Transcription is an obstacle to replication fork progression and a cause of genome instability. This instability is partially mediated by cotranscriptional R loops, believed to increase by suboptimal assembly of the nascent messenger ribonucleoprotein particle (mRNP). Aguilera and colleagues now show that yeast Npl3, the most abundant RNA-binding heterogeneous nuclear RNP (hnRNP), prevents R-loop-mediated genome instability. Importantly, replication stress in npl3 deletion cells confers hypersensitivity to DNA double-strand break-inducing agents, opening the possibility of using mRNP factors as therapeutic targets in anti-cancer treatment.
Keywords: R loops, transcription-associated genome instability, transcription–replication conflicts, Npl3, hnRNPs, DNA damage response
Abstract
Transcription is a major obstacle for replication fork (RF) progression and a cause of genome instability. Part of this instability is mediated by cotranscriptional R loops, which are believed to increase by suboptimal assembly of the nascent messenger ribonucleoprotein particle (mRNP). However, no clear evidence exists that heterogeneous nuclear RNPs (hnRNPs), the basic mRNP components, prevent R-loop stabilization. Here we show that yeast Npl3, the most abundant RNA-binding hnRNP, prevents R-loop-mediated genome instability. npl3Δ cells show transcription-dependent and R-loop-dependent hyperrecombination and genome-wide replication obstacles as determined by accumulation of the Rrm3 helicase. Such obstacles preferentially occur at long and highly expressed genes, to which Npl3 is preferentially bound in wild-type cells, and are reduced by RNase H1 overexpression. The resulting replication stress confers hypersensitivity to double-strand break-inducing agents. Therefore, our work demonstrates that mRNP factors are critical for genome integrity and opens the option of using them as therapeutic targets in anti-cancer treatment.
Eukaryotic RNA polymerase II (RNAPII)-driven transcription is a tightly regulated process that occurs coordinately with the processing and maturation of pre-mRNAs into mRNAs. The formation of an export-competent messenger ribonucleoprotein particle (mRNP) and its export to the cytoplasm through the nuclear pore complex (NPC) are also coupled to transcription (Kohler and Hurt 2007). A number of proteins have been described as participating in the integration of all these processes. This includes THO, a conserved protein complex containing Tho2, Hpr1, Mft1, Thp2, and Tex1 in Saccharomyces cerevisiae (Chavez et al. 2000; Pena et al. 2012), plus additional subunits in Drosophila and humans (Rehwinkel et al. 2004; Masuda et al. 2005). Together with the RNA-binding proteins Sub2/UAP56 and Yra1/REF1/Aly, THO forms a larger complex termed TREX (Strasser et al. 2002). Null mutations of THO/TREX components confer similar phenotypes of transcription impairment, mRNA export defects, and transcription-associated hyperrecombination (Luna et al. 2008), a phenomenon conserved in both Caenorhabditis elegans and human cells depleted of THO (Dominguez-Sanchez et al. 2011; Castellano-Pozo et al. 2012). THO/TREX represents a paradigmatic example of the connection between RNA metabolism and genome integrity that has been extended to other eukaryotic factors, although the molecular basis for this interconnection is not completely understood.
A key feature of THO/TREX mutants is their high levels of transcription-associated recombination (TAR). This has been shown to be dependent on the nascent RNA and the accumulation of cotranscriptional R loops (Huertas and Aguilera 2003), structures formed by a DNA–RNA hybrid and the displaced nontemplate DNA strand. Importantly, the connection between transcription, mRNA processing, and genome instability has been reported by proteins involved in different nuclear mRNA processes. Yeast mutants of the nuclear exosome (rrp6) and 3′ end processing factors (rna14 and rna15) are also affected in recombination and transcript accumulation, although to a lesser extent than THO mutants (Luna et al. 2005). Furthermore, the inactivation of the ASF/SF2 splicing factor in chicken DT40 and HeLa cells results in genome instability mediated by the formation of R loops (Li and Manley 2005). Finally, a number of RNA processing factors have been identified in global searches performed in yeast and human cells as proteins preventing different forms of R-loop-mediated genome instability (Paulsen et al. 2009; Wahba et al. 2011; Stirling et al. 2012).
In eukaryotes, the most abundant classes of RNA-binding proteins are the heterogeneous nuclear RNPs (hnRNPs) involved in multiple steps of mRNA processing and export (Dreyfuss et al. 2002) and the mammalian serine–arginine-rich (SR) proteins (SR family), which function in mRNA splicing, export, quality control, and translation (Shepard and Hertel 2009). A common feature for both protein families is the presence of one or more N-terminal RNA recognition motifs (RRMs), while their C terminus is composed of arginine and glycine-rich (RGG) regions in hnRNPs and multiple arginine and serine-rich (RS) domains in SR proteins. The most abundant hnRNP in yeast is Npl3, which is also considered an SR-like protein that shares structural homologies with both classes of RNA-binding proteins. Like mammalian SR proteins, Npl3 is a multifunctional protein involved in many processes of gene expression from transcription to splicing, mRNA export, and translation (Bucheli and Buratowski 2005; Dermody et al. 2008; Kress et al. 2008). It is an important mediator of RNA export that shuttles between the nucleus and the cytoplasm (Lee et al. 1996), regulated by phosphorylation via Sky1 kinase in the cytoplasm and dephosphorylation by Glc7 phosphatase in the nucleus, which is also important for interaction of Npl3 with the export receptor Mex67 (Gilbert and Guthrie 2004). Furthermore, Npl3 stimulates RNAPII elongation and is an antagonist of transcription termination (Bucheli and Buratowski 2005; Dermody et al. 2008). Indeed, Npl3 was found in a complex with RNAPII and is recruited to genes in a transcription-dependent manner (Lei et al. 2001). Interestingly, despite the general model that RNA-binding proteins control mRNP biogenesis and the ability of the nascent RNA to form R loops, no clear data reveal a role for Npl3 in genome instability that supports the idea of a failure in proper mRNA packaging that can cause RNA-dependent genome instability.
Genome instability associated with most mutations in RNA processing factors is manifested mainly by accumulation of double-strand breaks (DSBs), hyperrecombination, and high levels of plasmid or chromosome loss, implying an increase or defect in the processing of DNA breaks during replication (Aguilera and Garcia-Muse 2013). Consistently, some of these mutants show different degrees of sensitivity to DSB-inducing agents, such as methyl methane-sulfonate (MMS) or hydroxyurea (HU) (Gaillard et al. 2009). One particularly interesting DSB-inducing agent is trabectedin (ET-743), which has been shown to form covalent adducts with guanines located in the DNA minor groove in vitro (Pommier et al. 1996). The action mechanism of this drug is particularly intriguing, and different reports suggest that it might lead to the generation of replication- and transcription-dependent DSBs (Herrero et al. 2006; Guirouilh-Barbat et al. 2008), implying that it may be useful in identifying new factors involved in the maintenance of genome integrity during transcription. This is of particular interest given the potential anti-tumoral properties of trabectedin that might serve to define new targets in cancer treatment.
Here, after identifying Npl3 as a key factor for trabectedin resistance in S. cerevisiae, we show that npl3Δ cells have transcription-dependent genome instability phenotypes that are linked to a defect in replication progression. Such instability is at least in part mediated by R loops. Genome-wide chromatin immunoprecipitation (ChIP)–chip analyses show that Npl3 binds to highly RNAPII-transcribed genes and that its absence leads to R-loop-dependent genome-wide replication impairment, as detected by the accumulation of the Rrm3 helicase at transcribed genes. Our demonstration that an important and highly abundant RNA-binding hnRNP involved in mRNP processing and export, Npl3, has a key role in preventing cotranscriptional genome instability indicates a coordinated connection between RNA metabolism and DNA damage avoidance that may provide new perspectives in anti-tumor treatment.
Results
Hypersensitivity to DSB-inducing genotoxic agents of npl3Δ strains
Using a genome-wide screen in the yeast S. cerevisiae, we first identified the npl3Δ mutant as hypersensitive to trabectedin (ET-743), a DNA-binding drug that has been proposed as causing replication- and transcription-dependent DSBs (Fig. 1A; Herrero et al. 2006; Guirouilh-Barbat et al. 2008). Next, we examined the sensitivity of npl3Δ cells to other genotoxic agents that cause DNA lesions that can result in stalled replication forks (RFs) and/or DSBs. We found that npl3Δ cells were sensitive to MMS, phleomycin (Phl), UV radiation, and HU (Fig. 1A), suggesting a role of Npl3 in avoiding the incidence of DNA damage, in the activation of DNA damage checkpoints, and/or in DNA repair.
