The RNAi pathway is required for heterochromatin assembly, and loss of RNAi causes pericentric heterochromatin defects. Tadeo et al. show that deletion of telomere shelterin components in RNAi mutants restores pericentric heterochromatin. Shelterin component Poz1 mutant analysis reveals that defective telomere silencing, but not telomere length control, is critical for bypassing RNAi. Furthermore, heterochromatin protein Swi6 is redistributed to pericentric regions in RNAi mutants. Heterochromatin domains thus use multiple pathways to restrain Swi6 and avoid promiscuous heterochromatin formation.
Keywords: RNAi, heterochromatin assembly, telomere, shelterin, centromere
Abstract
The RNAi pathway is required for heterochromatin assembly at repetitive DNA elements in diverse organisms. In fission yeast, loss of RNAi causes pericentric heterochromatin defects, compromising gene silencing and chromosome segregation. Here we show that deletion of telomere shelterin components restores pericentric heterochromatin and its functions in RNAi mutants. We further isolated a separation-of-function mutant of Poz1 and revealed that defective telomere silencing, but not telomere length control, is critical for bypassing RNAi. Further analyses demonstrated that compromising shelterin-mediated heterochromatin assembly in RNAi mutants releases heterochromatin protein Swi6, which is redistributed to pericentric regions through RNAi-independent heterochromatin assembly pathways. Given the high mobility of Swi6 protein and that increased levels of Swi6 facilitates heterochromatin spreading as well as ectopic heterochromatin assembly, our results suggest that constitutive heterochromatin domains use multiple pathways to form high-affinity platforms to restrain Swi6, thus limiting its availability and avoiding promiscuous heterochromatin formation.
Eukaryotic genomes possess large stretches of repetitive DNA elements that form compact chromatin structures termed heterochromatin. These regions are relatively gene-poor but occupy crucial chromosome regions, such as centromeres and telomeres, which play critical roles in chromosome segregation and three-dimensional genome organization. Heterochromatin also inhibits transcription and recombination of the underlying DNA repeats, rendering it indispensable for maintaining genome integrity (Grewal and Jia 2007; Almouzni and Probst 2011).
Genomic DNA is folded with histones to form chromatin. Each chromosomal region is associated with distinct profiles of histone post-translational modifications, which play essential roles in regulating chromatin structure and function. Histones at heterochromatic regions are generally devoid of acetylation and are methylated at H3 Lys9 (H3K9me). H3K9me allows for the binding of heterochromatin protein 1 (HP1), which compacts chromatin through its self-association and the recruitment of other chromatin-modifying activities (Grewal and Jia 2007). Although heterochromatin has traditionally been considered silent, stable, and static, recent biochemical and imaging studies show that the binding of HP1 proteins to heterochromatin regions is surprisingly dynamic (Cheutin et al. 2003, 2004; Festenstein et al. 2003; Sadaie et al. 2008). Moreover, although heterochromatin normally excludes the transcriptional machinery, the DNA repeats within heterochromatin are transcribed, and the transcripts are processed by the RNAi pathway into siRNAs, which target histone-modifying complexes to repetitive DNA elements (Moazed, 2009; Lejeune and Allshire 2011; Castel and Martienssen 2013).
RNAi-dependent heterochromatin assembly is best studied in the fission yeast Schizosaccharomyces pombe (for review, see Moazed 2009; Lejeune and Allshire, 2011; Goto and Nakayama 2012; Castel and Martienssen 2013). In this organism, constitutive heterochromatin is localized at the centromeres, telomeres, and the silent mating type region, all of which share similar repetitive DNA elements composed of dg and dh repeats. The DNA repeats are transcribed by RNA polymerase II (Pol II) during the S phase of the cell cycle, producing dsRNAs with the help of a RNA-dependent RNA polymerase complex (RDRC). The ribonuclease Dicer (Dcr1) processes these dsRNAs into siRNAs, which are loaded onto the Argonaute siRNA chaperone complex (ARC) and then transferred to RNAi-induced transcriptional silencing complex (RITS). The Argonaute protein (Ago1) within RITS binds siRNAs and targets RITS to nascent RNA transcripts from repeat regions. RITS then recruits the CLRC complex, which contains the H3K9 methyltransferase Clr4. H3K9me recruits chromodomain proteins Swi6 and Chp2 together with their associated histone deacetylases and chromatin remodeling activities to achieve transcriptional silencing. H3K9me also stabilizes the binding of RITS to chromatin through the chromodomain protein Chp1, which in turn recruits RDRC to generate more dsRNAs and siRNAs, thus forming a self-reinforcing loop between siRNA production and heterochromatin assembly.
In addition to RNAi-based mechanisms, DNA-based mechanisms also function at repeat regions to recruit CLRC. For example, ATF/CREB family proteins Atf1 and Pcr1 cooperate with RNAi to establish heterochromatin at the silent mating type region (Jia et al. 2004; Kim et al. 2004). Similarly, telomere shelterin component Taz1 and the telomere-associated sequence (TAS) function together with RNAi to establish heterochromatin at telomeres (Kanoh et al. 2005). As a result, loss of RNAi severely affects pericentric heterochromatin functions but has little effect on heterochromatin structures at the silent mating type region or telomeres. However, even at pericentric regions, H3K9me and Swi6 are still present in RNAi mutants, albeit at lower levels, suggesting the existence of RNAi-independent mechanisms to establish heterochromatin (Sadaie et al. 2004). These pathways involve the Clr3 and Sir2 histone deacetylases since H3K9me levels are further reduced from pericentric regions in clr3Δ dcr1Δ and sir2Δ dcr1Δ cells, although Sir2 seems to function in a separate pathway from Clr3 (Yamada et al. 2005; Alper et al. 2013; Buscaino et al. 2013; Marina et al. 2013).
Interestingly, elimination of the Mst2 histone H3K14 acetyltransferase complex activity, the JmjC domain protein Epe1, or RNA quality control factor Mlo3 bypasses the requirement of RNAi for pericentric heterochromatin assembly (Trewick et al. 2007; Reddy et al. 2011; Reyes-Turcu et al. 2011). Although the mechanisms by which these mutants regulate heterochromatin assembly in the absence of RNAi differ, these results nonetheless indicate that RNAi is not obligatory for pericentric heterochromatin formation under certain conditions. To further understand heterochromatin assembly pathways at pericentric regions, we performed a high-throughput screen of the fission yeast deletion library for mutants that support pericentric heterochromatin assembly in the absence of RNAi. We discovered that loss of the telomere shelterin components involved in proper telomere maintenance also bypassed RNAi for pericentric heterochromatin formation. We further isolated a mutant of shelterin component Poz1 that specifically affected telomeric heterochromatin assembly without affecting telomere length and showed that loss of shelterin in an RNAi mutant background resulted in the redistribution of heterochromatin protein Swi6 from telomeric heterochromatin to pericentric regions through RNAi-independent heterochromatin assembly pathways involving Clr3 and Sir2. Furthermore, we showed that increasing overall Swi6 levels was sufficient to bypass RNAi for pericentric heterochromatin assembly. Our results demonstrate a dynamic equilibrium of different heterochromatin domains through the high mobility of Swi6 and suggest that constitutive heterochromatin regions, such as centromeres and telomeres, play important roles in balancing the availability of silencing factors to maintain the overall epigenetic landscape of the genome.
