to locally detect or perturb biological phenomena. Finally, many biomedical applications require the compatibility of artificial nanomaterials with biological systems. Artificial nanomaterials are often synthesized and fabricated under conditions that are highly toxic to cells and tissues. Thus, these artificial nanomaterials may not function as designed under complex physiological conditions. Maintaining the structure and function of both artificial and biological elements over extended time periods remains an ongoing area of research.
Moving forward, translating these proof-of-concept technologies from a controlled laboratory environment to widespread usage will require extensive testing and validation. In particular, biomedical diagnostics have to meet relatively high standards for performance and accuracy, especially compared with existing technologies. Another challenge is manufacturing, since nanotechnological systems are often synthesized or fabricated using highly specialized techniques that are not amenable for commercial scale-up. One approach, reminiscent of synthetic biology (Cheng and Lu 2012), is to begin standardization of nanotechnological “parts,” which would enable the ease of comparison between different studies as well as integration of these different modular components.
Finally, as artificial nanotechnologies become increasingly capable of interacting with each other and their biological counterparts, the resulting behaviors may be highly unpredictable and counterintuitive. For example, signaling and feedback mechanisms between the artificial and biological systems may generate emergent behaviors that would not be observed otherwise. Nonetheless, harnessing this complexity can enable cooperative systems capable of performing tasks beyond the individual functionalities of the constituent parts. For instance, under the proper conditions, biomimetic behaviors could occur, such as amplification, optimization, mapping, self-assembly, collective motion, synchronization, and decision-making. Ultimately, a systems approach (Csete and Doyle 2002) to model the interactions between artificial and biological systems may aid in understanding modularity, robustness, and fragility, which could inform biological investigations and guide new therapeutic strategies.
Acknowledgments
We apologize for those primary works that are not cited due to the scope of this review and space constraints. We thank S. Hauert and E. Seker for helpful comments, and M. Karabacak, G. Kwong, and A. Rotem for assistance with figures. I.Y.W. was a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2065-10). S.N.B. is a Howard Hughes Medical Investigator. Additional support was provided by the Lustgarten Foundation (to S.N.B.) and the National Institutes of Health (U54 CA151884, Massachusetts Institute of Technology-Harvard Center of Cancer Nanotechnology Excellence, to S.N.B., and P41-EB002503, BIoMEMS Resource Center, to M.T.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226837.113.
References
- Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret J-C, Marquez M, Klibanov AM, Griffiths AD, Weitz DA 2010. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci 107: 4004–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaye FA, Palmer AL, Sleiman HF 2008. Assembling materials with DNA as the guide. Science 321: 1795–1799 [DOI] [PubMed] [Google Scholar]
- Alivisatos P 2004. The use of nanocrystals in biological detection. Nat Biotechnol 22: 47–52 [DOI] [PubMed] [Google Scholar]
- Alivisatos AP, Andrews AM, Boyden ES, Chun M, Church GM, Deisseroth K, Donoghue JP, Fraser SE, Lippincott-Schwartz J, Looger LL, et al. 2013. Nanotools for neuroscience and brain activity mapping. ACS Nano 7: 1850–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG 2002. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics 1: 845–867 [DOI] [PubMed] [Google Scholar]
- Baker M 2012. Digital PCR hits its stride. Nat Methods 9: 541 [Google Scholar]
- Baker BM, Chen CS 2012. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J Cell Sci 125: 3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G, Mitragotri S, Tong S 2013. Multifunctional nanoparticles for drug delivery and molecular imaging. Annu Rev Biomed Eng 15: 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley H, Cremer PS 2001. Stochastic sensors inspired by biology. Nature 413: 226–230 [DOI] [PubMed] [Google Scholar]
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. 2008. The potential and challenges of nanopore sequencing. Nat Biotechnol 26: 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brolo AG 2012. Plasmonics for future biosensors. Nat Photonics 6: 709–713 [Google Scholar]