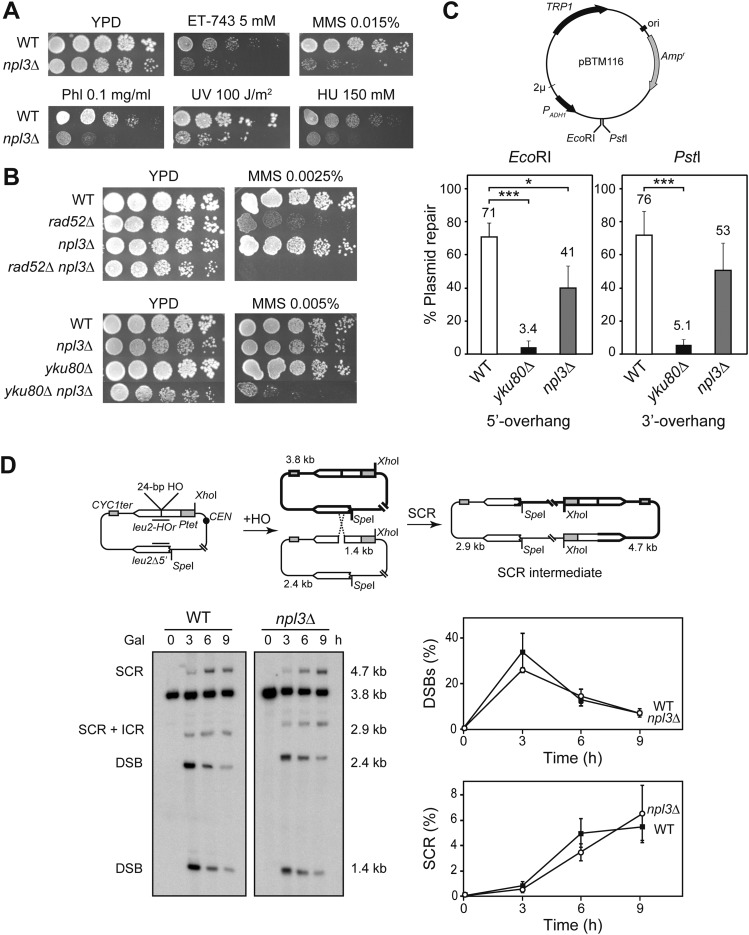
Figure 1.
DNA repair in npl3Δ cells. (A) Sensitivity of BY4741 (WT) and SC086 (npl3Δ) strains to trabectedin (ET-743), MMS, Phl, UV light, and HU. (B) Sensitivity of BY4741 (WT), SC086 (npl3Δ), SC063 (rad52Δ), SC087 (rad52Δ npl3Δ), SC064 (yku80Δ), and SC088 (yku80Δ npl3Δ) strains to MMS. (C) Plasmid relegation assay. (Top) Scheme of pBTM116 and enzymes used in the assay. (Bottom) Percentage of plasmid repair in SC071 (WT), SC073 (yku80Δ), and SC089 (npl3Δ) strains. (*) P < 0.05; (***) P < 0.001 (Student's t-test). (D) Repair of replication-borne HO-induced DSBs by SCR. (Top) Scheme of pRS316TINV plasmid carrying two inverted leu2 repeats. Fragments generated by HO cleavage and XhoI SpeI digestion, as detected by the LEU2 probe (represented as a black line), are indicated with their corresponding sizes. (Left) Physical analysis of HO-induced DSB formation and its repair kinetics in WS (WT) and WSNPL3-01 (npl3Δ) strains. (Right) Quantification of DSBs (1.4-kb plus 2.4-kb bands) and SCR (4.7-kb band) related to total plasmid DNA. Average and standard deviation of three independent experiments are shown.
DSB repair in npl3Δ cells
To test whether hypersensitivity to genotoxic agents of npl3Δ was due to a defect in DSB repair, we generated double mutants lacking, simultaneously, NPL3 and the key genes of the two major DSB repair pathways RAD52 and YKU80, which affect homologous recombination (HR) and nonhomologous end joining (NHEJ), respectively. As can be seen in Figure 1B, sensitivity of double mutants npl3Δ rad52Δ and npl3Δ yku80Δ to MMS was increased in a synergistic manner with respect to single mutants, suggesting that DSBs accumulate in npl3Δ cells and require HR and NHEJ for their repair.
To assay whether Npl3 was involved in one of the two pathways of DSB repair, NHEJ was first analyzed genetically as the ability of npl3Δ cells to repair a cut plasmid. In this assay, the 2μ-based plasmid pBTM116 was linearized by restriction enzyme digestions, generating 5′ or 3′ overhang ends (EcoRI and PstI, respectively), and introduced by transformation into cells (Fig. 1C). Since plasmid maintenance requires circularization by NHEJ, plasmid repair efficiency was calculated as the number of transformant colonies obtained with linearized plasmids relative to the uncut circular plasmid. As expected, yku80Δ cells showed a strong defect in DSB repair compared with wild type, while npl3Δ cells displayed a slight defect in the repair of EcoRI-induced DSBs and a nonsignificant effect in the repair of PstI-induced DSBs (41%–53%; P < 0.05 and P > 0.05, respectively) (Fig. 1C), indicating that Npl3 does not seem to have a relevant role in NHEJ that could explain the sensitivity of npl3Δ cells to genotoxic agents.
In S. cerevisiae, repair of DSBs occurs predominantly by HR using the sister chromatid as template. To test whether the increase in sensitivity to DSB-inducing agents of npl3Δ cells was due to an HR defect, we analyzed the involvement of Npl3 in sister chromatid recombination (SCR), since SCR is the preferential and major HR repair pathway (Kadyk and Hartwell 1992). We used the pTINV plasmid-borne recombinational system previously described (Gonzalez-Barrera et al. 2003). In this system, overexpression of the HO endonuclease from the GAL1 promoter generates a ssDNA break at the 24-base-pair (bp) mini-HO site located in one of two leu2 inverted repeats of the plasmid. During replication, the ssDNA break is converted to a DSB (Cortes-Ledesma and Aguilera 2006), which can be repaired by SCR, generating an intermediate molecule that can be visualized by restriction enzyme digestion followed by Southern (Fig. 1D).
We determined the kinetics of the appearance of replication-dependent DSBs as well as the kinetics of its repair at different time points after HO induction (Fig. 1D). Quantification of the different fragments generated showed a similar kinetics of DSB appearance, measured as the percentage of the 2.4-kb and 1.4-kb bands relative to total plasmid DNA in wild-type and npl3Δ cells. The value was maximum at 3 h after HO induction (∼30% of molecules were cut) and decreased to <10% later on. The kinetics of SCR, measured as the percentage of the 4.7-kb band relative to the total, was also similar in both wild-type and npl3Δ cells, reaching maximum levels (∼6%) at the 9-h time point (Fig. 1D). Therefore, Npl3 is not involved in the repair of replication-borne DSBs by SCR. It does not seem to play a relevant or direct role in DSB repair, suggesting that the sensitivity of DSB-inducing genotoxic agents of npl3Δ cells may be a consequence of replication stress induced by lack of Npl3.
Efficient replication progression through damaged DNA requires Npl3
In addition to the repair of DSBs, HR is also important for the repair and restart of stalled and blocked RFs (Aguilera and Garcia-Muse 2013). As DSB-inducing agents such as MMS can generate RF stalling, we reasoned that Npl3 could play a role in the stabilization of stalled RFs or in its progression through damaged DNA as a way to explain MMS or trabectedin sensitivity of npl3Δ cells. To test this possibility, we first monitored S-phase progression in wild-type and npl3Δ cells in the presence or absence of MMS by fluorescence-activated cell sorting analyses (FACS) (Fig. 2A). Cells were synchronized in G1 with α-factor and then released into fresh medium either with or without 0.033% MMS, a concentration that has been described to cause extensive fork stalling in wild-type cells (Tercero and Diffley 2001). We found that npl3Δ cells exhibited a delay in S-phase progression in the absence of MMS, and this delay was much more evident in the presence of the drug (Fig. 2A).