Results
A high-throughput screen for mutants that bypass the requirement of the RNAi machinery for pericentric heterochromatin assembly
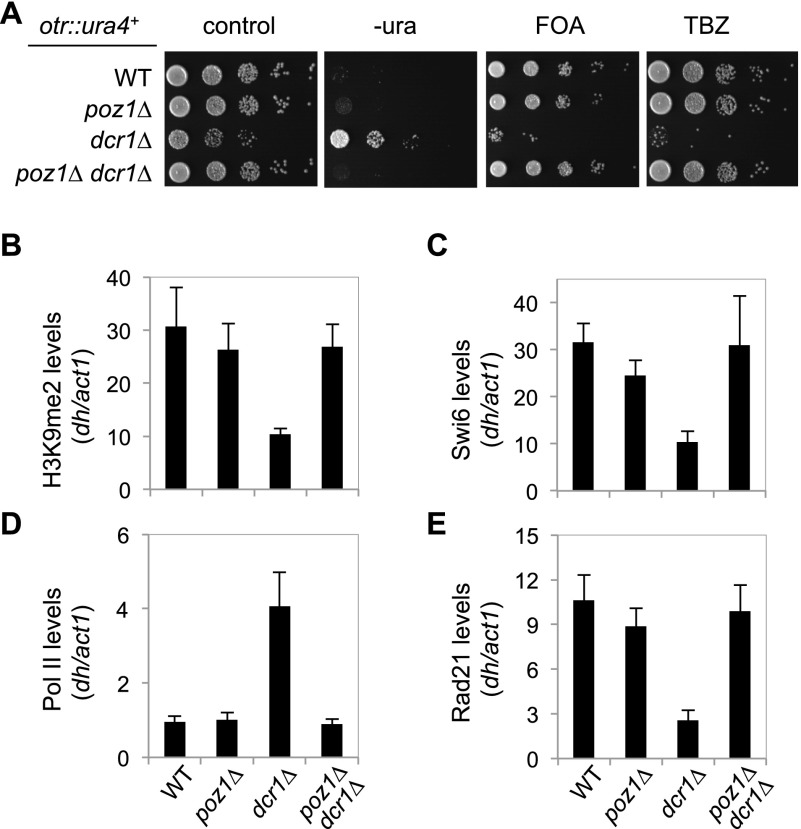
In order to understand the mechanism of RNAi and heterochromatin assembly, we performed a screen of the fission yeast deletion library for mutants that allow restoration of pericentric heterochromatin in the absence of RNAi function. We used a query strain containing a ura4+ reporter gene inserted at the outer repeat of centromere I (otr∷ura4+) (Supplemental Fig. S1A). In wild-type cells, this reporter gene is silenced due to the formation of heterochromatin. As a result, there is little growth on medium lacking uracil but robust growth on counterselective medium containing 5-fluoroorotic acid (FOA), which is toxic to cells expressing ura4+ (Fig. 1A). Mutations that affect heterochromatin structure at pericentric regions, such as dcr1Δ, resulted in strong cell growth on medium lacking uracil but little growth on medium containing FOA (Fig. 1A). Through two successive crosses, we introduced the otr∷ura4+ reporter and dcr1Δ into the deletion library and obtained haploid cells, each containing otr∷ura4+, dcr1Δ, and a single gene deletion (Supplemental Fig. S1B). The resulting cells were grown on medium containing FOA to measure cell growth (Supplemental Fig. S1C; Supplemental Table S1). Our screen identified mutants known to bypass RNAi for pericentric heterochromatin assembly, such as Mst2 complex components mst2Δ, nto1Δ, and ptf2Δ (Reddy et al. 2011; Wang et al. 2012), validating the effectiveness of our screen. Our screen also identified poz1Δ, a telomere shelterin component involved in telomere length control (Miyoshi et al. 2008), as showing the strongest rescue of heterochromatic silencing (Supplemental Fig. S1C; Supplemental Table S1). We also performed similar screens with an otr∷ade6+ reporter (Supplemental Fig. S2A). This reporter gene is silenced in wild-type cells, resulting in the formation of red colonies on low-adenine medium due to the accumulation of metabolic intermediates. In RNAi mutants, such as dcr1Δ, loss of pericentric heterochromatin resulted in the expression of Ade6, giving rise to white colonies (Supplemental Fig. S2A). We also identified poz1Δ as one of the strongest suppressors of dcr1Δ and ago1Δ with the otr∷ade6+ reporter (Supplemental Fig. S2B,C; Supplemental Table S1). Altogether, we identified a total of 33 gene deletions that bypassed RNAi in both otr∷ura4+ and otr∷ade6+ reporter screens.
Figure 1.
Elimination of Poz1 bypasses the requirement of Dicer for pericentric heterochromatin assembly and function. (A) Tenfold serial dilution analyses of the indicated yeast strains grown on the indicated medium to measure the expression of otr∷ura4+ and sensitivity to TBZ. (B–E) ChIP analysis of H3K9me2, Swi6, Pol II (Rpb1), and Rad21-Flag levels at pericentromeric dh repeats, normalized to act1. The numbers are averages of three experiments, and error bars represent standard deviation.
Elimination of Poz1 bypasses RNAi for pericentric heterochromatin functions
To confirm that loss of Poz1 can indeed restore pericentric heterochromatin in RNAi mutants, we constructed a poz1Δ dcr1Δ double mutant strain containing otr∷ura4+. Serial dilution analyses confirmed that poz1Δ dcr1Δ grew weakly on medium without uracil but robustly on medium containing FOA (Fig. 1A). Chromatin immunoprecipitation (ChIP) analyses showed that in poz1Δ dcr1Δ cells, heterochromatin hallmarks at pericentric dh repeats, such as H3K9me2 and Swi6, were restored to near wild-type levels (Fig. 1B,C). Moreover, ChIP analyses showed that Pol II was strongly excluded from pericentric repeats (Fig. 1D), suggesting that heterochromatin formed in poz1Δ dcr1Δ cells is capable of transcriptional gene silencing.
Pericentric heterochromatin is required for the accumulation of high levels of cohesins, which are critical for chromosome segregation during mitosis (Bernard et al. 2001; Nonaka et al. 2002; Yamagishi et al. 2008). When heterochromatin is compromised, such as in dcr1Δ, loss of cohesin results in defective attachment of centromeres by microtubules for biorientation. As a result, cells are very sensitive to the microtubule poison thiabendazole (TBZ) (Hall et al. 2003; Volpe et al. 2003). In poz1Δ dcr1Δ cells, both cohesin localization, as measured by ChIP analyses of cohesin subunit Rad21-Flag, and TBZ sensitivity were significantly rescued (Fig. 1A,E). Taken together, these results indicate that pericentric heterochromatin formed in poz1Δ dcr1Δ cells is functional in regulating chromosome segregation.