- Chauhan VP, Stylianopoulos T, Boucher Y, Jain RK 2011. Delivery of molecular and nanoscale medicine to tumors: Transport barriers and strategies. Annu Rev Chem Biomol Eng 2: 281–298 [DOI] [PubMed] [Google Scholar]
- Cheng AA, Lu TK 2012. Synthetic biology: An emerging engineering discipline. Annu Rev Biomed Eng 14: 155–178 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A 2012. Multifunctional nanoparticles: Cost versus benefit of adding targeting and imaging capabilities. Science 338: 903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CD, Linder V, Sia SK 2012. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 12: 2118–2134 [DOI] [PubMed] [Google Scholar]
- Choi Y, Kang T, Lee LP 2009. Plasmon resonance energy transfer (PRET)-based molecular imaging of cytochrome c in living cells. Nano Lett 9: 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csete ME, Doyle JC 2002. Reverse engineering of biological complexity. Science 295: 1664–1669 [DOI] [PubMed] [Google Scholar]
- Davis ME, Chen ZG, Shin DM 2008. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat Rev Drug Discov 7: 771–782 [DOI] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A 2010. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464: 1067–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMuth PC, Min Y, Huang B, Kramer JA, Miller AD, Barouch DH, Hammond PT, Irvine DJ 2013. Polymer multilayer tattooing for enhanced DNA vaccination. Nat Mater 12: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weer P 2000. A century of thinking about cell membranes. Annu Rev Physiol 62: 919–926 [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Irimia D, Tompkins RG, Toner M 2007. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci 104: 18892–18897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H, Douglas SM, Shih WM 2009. Folding DNA into twisted and curved nanoscale shapes. Science 325: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SM, Bachelet I, Church GM 2012. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335: 831–834 [DOI] [PubMed] [Google Scholar]
- Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R 2008. High-throughput electrophysiology: An emerging paradigm for ion-channel screening and physiology. Nat Rev Drug Discov 7: 358–368 [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Yamada KM 2005. Cell migration in 3D matrix. Curr Opin Cell Biol 17: 524–532 [DOI] [PubMed] [Google Scholar]
- Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu C-C, Gould J, Hood L, et al. 2008. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol 26: 1373–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE 2007. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol 2: 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MT, Rotem A, Heyman JA, Weitz DA 2012. Droplet microfluidics for high-throughput biological assays. Lab Chip 12: 2146–2155 [DOI] [PubMed] [Google Scholar]
- Hall AR, Scott A, Rotem D, Mehta KK, Bayley H, Dekker C 2010. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat Nanotechnol 5: 874–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson ME, Chapin SC, Doyle PS 2011. Hydrogel microparticles from lithographic processes: Novel materials for fundamental and applied colloid science. Curr Opin Coll Int Sci 16: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori SS, Gambhir SS 2011. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med 3: 109ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrkach J, Von Hoff D, Mukkaram Ali M, Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M, Horhota A et al. 2012. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med 4: 128ra139. [DOI] [PubMed] [Google Scholar]
- Jebrail MJ, Yang H, Mudrik JM, Lafrenière NM, McRoberts C, Al-Dirbashi OY, Fisher L, Chakraborty P, Wheeler AR 2011. A digital microfluidic method for dried blood spot analysis. Lab Chip 11: 3218–3224 [DOI] [PubMed] [Google Scholar]
- Jin H, Heller DA, Kalbacova M, Kim J-H, Zhang J, Boghossian AA, Maheshri N, Strano MS 2010. Detection of single-molecule H2O2 signalling from epidermal growth factor receptor using fluorescent single-walled carbon nanotubes. Nat Nanotechnol 5: 302–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy J, Ginzberg MB, Oh S, Lahav G, Kirschner MW 2013. Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature 494: 480–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Ong LL, Shih WM, Yin P 2012. Three-dimensional structures self-assembled from DNA bricks. Science 338: 1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Ghaffari R, Lu N, Rogers JA 2012a. Flexible and stretchable electronics for biointegrated devices. Annu Rev Biomed Eng 14: 113–128 [DOI] [PubMed] [Google Scholar]