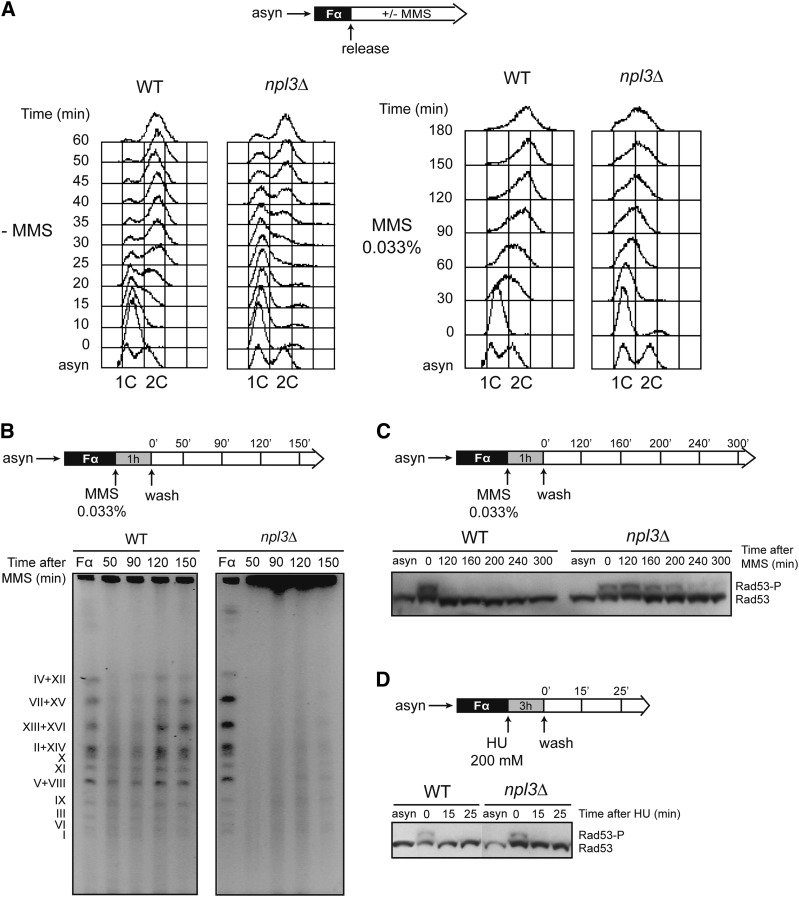
Figure 2.
Replication impairment and persistent checkpoint activation in response to MMS in npl3Δ cells. (A) S-phase progression of BY4741 (WT) and SC086 (npl3Δ) cells synchronized in G1 with α-factor and released in the absence (left) or presence (right) of 0.033% MMS. (B) Analysis of completion of replication by PFGE in BY4741 (WT) and SC086 (npl3Δ) strains at different time points after G1 synchronization, treatment with 0.033% MMS, and release into fresh medium. (C) Western against the Rad53 checkpoint kinase in BY4741 (WT) and SC086 (npl3Δ) strains. Cells were treated with MMS as in B. (D) Western against Rad53 in which G1-arrested cells were treated with 200 mM HU for 3 h, washed, and released into fresh medium. A diagram of each experiment is shown at the top of each panel.
We further analyzed completion of replication after treatment with MMS in wild-type and npl3Δ cells by pulsed field gel electrophoresis (PFGE). This technique allows the distinction of fully replicated linear chromosomes, which enter the gel, from actively replicating chromosomes, which are unable to enter the gel due to the branched replication intermediates and therefore remain in the well. Cells were synchronized in G1 with α-factor, treated with 0.033% MMS for 1 h, and then released into fresh medium without MMS. In both wild-type and npl3Δ cells, DNA was separated as individual chromosomes prior to MMS treatment, whereas most of the DNA was retained in the loading well 50 min after MMS washing and release into S phase. Interestingly, whereas chromosomal DNA re-entered the gel ∼2 h after MMS exposure in wild-type cells, most of the DNA from npl3Δ cells still remained in the well after 150 min (Fig. 2B). This result suggests that progression of the RF is affected in npl3Δ cells, possibly due to the accumulation of replication obstacles.
A defect in replication completion after MMS treatment could be the consequence of checkpoint activation in response to stalled or collapsed RFs. To test this possibility, we wondered whether the Rad53 DNA damage checkpoint effector kinase was activated after MMS treatment for a longer time in npl3Δ as compared with wild-type cells. We treated wild-type and npl3Δ cells as before and visualized both phosphorylated (active) and nonphosphorylated (inactive) forms of Rad53 by Western. As can be seen in Figure 2C, Rad53 remained active much longer in npl3Δ than in wild-type cells after MMS exposure, indicating that the absence of Npl3 activates the S-phase checkpoint in response to damage. However, after replication stress induced with HU, which depletes cells of dNTPs, Rad53 activation disappeared in npl3Δ cells as quickly as in wild-type cells (Fig. 2D), suggesting that DNA damage but not replication stress alone is toxic in the absence of Npl3.
DNA breaks and transcription-associated genetic instability in npl3Δ cells
Next, we tested whether the sensitivity of npl3Δ cells to MMS and their replication difficulties were due to a higher incidence of DSBs that could overcome the overall repair capacity of the cells. To assess this, we examined the formation of Rad52-YFP foci, which have been shown previously to be a reliable method in identifying recombinational repair centers (Lisby et al. 2001). We found that the percentage of cells exhibiting Rad52 foci was significantly higher in npl3Δ than in wild-type cells (8.4% vs. 3%; P < 0.05) (Fig. 3A), confirming that npl3Δ cells accumulate high levels of recombinogenic DNA breaks.
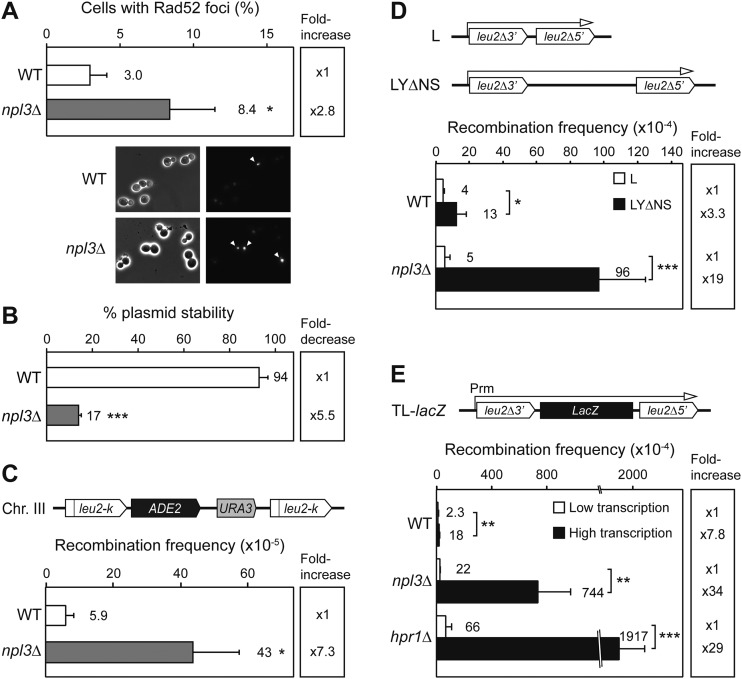
Figure 3.
Genetic instability and TAR in npl3Δ cells. (A) Spontaneous Rad52-YFP foci formation in W303-1A (WT) and WNPL3-1D (npl3Δ) strains and representative microscope images. Average and standard deviation of three to four independent experiments are shown. (B) Percentage of centromeric plasmid pRS316 stability in W303-1A (WT) and WNPL3-1D (npl3Δ) strains. (C) Recombination analysis of AYW3-1B (WT) and AYNPL3-1D (npl3Δ) strains carrying the chromosomal direct repeat system leu2-k∷ADE2-URA3∷leu2-k. (D) Recombination analysis of W303-1A (WT) and WNPL3-1D (npl3Δ) strains carrying L and LYΔNS plasmid systems. (E) Recombination analysis in the TL-lacZ plasmid system, whose transcription is regulated by the tet promoter, in the presence (Low transcription) or absence (High transcription) of 5 μg/mL doxycycline in W303-1A (WT), WNPL3-1D (npl3Δ), and U678-1C (hpr1Δ) strains. For B–E, average and standard deviation of three to four fluctuation tests consisting of the median value of six independent colonies for each one are shown. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 (Student's t-test). For recombination assays, a scheme of the system is shown at the top of each panel.
Since unrepaired DNA breaks can result in an increase in chromosome loss, we analyzed stability of a centromeric plasmid, pRS316, in wild-type and npl3Δ cells after growing in nonselective rich medium for the same number of generations. Figure 3B shows a significant decrease of more than fivefold in plasmid maintenance in npl3Δ cells (17% vs. 94% in wild type; P < 0.001), suggesting that Npl3 plays a general role in the maintenance of genome stability.
Next, we took advantage of the chromosomal leu2-k∷ADE2-URA3∷leu2-k system (Fig. 3C) to assay recombination in npl3Δ cells. This system is located in chromosome III and contains two 2.16-kb long direct repeats of the leu2-k allele separated by the ADE2 and URA3 genes. Deletions caused by recombination between the two repeats give rise to the loss of the URA3 marker, which can be scored in 5-FOA-containing medium. Consistently, quantification assays of Ura− cells revealed that recombination levels in npl3Δ were significantly higher than in wild-type cells (7.3-fold; P < 0.05) (Fig. 3C), confirming the important role of Npl3 in the maintenance of genome integrity.