Bypassing RNAi requires RNAi-independent heterochromatin assembly pathways at pericentric regions
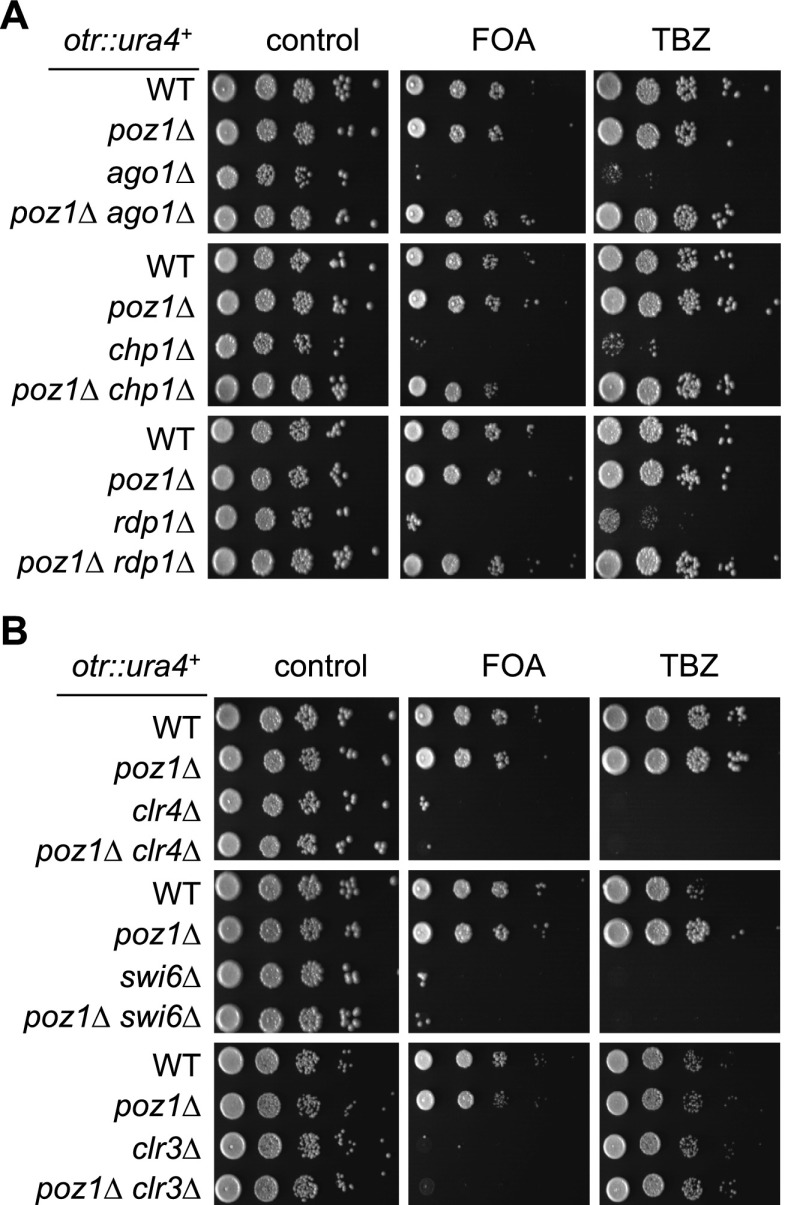
In fission yeast, the RNAi pathway involves several different complexes in addition to Dicer (Goto and Nakayama 2012). We also found that poz1Δ rescued silencing defects at otr∷ura4+ and TBZ sensitivity of all RNAi mutants tested, such as those in RITS (ago1Δ and chp1Δ) and RDRC (rdp1Δ) (Fig. 2A). However, poz1Δ could not rescue silencing defects and TBZ sensitivity of heterochromatin mutants involved in histone modifications or their recognition, such as clr4Δ, swi6Δ, or clr3Δ (Fig. 2B), suggesting that histone modifications are still essential for pericentric heterochromatin assembly in poz1Δ dcr1Δ cells.
Figure 2.
Elimination of Poz1 bypasses the entire RNAi pathway, but not histone modifications, for pericentric heterochromatin assembly and function. (A,B) Tenfold serial dilution analyses of the indicated yeast strains grown on the indicated medium to measure the expression of otr∷ura4+ and sensitivity to TBZ.
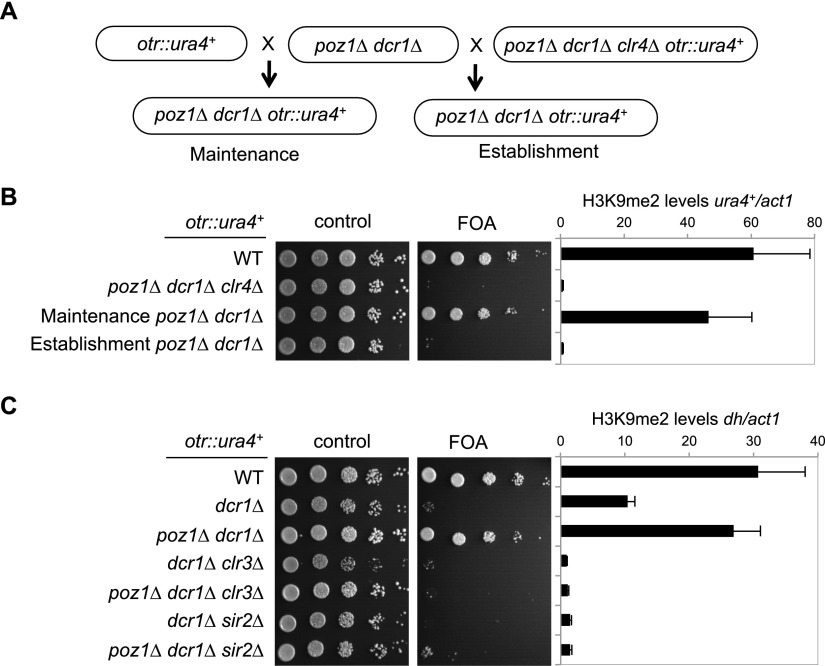
RNAi is required for both the establishment and maintenance of heterochromatin at pericentric regions (Sadaie et al. 2004). To distinguish whether poz1Δ dcr1Δ rescues heterochromatin maintenance or establishment, we introduced otr∷ura4+ in either a silenced state (from wild-type cells) or a desilenced state (from clr4Δ cells) into poz1Δ dcr1Δ cells by genetic crosses (Fig. 3A). Interestingly, poz1Δ dcr1Δ cells efficiently maintained silencing of otr∷ura4+ when it was inherited from a wild-type cell but failed to establish silencing when it was inherited from a clr4Δ cell (Fig. 3B). ChIP analyses of H3K9me2 also confirmed this conclusion (Fig. 3B).
Figure 3.
Elimination of Poz1 bypasses RNAi for pericentric heterochromatin maintenance but not for its establishment. (A) Schematic diagram of the experimental design to examine heterochromatin maintenance and establishment. (B,C, left) Tenfold serial dilution analyses of the indicated yeast strains to measure the expression of otr∷ura4+. (Right) ChIP analysis of H3K9me2 levels at ura4+ or pericentric dh repeats, normalized to act1. The numbers are averages of three experiments, and error bars represent standard deviation.