- Kim D-H, Provenzano PP, Smith CL, Levchenko A 2012b. Matrix nanotopography as a regulator of cell function. J Cell Biol 197: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong GA, von Maltzahn G, Murugappan G, Abudayyeh O, Mo S, Papayannopoulos IA, Sverdlov DY, Liu SB, Warren AD, Popov Y, et al. 2013. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat Biotechnol 31: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamon JH, Link DR, Egholm M, Rothberg JM 2006. Overview: Methods and applications for droplet compartmentalization of biology. Nat Methods 3: 541–543 [DOI] [PubMed] [Google Scholar]
- Lee H, Sun E, Ham D, Weissleder R 2008. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med 14: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS 2010. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods 7: 969–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JC 2010. Integrated process design for single-cell analytical technologies. AIChE J 56: 2496–2502 [Google Scholar]
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, et al. 2010. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci 107: 1864–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu C-C, Kwong GA, Radu CG, et al. 2011. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med 17: 738–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach AJ, Adeyiga OB, Di Carlo D 2013. Microfluidic sample preparation for diagnostic cytopathology. Lab Chip 13: 1011–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madou MJ. 2012. Fundamentals of microfabrication and nanotechnology. CRC Press, Boca Raton, FL. [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. 2008. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359: 366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin J, Quake SR 2007. Microfluidic large-scale integration: The evolution of design rules for biological automation. Annu Rev Biophys Biomol Struct 36: 213–231 [DOI] [PubMed] [Google Scholar]
- Merkel TJ, Jones SW, Herlihy KP, Kersey FR, Shields AR, Napier M, Luft JC, Wu H, Zamboni WC, Wang AZ, et al. 2011. Using mechanobiological mimicry of red blood cells to extend circulation times of hydrogel microparticles. Proc Natl Acad Sci 108: 586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitragotri S, Lahann J 2009. Physical approaches to biomaterial design. Nat Mater 8: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa NA, Jebrail MJ, Yang H, Abdelgawad M, Metalnikov P, Chen J, Wheeler AR, Casper RF 2009. Droplet-scale estrogen assays in breast tissue, blood, and serum. Sci Transl Med 1: 1ra2. [DOI] [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. 2007. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450: 1235–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M 2009. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater 8: 543–557 [DOI] [PubMed] [Google Scholar]
- Nelson CM, Bissell MJ 2006. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol 22: 287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P-I, Morgan B, Trautwein J et al. 2013. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 5: 179ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Millet LJ, Kim N, Li H, Jin X, Popescu G, Aluru NR, Hsia KJ, Bashir R 2010. Measurement of adherent cell mass and growth. Proc Natl Acad Sci 107: 20691–20696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash M, Gershenfeld N 2007. Microfluidic bubble logic. Science 315: 832–835 [DOI] [PubMed] [Google Scholar]
- Prausnitz MR, Mitragotri S, Langer R 2004. Current status and future potential of transdermal drug delivery. Nat Rev Drug Discov 3: 115–124 [DOI] [PubMed] [Google Scholar]
- Pregibon DC, Toner M, Doyle PS 2007. Multifunctional encoded particles for high-throughput biomolecule analysis. Science 315: 1393–1396 [DOI] [PubMed] [Google Scholar]
- Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF et al. 2012. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Sci Transl Med 4: 147ra112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund PWK 2006. Folding DNA to create nanoscale shapes and patterns. Nature 440: 297–302 [DOI] [PubMed] [Google Scholar]
- Ruoslahti E 2012. Peptides as targeting elements and tissue penetration devices for nanoparticles. Adv Mater 24: 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Kreutz J, Chiu DT 2013. The potential impact of droplet microfluidics in biology. Anal Chem 85: 3476–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC 2003. DNA in a material world. Nature 421: 427–431 [DOI] [PubMed] [Google Scholar]
- Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn D-R, Yoon M-H, Sutton A, Jorgolli M, Gertner RS, Gujral TS, et al. 2010. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc Natl Acad Sci 107: 1870–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Gaublomme JT, Wang L, Yosef N, Chevrier N, Andersen MS, Robinson JT, Pochet N, Neuberg D, Gertner RS, et al. 2012. Nanowire-mediated delivery enables functional interrogation of primary immune cells: Application to the analysis of chronic lymphocytic leukemia. Nano Lett 12: 6498–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H 2012. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med 18: 1835–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Votruba AR, Farokhzad OC, Langer R 2010. Nanotechnology in drug delivery and tissue engineering: From discovery to applications. Nano Lett 10: 3223–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih WM, Lin C 2010. Knitting complex weaves with DNA origami. Curr Opin Struct Biol 20: 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalak R, Branemark PI 1969. Deformation of red blood cells in capillaries. Science 164: 717–719 [DOI] [PubMed] [Google Scholar]
- Snijder B, Pelkmans L 2011. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol 12: 119–125 [DOI] [PubMed] [Google Scholar]
- Son S, Tzur A, Weng Y, Jorgensen P, Kim J, Kirschner MW, Manalis SR 2012. Direct observation of mammalian cell growth and size regulation. Nat Methods 9: 910–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone HA, Stroock AD, Ajdari A 2004. Engineering flows in small devices. Annu Rev Fluid Mech 36: 381–411 [Google Scholar]
- Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Floyd FP, Springer S, Irimia D, Nagrath S, Sequist LV, et al. 2010. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci 107: 18392–18397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E 2009. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 16: 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SP, Koutsonanos DG, del Pilar Martin M, Lee JW, Zarnitsyn V, Choi S-O, Murthy N, Compans RW, Skountzou I, Prausnitz MR 2010. Dissolving polymer microneedle patches for influenza vaccination. Nat Med 16: 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E 2009. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci 106: 16157–16162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Liu J, Dvir T, Jin L, Tsui JH, Qing Q, Suo Z, Langer R, Kohane DS, Lieber CM 2012. Macroporous nanowire nanoelectronic scaffolds for synthetic tissues. Nat Mater 11: 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toner M, Irimia D 2005. Blood-on-a-chip. Annu Rev Biomed Eng 7: 77–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDersarl JJ, Xu AM, Melosh NA 2012. Nanostraws for direct fluidic intracellular access. Nano Lett 12: 3881–3886 [DOI] [PubMed] [Google Scholar]
- Venkatesan BM, Bashir R 2011. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol 6: 615–624 [DOI] [PubMed] [Google Scholar]
- Vincent ME, Liu W, Haney EB, Ismagilov RF 2010. Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals. Chem Soc Rev 39: 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn G, Park J-H, Lin KY, Singh N, Schwöppe C, Mesters R, Berdel WE, Ruoslahti E, Sailor MJ, Bhatia SN 2011. Nanoparticles that communicate in vivo to amplify tumour targeting. Nat Mater 10: 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Byrne JD, Napier ME, DeSimone JM 2011a. More effective nanomedicines through particle design. Small 7: 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee Y-K, et al. 2011b. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed 50: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ota S, Guo B, Ryu J, Rhodes C, Xiong Y, Kalim S, Zeng L, Chen Y, Teitell MA, et al. 2011c. Subcellular resolution mapping of endogenous cytokine secretion by nano-plasmonic-resonator sensor array. Nano Lett 11: 3431–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler AR 2008. Chemistry. Putting electrowetting to work. Science 322: 539–540 [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG 2009. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov 8: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides GM 2003. The ‘right’ size in nanobiotechnology. Nat Biotechnol 21: 1161–1165 [DOI] [PubMed] [Google Scholar]
- Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE 2001. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng 3: 335–373 [DOI] [PubMed] [Google Scholar]
- Xie X, Xu AM, Leal-Ortiz S, Cao Y, Garner CC, Melosh NA 2013. Nanostraw-electroporation system for highly efficient intracellular delivery and transfection. ACS Nano 7: 4351–4358 [DOI] [PubMed] [Google Scholar]
- Yoo J-W, Irvine DJ, Discher DE, Mitragotri S 2011. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 10: 521–535 [DOI] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA 2011. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol 192: 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]