Npl3 has been described as playing an important role in RNAPII transcription and mRNA export to the cytoplasm. Consequently, we determined whether hyperrecombination was linked to transcription in npl3Δ cells. For this, we used different plasmid-borne recombination systems based on truncated repeats of 0.6 kb of the LEU2 gene located in direct orientation and separated by intervening sequences of increasing lengths (Fig. 3D,E). Recombination frequencies were measured as the frequency of Leu+ colonies. First, we analyzed the effect of the intervening regions located between the repeats and found that the hyperrecombination phenotype of npl3Δ cells was observed only when a long and G+C-rich sequence had to be transcribed (19-fold increase in LYΔNS vs. L system; 3.3-fold in wild type; P < 0.001 and P < 0.05, respectively) (Fig. 3D). These results are consistent with published data for other factors of the mRNP biogenesis pathway (Luna et al. 2008). Next, we generated a new recombination system in which the lacZ gene is located between the leu2 repeats, and transcription is regulated by tet promoter (TL-lacZ system). As expected, recombination was enhanced in the highly transcribed versus the low-transcribed system in wild-type cells (7.8-fold increase; P < 0.01), npl3Δ cells, and hpr1Δ cells (used as a positive control) (Fig. 3E). However, recombination levels were much higher in the mutant strains compared with wild type (34-fold and 29-fold increase in mutants, respectively; P < 0.01 and P < 0.001). Therefore, npl3Δ cells show a TAR phenotype similar to that observed in other mRNP biogenesis factors working at the interface transcription–mRNA export.
Suppression of npl3Δ genetic instability by RNase H1 overexpression
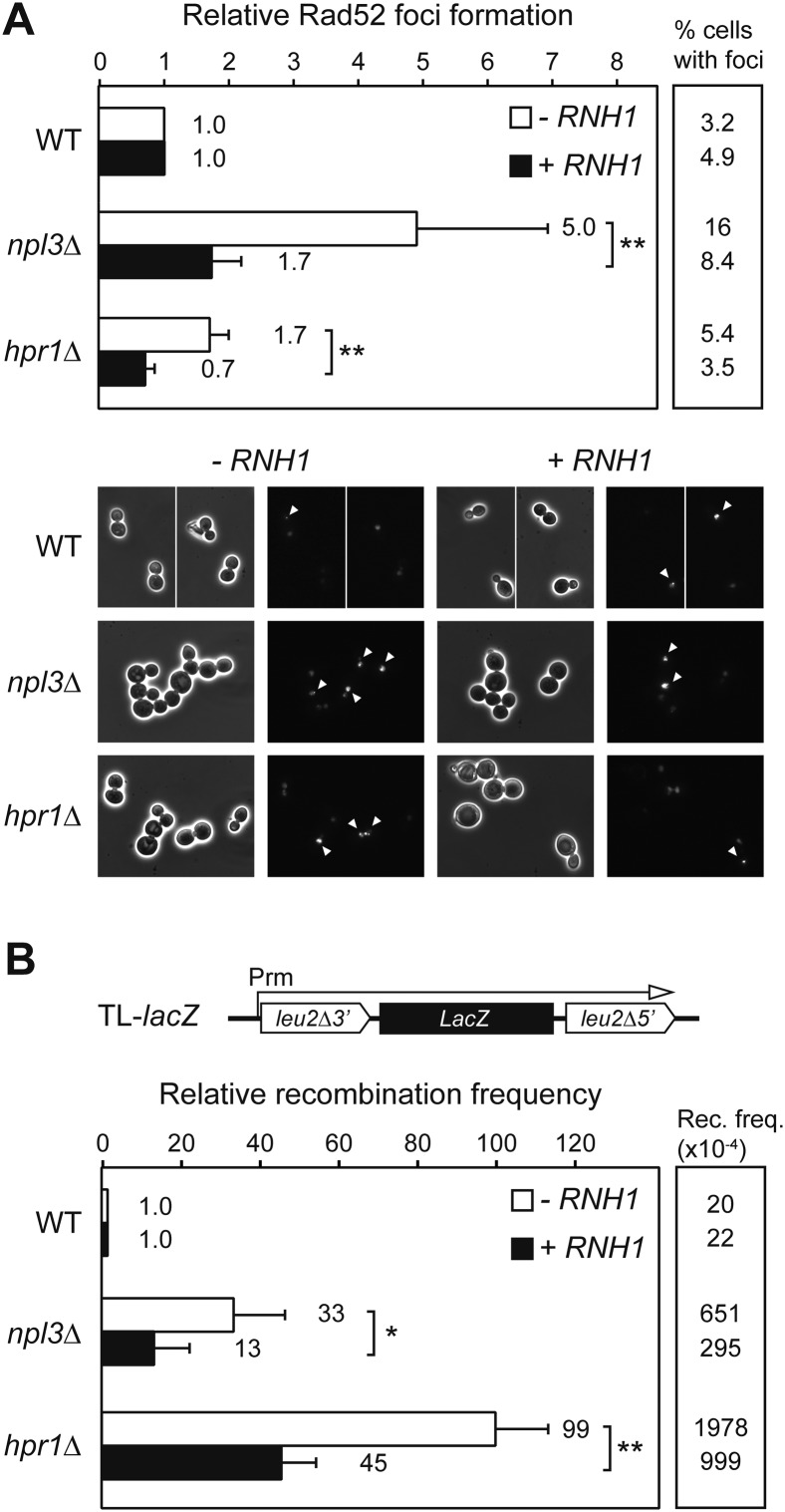
We next assayed whether the TAR phenotype of npl3Δ cells was mediated by R loops, as is the case of known mRNP biogenesis mutants. For this purpose, we overexpressed the RNase H1 gene under the tet promoter from a newly constructed plasmid in cells carrying the RAD52-YFP fusion and measured the formation of Rad52 foci. We observed an increase of Rad52 foci in npl3Δ cells (Figs. 3A, 4A). Importantly, Rad52 foci accumulation was partially but significantly suppressed by RNH1 overexpression (1.7-fold vs. fivefold increase; P < 0.01), similar to the hpr1Δ cells used as positive controls (0.7-fold vs. 1.7-fold increase; P < 0.01) (Fig. 4A).
Figure 4.
Suppression of Rad52 foci and hyperrecombination by overexpression of RNH1. (A) Rad52-YFP foci formation in W303-1A (WT), WNPL3-1D (npl3Δ), and U678-1C (hpr1Δ) strains carrying pCM189 (−RNH1) or pCM189RNH1 (+RNH1) in the absence of doxycycline. Representative microscope images are shown. Average and standard deviation of six independent experiments are shown normalized to wild-type levels. (B) Recombination analysis in the TL-lacZ system in W303-1A (WT), WNPL3-1D (npl3Δ), and U678-1C (hpr1Δ) strains under the same conditions as in A. Average and standard deviation of three to four fluctuation tests from six independent colonies for each one are shown normalized to wild-type levels. (*) P < 0.05; (**) P < 0.01 (Student's t-test).
Next, we did the same approach using the TL-lacZ recombination system (Fig. 3E) and found that hyperrecombination was also partially but significantly suppressed in npl3Δ and hpr1Δ cells (13-fold vs. 33-fold increase and 45-fold vs. 99-fold increase, respectively; P < 0.05 and P < 0.01) (Fig. 4B). We concluded that R loops have a significant impact over genetic stability in npl3Δ cells. We next wondered whether overexpression of AID, which stimulates recombination in R-loop-forming yeast strains (Gomez-Gonzalez and Aguilera 2007), also affected recombination in npl3Δ cells. As expected, AID overexpression from a plasmid-borne tet promoter produced a significant increase of Leu+ recombinants in the TL-lacZ system in all strains that was higher in npl3Δ and hpr1Δ cells (2.9-fold, 4.1-fold, and 4.3-fold increase, respectively; P < 0.001) (Supplemental Fig. S1). Therefore, our results indicate that npl3Δ-induced genome instability is mediated in large part by R loops, which contain a ssDNA fragment in the nontranscribed strand, a target of AID action.