In RNAi mutants, although silencing of reporter genes is completely lost, significant amounts of H3K9me are still retained at pericentric repeats (Fig. 3C; Sadaie et al. 2004). The histone deacetylases Clr3 and Sir2 function cooperatively with RNAi to establish heterochromatin at pericentric repeats, and H3K9me2 is completely abolished in clr3Δ dcr1Δ and sir2Δ dcr1Δ cells (Yamada et al. 2005; Alper et al. 2013; Buscaino et al. 2013; Marina et al. 2013). The results that poz1Δ dcr1Δ cells maintained pre-existing heterochromatin but failed to establish heterochromatin de novo suggest that the residual H3K9me in dcr1Δ cells is necessary for poz1Δ dcr1Δ cells to maintain pericentric heterochromatin. Indeed, neither poz1Δ dcr1Δ clr3Δ nor poz1Δ dcr1Δ sir2Δ cells can maintain silencing of otr∷ura4+ or H3K9me levels at pericentric repeats (Fig. 3C), suggesting that the RNAi-independent pathway is critical for the ability of poz1Δ dcr1Δ cells to maintain pericentric heterochromatin.
Elimination of other shelterin components also bypasses RNAi for pericentric heterochromatin assembly
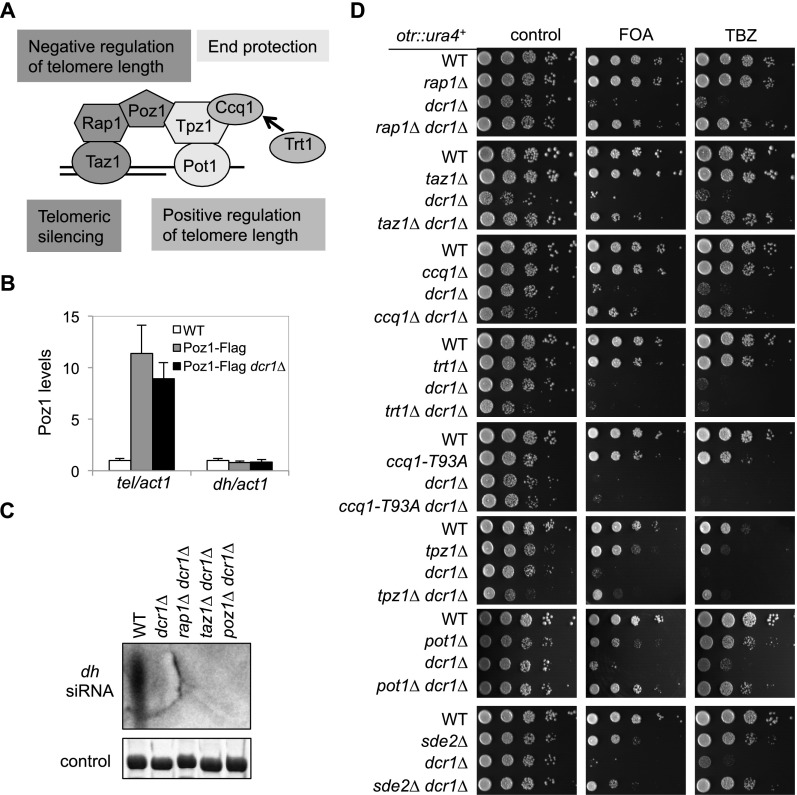
Poz1 is a member of the telomere-associated shelterin complex (Miyoshi et al. 2008) that protects chromosome ends from being recognized as DNA damage sites and prevents DNA loss during replication (Palm and de Lange 2008; Jain and Cooper 2010). Telomeric DNA consists of short tandem DNA repeats followed by a single-stranded overhang (Fig. 4A). In fission yeast, Taz1 (homolog of mammalian TRF1/2) binds to the dsDNA repeats (Cooper et al. 1997), and the Pot1–Tpz1 complex (homologous to mammalian POT1–TPP1) binds to the ssDNA overhang (Baumann and Cech 2001; Miyoshi et al. 2008). Rap1 and Poz1 form a bridge that connects Taz1 and Pot1–Tpz1 complexes (Miyoshi et al. 2008). Pot1–Tpz1 associates with Ccq1 to recruit telomerase Trt1, which uses associated RNA as a template to elongate the telomere overhang (Nakamura et al. 1997; Miyoshi et al. 2008; Tomita and Cooper 2008; Moser et al. 2011; Yamazaki et al. 2012). By ChIP analyses, we found that Poz1 was enriched at telomeres, as expected, but not detected at pericentric repeats, even in dcr1Δ cells (Fig. 4B). Furthermore, Northern blot analyses showed that no siRNAs derived from pericentric repeats were detected in poz1Δ dcr1Δ cells (Fig. 4C), suggesting that Poz1 regulates pericentric heterochromatin assembly indirectly through RNAi-independent mechanisms.
Figure 4.
Elimination of other shelterin components also bypasses RNAi for pericentric heterochromatin assembly. (A) Schematic diagram of the telomere complex. (B) ChIP analysis of Poz1-Flag levels at subtelomeric and pericentric regions. The numbers are averages of three experiments, and error bars represent standard deviation. (C) Northern blot analyses of siRNAs derived from pericentric dh repeats. (D) Tenfold serial dilutions of the indicated yeast strains were grown on the indicated medium to measure the expression of otr∷ura4+ and sensitivity to TBZ. The trt1Δ dcr1Δ and ccq1-T93A dcr1Δ cells were freshly germinated to avoid senescence-associated telomere rearrangement.
Since our screens of mutants that bypass RNAi also consistently identified rap1Δ (Supplemental Table S1), we tested whether other shelterin mutants behaved similarly. We found that rap1Δ, taz1Δ, and ccq1Δ also rescued RNAi mutants' defects in otr∷ura4+ silencing and TBZ sensitivity (Fig. 4D; Supplemental Fig. S3). Similarly, rap1Δ dcr1Δ clr3Δ and taz1Δ dcr1Δ clr3Δ failed to maintain pericentric heterochromatin silencing (Supplemental Fig. S4), indicating the requirement of RNAi-independent pericentric heterochromatin pathways. In contrast, trt1Δ and a ccq1-T93A mutant, which attenuated recruitment of Trt1 to telomeres (Moser et al. 2011; Yamazaki et al. 2012), had no effect on this process (Fig. 4D). We reasoned that the different behavior of ccq1Δ and ccq1-T93A is because Ccq1 is also part of the SHREC complex involved in telomere silencing (Sugiyama et al. 2007; Motamedi et al. 2008), which is not affected by the ccq1-T93A mutation (Moser et al. 2011; Yamazaki et al. 2012). Consistent with this idea, we also found that sde2Δ, which resulted in selective loss of telomere silencing (Sugioka-Sugiyama and Sugiyama 2011), was able to bypass RNAi for pericentric heterochromatin formation (Supplemental Table S1; Fig. 4D). Pot1 and Tpz1 are required for telomere end protection. As a result, pot1Δ or tpz1Δ cells lose telomeres quickly and can only survive by circularizing all three chromosomes (Baumann and Cech 2001; Miyoshi et al. 2008). Nonetheless, pericentric heterochromatin was efficiently maintained in pot1Δ dcr1Δ and tpz1Δ dcr1Δ cells, as indicated by otr∷ura4+ silencing and TBZ sensitivity (Fig. 4D).