Suppression of npl3Δ phenotypes by overexpression of hnRNPs
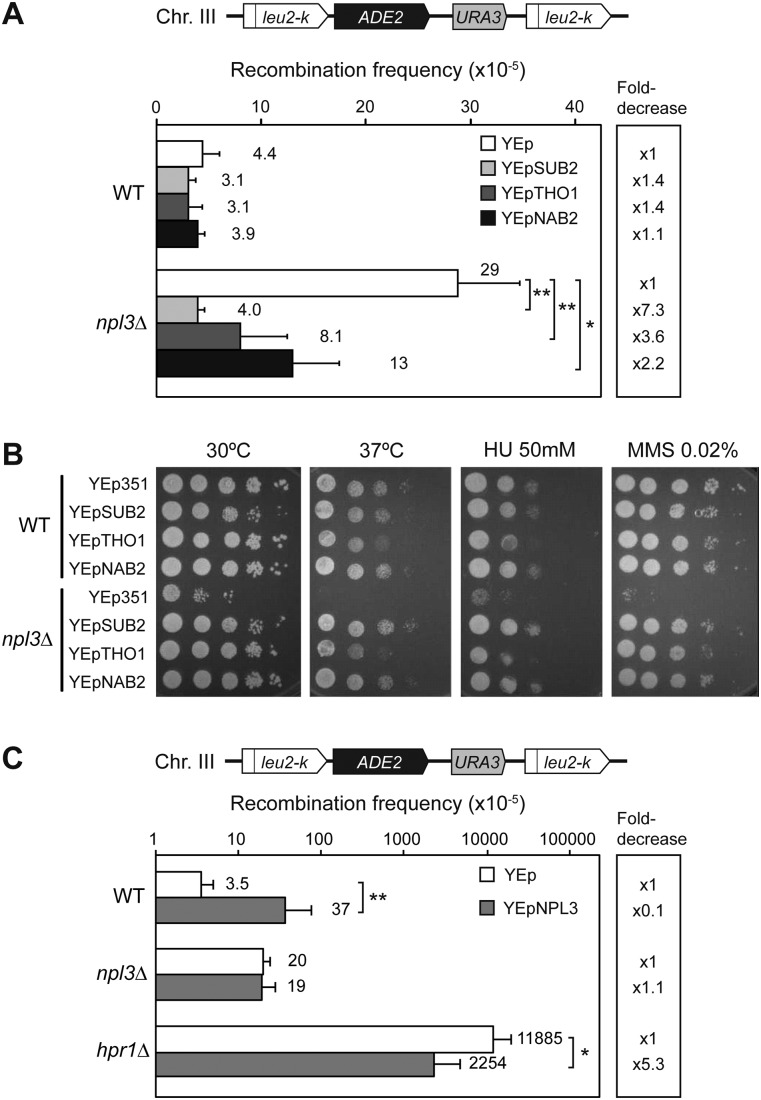
Overexpression of several hnRNPs and RNA biogenesis factors has been shown to suppress the transcription and recombination-related phenotypes of transcription–RNA export mutants. Thus, overexpression of SUB2, THO1, and NAB2 suppresses mutations of either THO or THSC (Rondon et al. 2010). We wondered whether overexpression of these genes also suppressed npl3Δ phenotypes. Using the chromosomal leu2-k∷ADE2-URA3∷leu2-k recombination system, we assayed the effect of overexpression of SUB2, THO1, and NAB2 carried in multicopy plasmids in both wild-type and npl3Δ cells. Overexpression of these RNA factors did not have any effect on recombination levels in wild-type cells. The recombination levels of npl3Δ cells transformed with the empty vector were higher than in wild type. Interestingly, overexpression of SUB2, THO1, and NAB2 suppressed this hyperrecombination phenotype either completely or partially (7.3-fold, 3.6-fold, and 2.2-fold decrease, respectively; P < 0.01-0.05) (Fig. 5A). We then analyzed whether overexpression of these proteins also suppressed the temperature, HU, and MMS sensitivities of npl3Δ by serial dilution experiments. A complete or nearly complete suppression of the three phenotypes was observed by overexpressing the three genes SUB2, THO1, and NAB2 (Fig. 5B). Altogether, these results are consistent with the idea that Npl3 is an mRNP biogenesis factor with a function related to those hnRNPs involved in mRNP assembly and export.
Figure 5.
Effect of multicopy SUB2, THO1, NAB2, and NPL3 overexpression over different strains. (A) Recombination analysis of AYW3-1B (WT) and AYNPL3-1D (npl3Δ) strains carrying the chromosomal direct repeat system leu2-k∷ADE2-URA3∷leu2-k and multicopy plasmids YEp351 (empty vector, YEp), YEpSUB2 (SUB2), YEpTHO1 (THO1), and YEpNAB2 (NAB2). Average and standard deviation of three to four fluctuation tests from six independent colonies for each one is shown. (B) Suppression of temperature, HU, and MMS sensitivity phenotypes by YEpSUB2, YEpTHO1, and YEpNAB2 in serial dilutions of AYW3-1B (WT) and AYNPL3-1D (npl3Δ) cells. (C) Recombination analysis of AYW3-1B (WT), AYNPL3-1D (npl3Δ), and AYW3-3C (hpr1Δ) strains carrying the chromosomal direct repeat system leu2-k∷ADE2-URA3∷leu2-k and multicopy plasmids YEp351 (empty vector, YEp) or YEpNPL3 (NPL3). Average and standard deviation of six fluctuation tests from six independent colonies for each one are shown. (*) P < 0.05; (**) P < 0.01 (Student's t-test).
Reciprocally, we analyzed the effect of multicopy NPL3 on recombination in the same chromosomal system. We found that multicopy NPL3 increased recombination in wild-type cells 10-fold (P < 0.01), whereas in npl3Δ cells, it did not enhance recombination further, suggesting that Npl3 levels have to be tightly regulated (Fig. 5C). Interestingly, overexpression of NPL3 in hpr1Δ cells showed a significant reduction of hyperrecombination (5.3-fold decrease; P < 0.05) (Fig. 5C), indicating that Npl3 can partially compensate the loss of Hpr1, which is consistent with the idea that Npl3 is a structural component of the mRNP.
Genome-wide distribution of Npl3 is enriched toward the 3′ end of transcribed genes
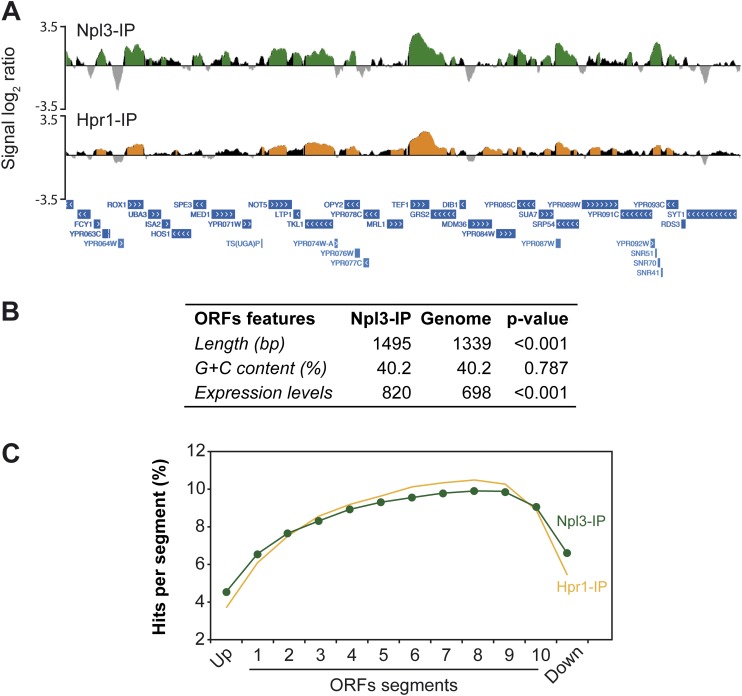
To gain insight into the Npl3 function all over the genome, we performed ChIP–chip experiments using an Npl3-MYC fusion protein. Data were subjected to computational analysis to obtain a genomic map distribution (Fig. 6A; Supplemental Fig. S2A) that was compared with previously published Hpr1-Flag ChIP–chip data and used as a control of a cotranscriptional mRNP biogenesis factor (Gomez-Gonzalez et al. 2011). Statistical analysis revealed that 82.1% of Npl3 clusters and 83.8% of Hpr1 peaked inside ORFs. Whereas Npl3 was significantly enriched in 4769 ORFs, Hpr1 was found in 4324, most of them being coincident (73%) (Fig. 6B; Supplemental Figs. S2A, S5), indicating that Npl3 binds to the same genes as THO does, in general, and that they correspond to actively transcribed genes. Additionally, we analyzed the structural and functional features of Npl3-bound genes and found that they were longer and more expressed than the genome average (Fig. 6B). Therefore, Npl3 binds to actively transcribed chromatin, consistent with its role in mRNA biogenesis.
Figure 6.
Npl3 and Hpr1 recruitment to highly transcribed ORFs. (A) Genomic view of Npl3-MYC (YAM535) and Hpr1-Flag (SYHPR1) recruitment. A fragment of chromosome XVI is plotted with the signal log2 ratio values. Green (Npl3-IP) and orange (Hpr1-IP) histograms represent the significant clusters. SGD features are represented below as blue bars with white arrows according to the direction of transcription. (B) Table showing the statistical analysis of length, G+C content, and expression levels of the genes mapped by Npl3 clusters. P-value was calculated by Mann-Whitney's U-test. (C) Composite profile of Npl3 and Hpr1 occupancy detected by ChIP–chip across the average ORF plotted as Npl3 or Hpr1 percentage of ChIP clusters per segment.
We then analyzed the distribution of Npl3 along the length of all ORFs by subdividing them into 10 segments independent of ORF size, including additional upstream and downstream segments of the same length. The percentage of clusters mapping on each segment along a given ORF determined the occupancy of Npl3. Interestingly, Npl3 binding increases gradually toward the 3′ end of genes (Fig. 6C) regardless of gene length (Supplemental Fig. S2B) and similar to Hpr1 and other factors previously shown to be involved in transcription elongation (Kim et al. 2010; Mayer et al. 2010; Gomez-Gonzalez et al. 2011). The result suggests that Npl3 binds during transcription, reaching a higher accumulation as either the nascent RNA grows and/or the RNAPII approaches the 3′ end pause.
Genome-wide DNA replication impairment in npl3Δ cells detected by Rrm3 distribution
Results obtained so far suggest that the absence of Npl3 causes a defect in mRNP biogenesis at the site of transcription that may cause DNA replication impairment and a DNA damage response deregulation responsible for the different forms of genome instability observed. To assay whether NPL3 depletion has any effect on replication progression, we performed ChIP–chip with an Rrm3-Flag fusion protein. Rrm3 is a helicase required for the progression of the RF through obstacles in the DNA, and its accumulation at specific DNA sites has been used to identify RF pauses or stalls, which have been shown to occur at a high proportion in transcribed DNA regions in wild-type cells (Ivessa et al. 2003; Azvolinsky et al. 2009). In addition, we showed that Rrm3 is highly accumulated at transcribed genes in hpr1Δ cells (Gomez-Gonzalez et al. 2011).
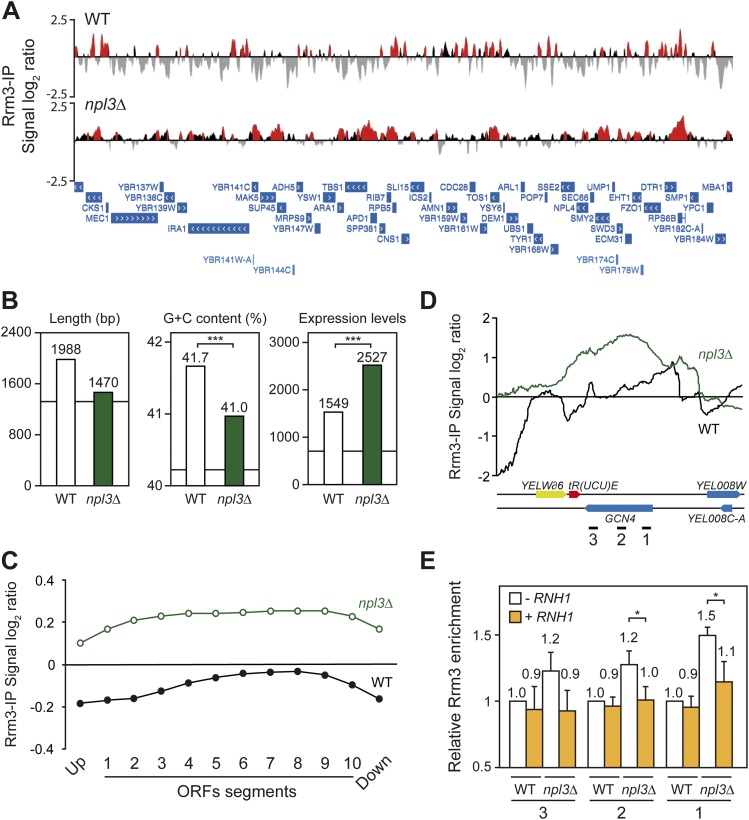
We found that clusters of Rrm3 accumulation were distributed all over the genome in npl3Δ cells (Supplemental Fig. S3) and that such clusters were significantly longer in npl3Δ cells (average size of 656 bp vs. 475 bp in wild type; P < 0.01), with a total extension of 3.12 Mb vs. 2.50 Mb in wild-type cells (26% and 21% of the genome, respectively) (Fig. 7A), consistent with the idea that replication obstacles are stronger or more abundant in npl3Δ cells. Indeed, the percentage of clusters mapping at ORFs was also higher in the mutant (83.6% vs. 76.0%, respectively), consistent with the effect on replication being preferentially observed at transcribed regions and with the role of Npl3 promoting RNAPII transcription. Furthermore, both clusters of Rrm3 and Npl3 were significantly enriched at introns, snRNAs, snoRNAs, and RNAPIII-transcribed genes, including tRNAs (Supplemental Fig. S4; Supplemental Table S1), suggesting an additional role of Npl3 facilitating RF progression through these regions. Additionally, we found that Rrm3 bound to centromeres in npl3Δ cells, but not Npl3.
Figure 7.
Rrm3 genome-wide recruitment in wild-type and npl3Δ cells. (A) Genomic view of Rrm3 recruitment in WRBb-9B (WT) and N3RBb-4B (npl3Δ) cells. A fragment of chromosome II is plotted with the signal log2 ratio values. Red histograms represent the significant clusters. SGD features are represented below as blue bars with white arrows according to the direction of transcription. (B) Statistical analysis of length, G+C content, and expression values of the top 500 genes showing significant Rrm3 recruitment in wild-type and npl3Δ cells. (***) P < 0.001 (Mann-Whitney's U-test). (C) Composite profile of Rrm3 occupancy detected by ChIP–chip across the average ORF plotted as signal log2 ratio average per each segment. (D) Detailed analysis of ChIP–chip data of Rrm3-Flag at the GCN4 region. (E) Specific Rrm3-Flag ChIP analysis using RT-qPCR of three regions (depicted as black lines in D with numbers 1–3) of GCN4 in WRBb-9B (WT) and N3RBb-4B (npl3Δ) cells carrying pCM184 (−RNH1) or pCM184RNH1 (+RNH1) without doxycycline. Data were normalized to wild-type levels without RNH1 overexpression. Average and standard deviation of three independent experiments are shown. (*) P < 0.05 (Student's t-test).
There was a 52% coincidence between the ORFs recruiting Rrm3 in wild-type and npl3Δ cells, the ORFs showing a high overlap with Npl3-bound ORFs (Supplemental Fig. S5). To determine the main functional and structural features of the genes to which Rrm3 preferentially binds, we selected the top 500 ORFs with the highest signal log2 ratio average in a sliding window of 200 bp. Statistical analysis revealed that, on average, these were long, GC-rich, and highly transcribed genes in both wild-type and npl3Δ cells, indicating that RNAPII-transcribed genes are more prone to affect RF progression (Fig. 7B). Interestingly, top Rrm3-bound genes were more expressed in npl3Δ cells, suggesting that replication obstacles occur preferentially at the highest transcribed genes in the absence of Npl3 (Fig. 7B), these genes preferentially being shorter all over the genome (Marin et al. 2003). In this sense, although gene ontology analysis revealed that genes involved in ribosome biogenesis or intron-containing genes were significantly overrepresented in npl3Δ cells, this is consistent with the fact that ribosome biogenesis genes are highly transcribed and represent the major class of intron-containing genes in yeast.
It is worth noting that Rrm3-binding profiles showed an increase toward the 3′ ends of genes in both wild-type and npl3Δ cells, with a high decrease in the regions immediately upstream of and downstream from them (Supplemental Fig. S6A), consistent with the pattern of Npl3 binding to transcribed genes (Fig. 6C). We also found a slight displacement to the 5′ end of long genes in the Rrm3 profile in the mutant that correlates with the Npl3 profile (Supplemental Figs. S2B, S6B), suggesting that Npl3 helps in preventing or resolving RF progression impairment all over the gene, especially at long genes. Additionally, we measured the Rrm3 signal log2 ratio average along all ORF segments and found a notable increase in npl3Δ cells (Fig. 7C), which indicates that replication obstacles are stronger in RNAPII genes when Npl3 is absent.