The ability of shelterin to regulate telomeric heterochromatin is key to bypassing RNAi
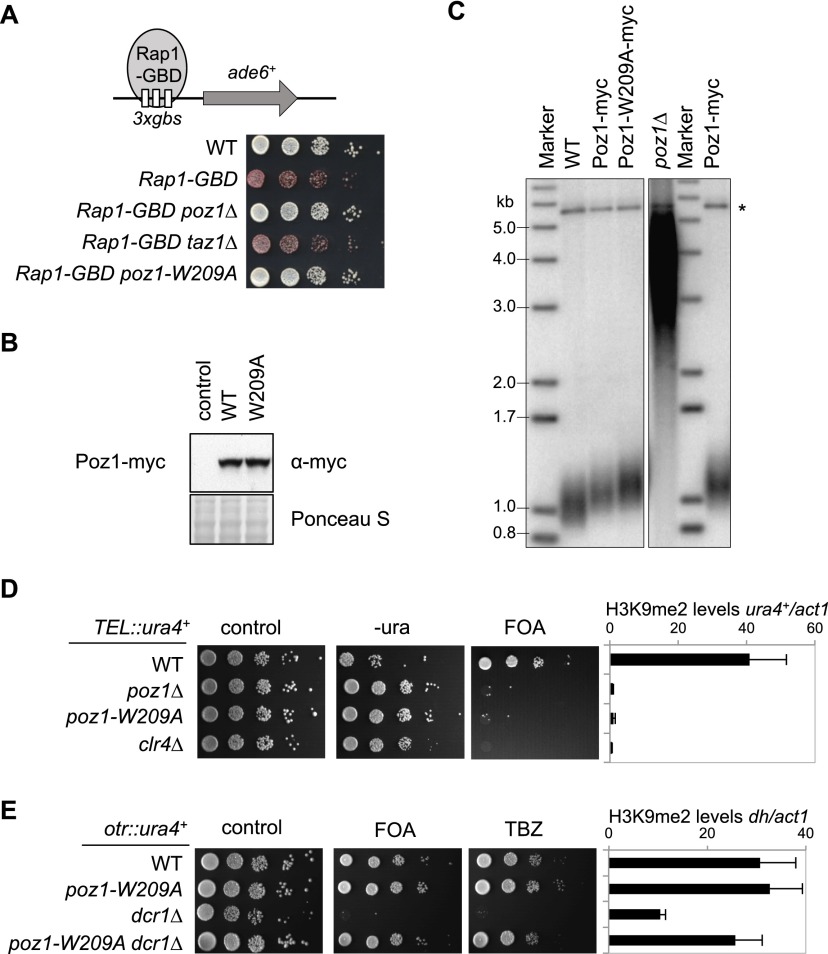
One of the common phenotypes of the telomere-related mutants that bypass RNAi is that they all affect telomere silencing (Cooper et al. 1997; Nimmo et al. 1998; Kanoh and Ishikawa 2001; Sugiyama et al. 2007; Moser et al. 2009). However, the mechanism by which shelterin nucleates heterochromatin assembly is unknown. Because Taz1 and Pot1 contribute independently to shelterin recruitment, it is difficult to examine the sequence by which these factors are recruited at native telomeres. We constructed Rap1 fused with a Gal4 DNA-binding domain (Rap1-GBD) at its endogenous chromosome location. Rap1-GBD is sufficient to induce ectopic heterochromatin assembly at a reporter gene with adjacent Gal4-binding sites (3xgbs-ade6+) under the condition of mild Swi6 overexpression (Supplemental Fig. S5). Silencing depends on Poz1 but not Taz1 (Fig. 5A), suggesting that Poz1 functions downstream from Taz1 and Rap1 in telomeric heterochromatin assembly.
Figure 5.
Loss of shelterin-mediated silencing at telomeres is required for bypassing RNAi for pericentric heterochromatin assembly. (A) Tenfold serial dilutions of the indicated yeast strains were grown on a low-adenine medium to measure the expression of 3xgbs-ade6+. (B) Western blot analyses were performed to measure Poz1-myc protein levels. (C) Southern blot analyses were performed to measure telomere length. The genomic DNA was digested with EcoRI, and the telomeric repeat sequence was used as a probe. (*) Control pol1 fragment. (D,E) Tenfold serial dilutions of the indicated yeast strains were grown on the indicated medium to measure the expression of TEL∷ura4+ and ChIP analysis of H3K9me2 levels at ura4+ or pericentric dh repeat, normalized to act1. The numbers are averages of three experiments, and error bars represent standard deviation.
To distinguish the function of telomere length control and telomeric silencing in bypassing RNAi, we mutated seven conserved residues of Poz1 individually at the endogenous poz1 locus and screened for mutations that specifically affected telomeric silencing without affecting telomere length homeostasis (Supplemental Fig. S6). In the end, we isolated a W209A mutation that had no effect on Poz1 protein levels, as indicated by Western blot analysis (Fig. 5B), and had little effect on telomere length homeostasis, as indicated by Southern blot analyses (Fig. 5C). However, it caused defects in Rap1-GBD-mediated heterochromatin assembly, like in poz1Δ cells (Fig. 5A). We also tested the effect of poz1-W209A on silencing near telomeres. Since multiple pathways cooperatively regulate telomeric heterochromatin assembly, including shelterin, RNAi (through a repeat sequence within the tlh1+ gene), and an unknown mechanism at TAS (Kanoh et al. 2005), we used a ura4+ reporter inserted near telomeric repeats on a minichromosome (TEL∷ura4+) (Nimmo et al. 1994) that lacks the tlh1+ gene and TAS, making the role of shelterin on silencing more apparent (Supplemental Fig. S7). Indeed, poz1-W209A resulted in a strong loss of silencing at this reporter, concomitant with the loss of H3K9me at this location, to a degree similar to poz1Δ and clr4Δ cells (Fig. 5D). Remarkably, poz1-W209A also rescued pericentric heterochromatin assembly in dcr1Δ cells (Fig. 5E), as indicated by the restoration of silencing at the otr∷ura4+ reporter and H3K9me at pericentric dh repeats. Thus, compromised telomeric silencing is the reason why loss of shelterin rescues pericentric heterochromatin assembly defects associated with RNAi mutants.
Shelterin and RNAi modulate the distribution of Swi6 between pericentric regions and telomeres
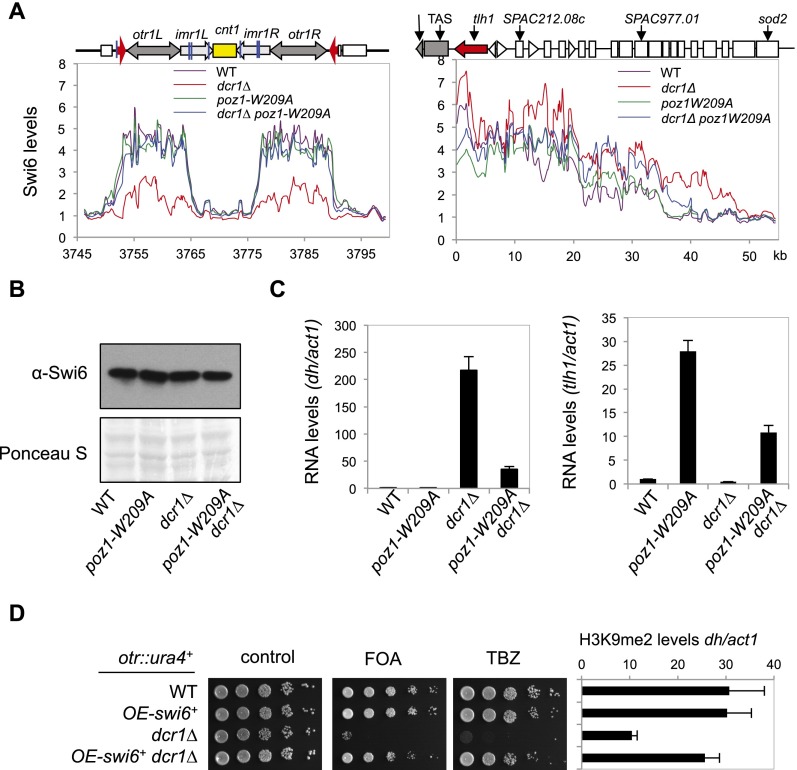
We then performed ChIP–chip analysis of Swi6 with an Agilent microarray. We designed additional probes that provide high coverage of centromere I, which was originally absent in the array due to its repetitive nature. In dcr1Δ cells, Swi6 levels were lower, but not abolished, at the entire pericentric region, as expected (Fig. 6A). Unexpectedly, Swi6 levels increased significantly across the subtelomeric heterochromatin domain, with significant spreading into euchromatic regions (Fig. 6A). In poz1-W209A dcr1Δ cells, Swi6 is not only restored to wild-type levels at pericentric regions but also reduced at the subtelomere regions compared with dcr1Δ. The total levels of Swi6 remain constant in these cells (Fig. 6B), suggesting that the differences in Swi6 localization are the result of Swi6 redistribution at these regions.
Figure 6.
Shelterin and RNAi affect the distribution of Swi6 between pericentric and telomeric regions. (A) ChIP–chip analyses of Swi6 levels at centromere I (left panel) and the left side of telomere I (right panel). (B) Western blot analyses were performed to measure Swi6 protein levels. (C) RT–PCR analyses were performed to measure pericentric dh (left) or telomeric tlh1 (right) transcript levels, normalized to act1. The numbers are averages of three experiments, and error bars represent standard deviation. (D) Tenfold serial dilutions of the indicated yeast strains were grown on the indicated medium to measure the expression of otr∷ura4+ and ChIP analysis of H3K9me2 levels at ura4+ or pericentric dh repeat, normalized to act1. The numbers are averages of three experiments, and error bars represent standard deviation. The OE-swi6+ strains also contain a wild-type copy of swi6+.
To examine whether this redistribution of Swi6 has any physiological consequences, we measured RNA transcript levels derived from the pericentric dh repeat and subtelomeric tlh1. In dcr1Δ cells, high levels of dh repeat transcripts were detected, and in poz1-W209A dcr1Δ cells, dh transcript levels decreased significantly, consistent with our results that these cells form normal pericentric heterochromatin. In contrast, tlh1 transcripts levels were high in poz1-W209A cells but significantly reduced in poz1-W209A dcr1Δ cells (Fig. 6C). Thus, poz1-W209A and dcr1Δ mutually suppressed the other's phenotypes.
If mobilization of silencing proteins from telomeres is the reason that shelterin mutants bypass RNAi for pericentric heterochromatin assembly, we expect that simply increasing the concentration of silencing proteins is sufficient to bypass RNAi. As expected, introducing the OE-swi6+ transgene, which resulted in about fourfold excess Swi6 protein in the cells (Supplemental Fig. S5), was sufficient to rescue silencing defects and TBZ sensitivity of all RNAi mutants tested, such as dcr1Δ, ago1Δ, and rdp1Δ (Fig. 6D; Supplemental Fig. S8A). The OE-swi6+ transgene also complemented swi6Δ but could not rescue the silencing phenotype and TBZ sensitivity of clr4Δ (Supplemental Fig. S8A). Moreover, OE-swi6+ dcr1Δ clr3Δ and OE-swi6+ dcr1Δ sir2Δ failed to maintain pericentric silencing (Supplemental Fig. S4), suggesting that the ability of Swi6 overexpression to rescue RNAi defects also depends on RNAi-independent heterochromatin assembly pathways at pericentric regions. We obtained similar results with cells containing one additional copy of Swi6 inserted at the ars1 locus driven under the endogenous swi6 promoter, which is expected to increase Swi6 levels by twofold (Supplemental Fig. S8B; Sadaie et al. 2008).
We also tested whether overexpression of other heterochromatin factors could also bypass RNAi. Chp2 is another HP1 homolog and is enriched at all heterochromatin domains (Thon and Verhein-Hansen 2000; Sadaie et al. 2004; Motamedi et al. 2008; Fischer et al. 2009). However, incorporating an additional copy of chp2+ under the control of its endogenous promoter into the ars1 locus (Sadaie et al. 2008) could not rescue dcr1Δ (Supplemental Fig. S8B). Moreover, introducing Chp2 on a multicopy plasmid driven under an inducible nmt1 promoter could not rescue dcr1Δ either (Supplemental Fig. S8C). Similar results were obtained with overexpression of Clr4 or Clr3 (Supplemental Fig. S8C). However, we caution that both Clr4 and Clr3 are present in multisubunit complexes, so their overexpression might not result in significant up-regulation of their enzymatic activities in vivo. Nonetheless, the different results obtained with HP1 homologs suggest that Swi6, but not Chp2, plays a major role in balancing the strength of different heterochromatin domains.