Finally, microarray analysis revealed that gene expression profiles of wild-type and npl3Δ cells were quite similar (Pearson's R correlation coefficient of 0.978), indicating that the absence of Npl3 does not have a significant impact on global expression levels. Nevertheless, down-regulated genes in npl3Δ cells were longer, G+C-richer, and better expressed than up-regulated genes (Supplemental Fig. S7A), consistent with the results obtained in hpr1Δ, tho2Δ, and sub2Δ cells (Gomez-Gonzalez et al. 2011). Furthermore, we found a good correlation between Rrm3 binding and expression levels in the 2758 genes accumulating Rrm3 in both the wild-type and the npl3Δ cells in the mutant (Pearson's R correlation coefficient of 0.59234) that was clearly higher than that of wild-type cells (0.24348, Supplemental Fig. S7B), supporting the idea that replication obstacles occur preferentially at highly transcribed genes in npl3Δ cells.
Rrm3 accumulation at transcribed ORFs in npl3Δ cells is R-loop-dependent
We showed that R-loop formation at transcribed genes is in part responsible for the genetic instability phenotypes of npl3Δ cells. If this instability were mediated by RF progression impairment, we would expect that R loops could also contribute to the replication impairment caused by the absence of Npl3. To test this, we performed specific Rrm3-Flag ChIP RT-qPCR experiments in selected genes with high levels of Rrm3 recruitment as determined by the ChIP–chip, such as GCN4 (Fig. 7D). As can be seen in Figure 7E, there was a significant enhancement of Rrm3 recruitment in this gene in npl3Δ cells, validating the ChIP–chip results. Importantly, overexpression of RNase H1 significantly reduced the levels of Rrm3 to those of the wild type (P < 0.05), suggesting that the genome-wide effect of Npl3 in preventing transcription-dependent replication impairment is linked to cotranscriptional R-loop formation.
Discussion
In this study, we show that the RNA-binding hnRNP Npl3 has a function in preventing transcription-dependent genome instability. Moreover, loss of Npl3 results in increased sensitivity to DSB-inducing agents and genome-wide replication impairment mediated by the formation of R loops. Our results not only provide a new link between mRNP biogenesis and the prevention of R-loop-mediated transcription–replication collisions but support a model in which a number of RNA-binding hnRNPs prevent RNA-mediated replication stress and open the possibility of using them as a therapeutic target in anti-cancer treatment.
We identified npl3Δ as a mutant hypersensitive to trabectedin (ET-743). The action mechanism of this anti-tumor drug remains poorly understood, but it has been shown that in vitro, it forms covalent adducts with guanines located in the DNA minor groove (Pommier et al. 1996). Addition of trabectedin leads to the generation of DSBs, the activation of the DNA damage checkpoint, and the recruitment of proteins of the nucleotide excision repair and HR pathways in Schizosaccharomyces pombe (Herrero et al. 2006; Guirouilh-Barbat et al. 2008). Interestingly, it is worth noting that trabectedin binding to the DNA leads to the formation of a complex that resembles a DNA–RNA hybrid (Marco et al. 2002). npl3Δ cells were also hypersensitive to other DNA-damaging agents, including MMS, Phl, HU, and UV light, all of which lead directly or indirectly to RF stalling and/or DSBs. However, the ability of npl3Δ cells to repair DSBs by SCR or NHEJ is not affected or is poorly affected (Fig. 1), suggesting that the lack of Npl3 generates replication stress that results in the accumulation of recombinogenic DNA breaks, which is strengthened by genotoxic agents. In fact, sensitivity of npl3Δ cells to MMS was increased synergistically in npl3Δ rad52Δ and npl3Δ yku80Δ cells, in agreement with a previous genome-wide analysis that reported a synthetic growth defect for npl3Δ rad52Δ double mutants (Pan et al. 2006). Interestingly, npl3Δ cells are not able to complete replication after treatment with MMS, leading to activation of the DNA damage response to prevent the collapse of putatively stalled RFs. Altogether, these data suggest that npl3Δ cells are under permanent replication stress.
In the absence of induced DNA damage, npl3Δ cells accumulate DNA breaks monitored as Rad52 foci and show plasmid instability and transcription-dependent hyperrecombination (Fig. 3), which is consistent with the identification of Npl3 in a screening for mutants involved in yeast minichromosome maintenance (Wahba et al. 2011). Importantly, hyperrecombination in the absence of Npl3 is transcription-dependent, according to the role of Npl3 in transcription (Lei et al. 2001), and can be partially suppressed by the overexpression of RNase H1, which can also suppress Rad52 foci accumulation (Fig. 4), indicating that R loops are at least in part responsible for the genetic instability generated in npl3Δ cells. Additionally, human AID overexpression exacerbates recombination, consistent with its enhanced action on the displaced ssDNA of cotranscriptional R loops (Chaudhuri et al. 2003; Gomez-Gonzalez and Aguilera 2007). R loops form naturally during specific cellular processes, including Escherichia coli plasmid replication, mitochondrial DNA replication, and immunoglobulin class switching (Aguilera and Garcia-Muse 2012). However, they can also be generated as transcriptional by-products that compromise genome integrity, as shown for cells (from yeast to humans) depleted of a number of conserved proteins involved in RNA metabolism, including the THO–TREX complex (Huertas and Aguilera 2003; Dominguez-Sanchez et al. 2011; Castellano-Pozo et al. 2012), the DNA–RNA helicase Sen1/Senataxin (Mischo et al. 2011), topoisomerase I (Tuduri et al. 2009; El Hage et al. 2010), or the ASF/SF2 splicing factor (Li and Manley 2005). In addition, several screenings in yeast and human cells have revealed that the genome instability generated by the absence of a great number of RNA processing factors is also R-loop-dependent (Paulsen et al. 2009; Wahba et al. 2011; Stirling et al. 2012).
An enhancement of R-loop formation or stabilization might explain the transcription-dependent and R-loop-dependent replication collisions and genome instability of npl3Δ cells, consistent with its main putative role in mRNP assembly, although we cannot discard an indirect contribution of a defect in mRNA export given the interaction of Npl3 with the Mex67 export factor, whose mutations also lead to genetic instability (Gilbert and Guthrie 2004). Importantly, however, overexpression of SUB2, THO1, and NAB2 is able to suppress npl3Δ phenotypes (Fig. 5), suggesting an interchangeable role of these proteins that rely on their ability to bind nascent RNA. Consistent with the functional interrelationship between Npl3 and THO, Npl3 overexpression is able to partially suppress the phenotypes of hpr1Δ. Indeed, the fact that overexpression of NPL3 leads to hyperrecombination in wild-type cells, as has been also shown for SUB2 overexpression (Fan et al. 2001), suggests that the proper stoichiometry of proteins involved in mRNP assembly is key to preventing RNA processing defects from compromising genome integrity. However, we cannot discard that the inhibition of transcription termination reported by excess of Npl3 (Bucheli and Buratowski 2005) could be one of the causes of this instability.
Many transcription elongation and RNA processing factors, including THO–TREX, have been shown to be recruited along transcribed ORFs in a gradient profile that increases toward the 3′ end of genes all over the genome (Kim et al. 2010; Mayer et al. 2010; Gomez-Gonzalez et al. 2011). This, together with the 3′ end processing defects of THO mutants (Saguez et al. 2008) as well as the transcription termination role of Sen1 RNA–DNA helicase (Kawauchi et al. 2008), suggests that termination may be one of the most sensitive steps connected to RNA-mediated genome instability. Notably, Npl3 is also distributed along transcribed ORFs according to an increasing pattern toward the 3′ end (Fig. 6) and causes a preferential reduction in RNA accumulation of long and highly transcribed genes in concordance with the features of the genes to which it is preferentially bound. These results confirm that Npl3 has a main role in cotranscriptional mRNP assembly.
Given the double effect of npl3Δ on gene expression and genome instability, the main question remaining is whether both are interconnected. It is known from many reports from bacteria to human cells that transcription stimulates recombination (Gaillard et al. 2013). Consistent with recombination being the main pathway of repair of DSBs generated during replication, it is believed that most TAR events arise during replication as a result of transcription–replication collisions that cause the stalling and collapse of forks (Azvolinsky et al. 2009; Bermejo et al. 2009). Here we show that, indeed, npl3Δ cells have a replication delay and that most replication obstacles occur at transcribed genes in these cells, visualized as the genome-wide distribution of Rrm3, a DNA helicase required for RFs to bypass obstacles (Ivessa et al. 2003; Azvolinsky et al. 2009). As Npl3-bound genes, Rrm3-bound genes are also long and highly expressed (Fig. 7), and there is a high enrichment of Rrm3 in ribosomal protein genes, the most highly transcribed class of genes in yeast. This is consistent with a previous study showing that Npl3 is preferentially associated with ribosomal protein mRNAs and other highly transcribed transcripts (Kim Guisbert et al. 2005) and the reported role of Npl3 mediating nuclear export of large ribosomal subunits (Hackmann et al. 2011). Rrm3 is preferentially observed in a gradient manner that increases toward the 3′ end of RNAPII genes, following a pattern similar to that previously shown for Rrm3 in THO mutants (Gomez-Gonzalez et al. 2011), which is partially suppressed by RNase H1 overexpression (Fig. 7E). Therefore, cotranscriptional R loops in npl3Δ cells constitute a major cause of RF stalling responsible for recombination-mediated genome instability.
Consequently, we propose a model in which Npl3 acts as a key player in coupling transcription of RNAPII genes with mRNP formation and export, thus facilitating replication through highly transcribed regions (Supplemental Fig. S8). In the absence of Npl3, the mRNP assembly would be suboptimal, and the nascent mRNA may hybridize more stably with the transcribed DNA strand, forming R loops that would constitute roadblocks to replication and cause fork stalling and breaks that would demand HR for fork restart and survival. In summary, our results show that Npl3, the most abundant RNA-binding hnRNP, is necessary to prevent R-loop-mediated transcription–replication conflicts and genome instability. Given the relevance of genome instability as a common feature of tumoral cells (Bartkova et al. 2005; Gorgoulis et al. 2005), the R-loop-like structure of the trabectedin–DNA complex (Marco et al. 2002), and the importance of Npl3 for resistance to this anti-tumor drug, it would be interesting to explore whether deregulation of RNA-binding proteins leading to the formation of R loops in human cells could be used to identify new anti-tumoral targets in cancer treatment.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids used in this study are listed in Supplemental Tables S2 and S3.
Physical analysis of SCR
SCR assays were carried out essentially as described (see the Supplemental Material; Gonzalez-Barrera et al. 2003).
ChIP analyses
Recruitment of Rrm3 to chromatin was determined by ChIP analyses on the endogenous GCN4 gene as previously described (see the Supplemental Material; Hecht et al. 1999).
Microarray gene expression analysis
Microarray determination of total RNA was performed using the Affymetrix platform (see the Supplemental Material) as previously described (see the Supplemental Material; Gomez-Gonzalez et al. 2011). The expression data can be accessed at Gene Expression Omnibus (GSE50187 and GSE50186).
ChIP–chip experiments
S. cerevisiae oligonucleotide tiling microarrays were provided by Affymetrix. The high-density oligonucleotide arrays used were able to analyze yeast chromosomes at a 300-bp resolution, each of the 300-bp regions being covered by at least 60 probes. ChIP–chip of asynchronously growing cells was carried out as described (see the Supplemental Material; Katou et al. 2006; Bermejo et al. 2009). The ChIP–chip data can be accessed at Gene Expression Omnibus (GSE50187 and GSE50185).
Statistical analysis of genome-wide data
Microarray data were normalized by RMA (robust microarray average) and statistically analyzed by LIMMA (linear models for microarray analysis), comparing the mutant expression profile with its isogenic wild-type strain. The genes showing at least a 1.5-fold expression change with a P-value < 0.01 with a false discovery rate (FDR) correction were considered as altered. ChIP–chip data were analyzed using the Tiling Array suite (TAS) software from Affymetrix. For each probe position, TAS produces the signal and the change P-value, taking into account the probes localized within a given bandwidth around the inspected probe. Protein chromosomal distribution was then analyzed by detecting binding clusters, which were defined as ranges within the chromosome respecting the following conditions: estimated signal (IP/SUP-binding ratio) positive in the whole range, P-value < 0.01, minimum run of 100 bp, and maximum gap of 250 bp. The results were visualized with the University of California at Santa Cruz Genome Browser, developed and maintained by the Genome Bioinformatics Group (Center for Biomolecular Science and Engineering at the University of California at Santa Cruz; http://genome.ucsc.edu). For statistical analysis of the functional and structural features of the genes, expression levels were taken from microarray of wild-type cells. Distribution of binding sites along genes was carried out as previously described (Gomez-Gonzalez et al. 2011).
Miscellanea
Analysis of sensitivity to genotoxic agents, PFGE, Western analyses, Southerns, FACS using a FACScalibur Becton Dickinson machine, and yeast cultures were performed using standard procedures. Genetic assays of NHEJ, plasmid maintenance, recombination, and Rad52 foci are described in the Supplemental Material.
Acknowledgments
We thank S. Fields and A. McBride for reagents, P. Domínguez (Microscopy Unit, CABIMER) for technical assistance in fluorescence microscopy, E. Andújar and M. Pérez-Alegre (Genomics Unit, CABIMER) for technical support with microarray and ChIP–chip experiments, U. Galindo for technical assistance, and D. Haun for style supervision. Research was funded by grants from the Spanish Ministry of Economy and Competitiveness (Consolider 2010 CSD2007-0015 to A.A. and S.M., BFU2010-16372 to A.A., and BFU2011-28274 to S.M.), the Junta de Andalucía (CVI4567 to A.A.), and the European Union (FEDER). J.M.S.-P. was the recipient of a JAE predoctoral training grant from the Spanish Research Council (CSIC). A.B.H. was supported by a post-doctoral grant from the Asociación Española Contra el Cáncer (AECC).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.229880.113.
References
- Aguilera A, Garcia-Muse T 2012. R loops: From transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T 2013. Causes of genome instability. Annu Rev Genet 47: 19–50 [DOI] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, et al. 2009. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell 138: 870–884 [DOI] [PubMed] [Google Scholar]
- Bucheli ME, Buratowski S 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J 24: 2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Garcia-Muse T, Aguilera A 2012. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep 13: 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422: 726–730 [DOI] [PubMed] [Google Scholar]
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ledesma F, Aguilera A 2006. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep 7: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME 2008. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 3: e3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7: e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Merker RJ, Klein HL 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol Cell Biol 21: 5459–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, et al. 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: A new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet 5: e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Herrera-Moyano E, Aguilera A 2013. Transcription-associated genome instability. Chem Rev. doi: 10.1021/cr400017y [DOI] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 13: 201–212 [DOI] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Aguilera A 2007. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci 104: 8409–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 30: 3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Barrera S, Cortes-Ledesma F, Wellinger RE, Aguilera A 2003. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol Cell 11: 1661–1671 [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913 [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Redon C, Pommier Y 2008. Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11-Rad50-Nbs1 complex. Mol Biol Cell 19: 3969–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackmann A, Gross T, Baierlein C, Krebber H 2011. The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep 12: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M 1999. Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol Biol 119: 469–479 [DOI] [PubMed] [Google Scholar]
- Herrero AB, Martin-Castellanos C, Marco E, Gago F, Moreno S 2006. Cross-talk between nucleotide excision and homologous recombination DNA repair pathways in the mechanism of action of antitumor trabectedin. Cancer Res 66: 8155–8162 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kaneshiro K, Aburatani H, Shirahige K 2006. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol 409: 389–410 [DOI] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ 2008. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev 22: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Guisbert K, Duncan K, Li H, Guthrie C 2005. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. RNA 11: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C 2008. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell 32: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev 10: 1233–1246 [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell 18: 711–722 [DOI] [PubMed] [Google Scholar]
- Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A 2008. Biogenesis of mRNPs: Integrating different processes in the eukaryotic nucleus. Chromosoma 117: 319–331 [DOI] [PubMed] [Google Scholar]
- Marco E, Garcia-Nieto R, Mendieta J, Manzanares I, Cuevas C, Gago F 2002. A 3.(ET743)–DNA complex that both resembles an RNA–DNA hybrid and mimicks zinc finger-induced DNA structural distortions. J Med Chem 45: 871–880 [DOI] [PubMed] [Google Scholar]
- Marin A, Gallardo M, Kato Y, Shirahige K, Gutierrez G, Ohta K, Aguilera A 2003. Relationship between G+C content, ORF-length and mRNA concentration in Saccharomyces cerevisiae. Yeast 20: 703–711 [DOI] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena A, Gewartowski K, Mroczek S, Cuellar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. 2012. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J 31: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW 1996. DNA sequence- and structure-selective alkylation of guanine N2 in the DNA minor groove by ecteinascidin 743, a potent antitumor compound from the Caribbean tunicate Ecteinascidia turbinata. Biochemistry 35: 13303–13309 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Aguilera A 2010. The interface between transcription and mRNP export: From THO to THSC/TREX-2. Biochim Biophys Acta 1799: 533–538 [DOI] [PubMed] [Google Scholar]
- Saguez C, Schmid M, Olesen JR, Ghazy MA, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH 2008. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell 31: 91–103 [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2009. The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 44: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]