Discussion
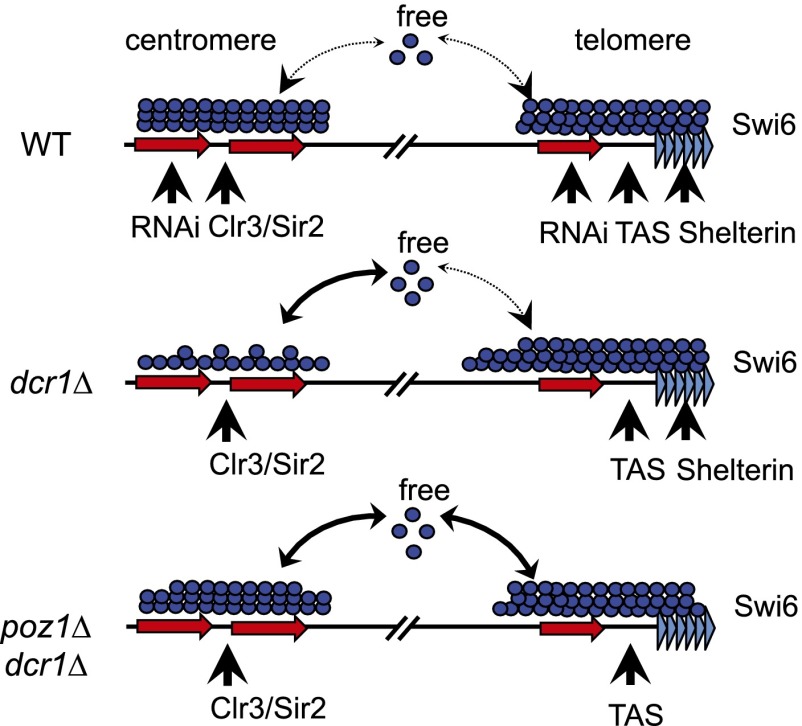
RNAi plays essential roles in heterochromatin assembly in diverse organisms. In fission yeast, loss of RNAi selectively impairs heterochromatin assembly at pericentric regions. We found that elimination of telomere shelterin components bypassed the requirement of RNAi for pericentric heterochromatin assembly. The separation-of-function mutant of Poz1 allowed us to demonstrate that loss of shelterin and RNAi results in the release from telomeres of silencing factors such as Swi6, which are then competitively recruited to pericentric regions through RNAi-independent heterochromatin assembly pathways (Fig. 7). In wild-type cells, the histone deacetylase Clr3/Sir2 functions cooperatively with RNAi to regulate pericentric heterochromatin assembly, whereas shelterin and TAS sequences function together with RNAi for telomeric heterochromatin assembly. These parallel pathways form high-affinity platforms for the recruitment of heterochromatin protein Swi6. In dcr1Δ cells, pericentric heterochromatin is severely compromised, and the Clr3/Sir2 pathway recruits Swi6 at a lower efficiency. The relatively higher affinity of telomeric heterochromatin, which is due to the presence of shelterin and TAS-mediated heterochromatin assembly pathways, absorbs the extra Swi6, resulting in higher Swi6 levels at telomeres and the spreading of Swi6 into euchromatin at subtelomeric regions. In poz1Δ dcr1Δ cells, telomeric heterochromatin assembly pathways are further compromised, and the Clr3/Sir2 pathway at pericentric regions is able to compete with TAS sequences for Swi6. Increased Swi6 might facilitate the recruitment of CLRC and the restoration of H3K9me at pericentric heterochromatin. The subtelomere region's lower affinity for Swi6 in poz1Δ dcr1Δ cells results in a reduction of Swi6 levels and heterochromatin spreading compared with dcr1Δ cells. We suspect that the ability of pot1Δ and tpz1Δ to bypass dcr1Δ is due to the loss of telomere and subtelomere sequences, which consequently affects silencing at telomeres. Such a model is consistent with recent imaging and biochemical studies showing that HP1/Swi6 proteins are highly mobile on chromatin (Cheutin et al. 2003, 2004; Festenstein et al. 2003; Sadaie et al. 2008).
Figure 7.
A model for shelterin and RNAi in regulating heterochromatin assembly.
Our results that increasing Swi6 levels bypasses RNAi for pericentric heterochromatin assembly suggest that the availability of heterochromatin proteins in the nucleus, such as Swi6, is limiting. Sustaining low Swi6 availability is essential for maintaining the epigenetic landscape of the genome, as higher levels of Swi6 have been associated with increased efficiency of heterochromatin assembly at endogenous and ectopic sites. For example, artificial targeting of Clr4 to DNA fails to induce heterochromatin assembly unless Swi6 is overexpressed (Supplemental Fig. S5B) or endogenous heterochromatin structures are compromised to release silencing proteins (Kagansky et al. 2009). In addition, exogenously introduced siRNAs fail to establish heterochromatin unless Swi6 is overexpressed (Iida et al. 2008). Moreover, overexpression of Swi6 increases the conversion rate of a less stable heterochromatin domain at the mating type region (Nakayama et al. 2000) and enhances the spreading of heterochromatin when chromatin boundaries are compromised (Noma et al. 2001; Wang et al. 2013). Thus, the multiple pathways that regulate heterochromatin assembly at constitutive heterochromatin regions, such as telomeric, pericentric, and the silent mating type regions, might be key to the formation of higher-affinity platforms for heterochromatin proteins, thus balancing the need for proper heterochromatin assembly at these regions and preventing heterochromatin assembly at cryptic sites. When any of these mechanisms are compromised, Swi6 redistributes according to the strength of heterochromatin assembly pathways at different regions and is more likely to be recruited to sites that otherwise cannot form heterochromatin.
Heterochromatin is mainly present at pericentric and subtelomeric regions in diverse organisms. Although the role of pericentric heterochromatin in regulating chromosome segregation has been well studied, the functions of telomeric heterochromatin are less clearly defined. In certain organisms such as Drosophila, telomeres are composed of retrotransposons instead of telomeric repeats, which are packaged into heterochromatin to recruit end protection factors (Mason et al. 2008). In fission yeast, amplification of subtelomere heterochromatin is also associated with cell survival in the absence of telomerase (Jain et al. 2010). Certain heterochromatin factors, such as Clr4 and Rik1, also regulate telomere clustering during meiosis (Tuzon et al. 2004). However, heterochromatin factors have no effect on telomere length control and do not protect against telomere end fusions (Tuzon et al. 2004). Compared with other heterochromatin regions, the telomere and subtelomere regions are unique in buffering the changes in heterochromatin levels because there are no defined heterochromatin boundaries, and a large transition zone is formed, with Swi6 levels gradually declining (Fig. 6A). In contrast, at centromeres and the silent mating type region, silencing factor concentrations are high, and well-defined heterochromatin boundaries prevent spreading (Cam et al. 2005). Thus, these regions might already be saturated by silencing factors, and the buffering capacity is limited. This is exemplified in dcr1Δ, in which Swi6 protein levels were significantly increased across the entire subtelomeric region, with significant amounts of spreading into euchromatic regions, whereas in poz1Δ cells, there were little changes of Swi6 levels at pericentric regions but enhanced heterochromatin assembly by GBD-Clr4 (Fig. 6A; Supplemental S5B). Conversely, cells containing the minichromosome Ch10 (Niwa et al. 1989), which is mainly composed of pericentric repeats and centromere and telomere sequences, show reduced levels of Swi6 selectively at subtelomeric regions (Chikashige et al. 2007). It is worth noting that the budding yeast, which uses Sir2/3/4 proteins to assemble heterochromatin instead of the H3K9me-HP1 system, also uses telomeres to limit silencing factors, and releasing these factors from telomeres similarly increases silencing at internal sites (Maillet et al. 1996; Marcand et al. 1996; Taddei et al. 2009). Thus, the sequestration of silencing factors through constitutive heterochromatin domains might be especially important for genomes that are highly active to prevent ectopic heterochromatin assembly in order to maintain the epigenetic landscape.
Materials and methods
Fission yeast strains and genetic analyses
Yeast strains containing poz1Δ, rap1Δ, and trt1Δ were derived from the Bioneer fission yeast deletion library, verified via PCR, and backcrossed. The strain containing OE-swi6+ was constructed by integrating a PCR fragment containing the ade6 promoter, swi6 coding region, and 3′ untranslated region and a hphMX6 cassette into the endogenous ade6+ locus. Poz1 mutants were constructed by integrating a PCR fragment containing the mutations, a myc tag, and a KanMX6 cassette into the endogenous poz1+ locus. Rap1-Flag and taz1Δ were constructed by a PCR-based module method. Genetic crosses were used to construct all other strains. Strains containing tpz1Δ or pot1Δ were first generated in diploid cells, followed by tetrad dissection to obtain desired genotypes. For serial dilution plating assays, 10-fold dilutions of a log-phase culture were plated on the indicated medium and grown for 3 d at 30°C.
Screens for mutations that suppress the silencing defects of RNAi mutants
Query strains were mated with the fission yeast deletion library with the aid of the Singer RoToR HDA pinning robot as previously described (Roguev et al. 2007). The fission yeast deletion library was constructed with a KanMX4 cassette that confers resistance to geneticin (Bioneer). Two reporter strains (otr∷ura4+ or otr∷ade6+) were constructed by inserting a NatMX6 cassette, which confers resistance to nourseothricin, ∼400 base pairs (bp) to the right side boundary of centromere I heterochromatin. The dcr1Δ and ago1Δ query strains were constructed with an hphMX6 cassette, conferring resistance to hygromycin. The otr∷ura4+ or otr∷ade6+ reporter and dcr1Δ were sequentially introduced into the deletion library, and the resulting haploid cells were selected and pinned to selective medium to measure cell growth or colony color.
ChIP analysis
ChIP analyses were performed as described previously (Hou et al. 2010). Antibodies used were H3K9me2 (Abcam), Swi6 (Reddy et al. 2011), Pol II (Covance), myc (Covance), and Flag (Sigma). Quantitative real-time PCR (qPCR) was performed with Maxima SYBR Green qPCR master mix (Fermentas) in an ABI 7300 Real-Time PCR System. DNA serial dilutions were used as templates to generate a standard curve of amplification for each pair of primers, and the relative concentration of target sequence was calculated accordingly. An act1 fragment was used as a reference to calculate the enrichment of ChIP over whole-cell extract for each target sequence. ChIP–chip analysis was performed according to the Agilent Yeast ChIP-on-chip analysis protocol. The microarray used was an Agilent S. pombe Whole-Genome ChIP-on-chip microarray (G4810A) with additional probes that encompass centromeres, which were originally absent from the array due to the repetitive nature of these DNA sequences.
RNA analyses
Total cellular RNA was isolated from log-phase cells using a MasterPure Yeast RNA purification kit (Epicentre) according to the manufacturer's protocol. Quantification with real-time RT–PCR was performed with Power SYBR Green RNA-to-CT One-Step kit (Applied Biosystems). RNA serial dilutions were used as template to generate the standard curve of amplification for each pair of primers, and the relative concentration of target sequence was calculated accordingly. An act1 fragment served as a reference to normalize the concentration of samples. The concentration of each target gene in wild type was arbitrarily set to 1 and served as reference for other samples. Northern blot of siRNAs was performed as described previously (Reddy et al. 2011).
Telomere length analysis
For poz1 mutant and poz1Δ strains, a single colony from the first restreak after the mutation or deletion was grown overnight. Genomic DNA was isolated, digested with EcoRI, and separated on a 1% agarose gel. Hybridization with radiolabeled telomeric DNA and pol1 (control) probes was performed as previously described (Jun et al. 2013).
Acknowledgments
We thank Junichi Nakayama, Robin Allshire, Michael Keogh, and Toru Nakamura for yeast strains, plasmids, and antibodies, and Allison Cohen for technical assistance. This work was supported by National Institutes of Health (NIH) grants R01-GM085145 (to S.J.) and R01-GM098943 to (F.Q.). X.T. was supported by the Fulbright Scholar Program. B.R and S.P.K. were supported by NIH training grant T32-GM008798. F.Q. was supported by a Basil O'Connor Starter Scholar Research Award from March of Dimes and a Beginning Grant-in-aid from the American Heart Association.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226118.113.
References
- Almouzni G, Probst AV 2011. Heterochromatin maintenance and establishment: Lessons from the mouse pericentromere. Nucleus 2: 332–338 [DOI] [PubMed] [Google Scholar]
- Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF 2013. Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. EMBO J 32: 2321–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC 2013. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J 32: 1250–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA 2013. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- Cheutin T, Gorski SA, May KM, Singh PB, Misteli T 2004. In vivo dynamics of Swi6 in yeast: Evidence for a stochastic model of heterochromatin. Mol Cell Biol 24: 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Okamasa K, Yamane M, Nakayama J, Niwa O, Haraguchi T, Hiraoka Y 2007. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct Funct 32: 149–161 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299: 719–721 [DOI] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI 2009. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci 106: 8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto DB, Nakayama JI 2012. RNA and epigenetic silencing: Insight from fission yeast. Dev Growth Differ 54: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci 100: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR 3rd, Jia S 2010. Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J Biol Chem 285: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Nakayama J, Moazed D 2008. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell 31: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP 2010. Telomeric strategies: Means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227 [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Jun HI, Liu J, Jeong H, Kim JK, Qiao F 2013. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via Ccq1. Genes Dev 27: 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC 2009. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F 2005. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819 [DOI] [PubMed] [Google Scholar]
- Kim HS, Choi ES, Shin JA, Jang YK, Park SD 2004. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem 279: 42850–42859 [DOI] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC 2011. Common ground: Small RNA programming and chromatin modifications. Curr Opin Cell Biol 23: 258–265 [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM 1996. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Marcand S, Buck SW, Moretti P, Gilson E, Shore D 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev 10: 1297–1309 [DOI] [PubMed] [Google Scholar]
- Marina DB, Shankar S, Natarajan P, Finn KJ, Madhani HD 2013. A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev 27: 1851–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Frydrychova RC, Biessmann H 2008. Drosophila telomeres: An exception providing new insights. Bioessays 30: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F 2008. Fission yeast Pot1–Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM 2009. Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet 5: e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101: 307–317 [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J 13: 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828 [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M 1989. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J 8: 3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293: 1150–1155 [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4: 89–93 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bahler J, Smith GR, Jia S 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev 25: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI 2011. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18: 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ 2007. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods 4: 861–866 [DOI] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J 2008. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol 28: 6973–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka-Sugiyama R, Sugiyama T 2011. Sde2: A novel nuclear protein essential for telomeric silencing and genomic stability in Schizosaccharomyces pombe. Biochem Biophys Res Commun 406: 444–448 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM 2009. The functional importance of telomere clustering: Global changes in gene expression result from SIR factor dispersion. Genome Res 19: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155: 551–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Cooper JP 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC 2007. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J 26: 4670–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Borgstrom B, Weilguny D, Egel R, Cooper JP, Nielsen O 2004. The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J Cell Biol 165: 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146 [DOI] [PubMed] [Google Scholar]
- Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez-Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, et al. 2012. Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J Biol Chem 287: 4386–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tadeo X, Hou H, Tu PG, Thompson J, Yates JR 3rd, Jia S 2013. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev 27: 1886–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y 2008. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455: 251–255 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]