Abstract
Little is known about the genes regulating disease severity and joint damage in rheumatoid arthritis (RA). In the present study we analyzed the gene expression characteristics of synovial tissues from four different strains congenic for non-MHC loci that develop mild and nonerosive arthritis compared with severe and erosive DA rats. DA.F344(Cia3d), DA.F344(Cia5a), DA.ACI(Cia10), and DA.ACI(Cia25) rats developed mild arthritis compared with DA. We found 685 genes with significantly different expression between congenics and DA, independent of the specific congenic interval, suggesting that these genes represent a new nongenetic core group of mediators of arthritis severity. This core group includes genes not previously implicated or with unclear role in arthritis severity, such as Tnn, Clec4m, and Spond1 among others, increased in DA. The core genes also included Scd1, Selenbp1, and Slc7a10, increased in congenics. Genes implicated in nuclear receptor activity, xenobiotic and lipid metabolism were also increased in the congenics, correlating with protection. Several disease mediators were among the core genes reduced in congenics, including IL-6, IL-17, and Ccl2. Analyses of upstream regulators (genes, pathways, or chemicals) suggested reduced activation of Stat3 and TLR-related genes and chemicals in congenics. Additionally, cigarette smoking was among the upstream regulators activated in DA, while p53 was an upstream regulator activated in congenics. We observed congenic-specific differential expression and detection in each individual strain. In conclusion, this new nongenetically regulated core genes of disease severity or protection in arthritis should provide new insight into critical pathways and potential new environmental risk factor for arthritis.
Keywords: autoimmunity, synovitis, erosion, environment, innate
rheumatoid arthritis (ra) affects 0.5–1% of the population. New insights about the inflammatory process in the joint and disease pathogenesis have led to the development of new treatments during the past decade with improved disease control (20, 37, 46). However, long-term remission is still rarely achieved (48), highlighting the need for better treatments.
Disease severity strongly correlates with joint damage and mortality in RA (36). Yet, little is known about the genes involved in the regulation of these disease characteristics, and there has been no genome-wide genetic association study looking at RA disease severity or joint damage. We have identified several quantitative trait loci (QTL) regulating the severity of rodent models of RA (4, 7, 12, 23, 24, 26). Cia3d, Cia5a, Cia10, and Cia25 are disease severity QTL that regulate clinical severity, pannus formation, and bone and cartilage erosions in collagen- and pristane-induced arthritis (CIA and PIA) (4, 6, 7, 13, 17, 33).
We have previously generated congenic strains where a chromosomal segment derived from an arthritis-resistant strain (ACI or F344) was introgressed into the genome of the arthritis-susceptible strain DA (2, 4, 6, 7). Each single locus congenic strain was significantly protected developing mild and nonerosive disease. The magnitude of the disease protection was similar in most congenic strains, suggesting that the arthritis genes located in each interval regulate different but similarly critical processes for disease pathogenesis. Therefore, identifying these genes and understanding the cellular and molecular processes regulated by them have the potential to generate new and perhaps better targets for therapy and prognostic biomarkers.
In the present study we used synovial tissues from four different congenic strains and DA rats from two different sources to identify core arthritis severity genes that have similarly differential expression in all congenics independently of their genetic differences (the congenic interval) compared with DA, as well as genes uniquely regulated by the arthritis genes located within the QTL interval. Our results also demonstrate that each congenic operates as a phenocopy where the clinical and histologic disease severity is similarly suppressed, with consequences of genetic differences detected only at the molecular level. These observations raise the possibility of a similar scenario occurring in RA and other complex traits regulated by several genes, where patients develop a similar disease syndrome, but different genes may have a more central role in one case but not in the other, depending on the genetic make-up of that individual. Our results also point to genes, gene pathways, and chemicals that had not been previously considered to have a role in arthritis pathogenesis.
METHODS
Animals
Arthritis-susceptible DA rats from two different origins were studied. DA/BklArbNsi (DA/Arb) rats were originally purchased from Bantin & Kingman (Fremont, CA), maintained at Arthritis and Rheumatism Branch, National Institute of Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, and then transferred to the Feinstein Institute for Medical Research (FIMR, formerly North Shore-LIJ Research Institute). DA/Hsd rats were purchased from Harlan (Indianapolis, IN).
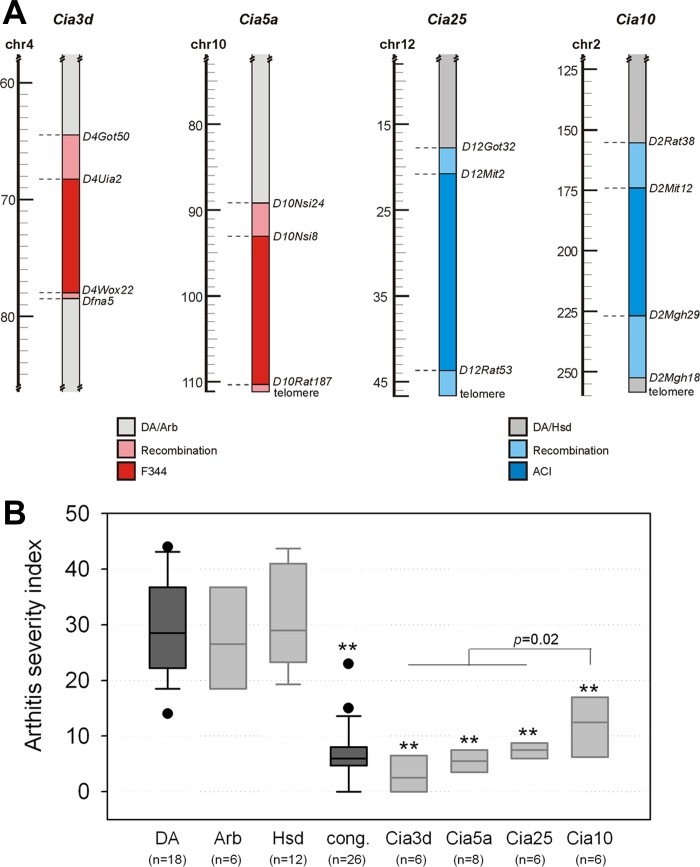
All four arthritis-protected congenic strains shared the DA genetic background, including the MHC region and differed only at their nonoverlapping congenic loci. DA.F344(Cia3d) (referred to as Cia3d) (4) and DA.F344(Cia5a) (referred to as Cia5a) (7) congenic rats have a DA/Arb genetic background and F344 alleles at Cia3d on chromosomes 4 and Cia5a on chromosome 10. DA.ACI(Cia10) (6) and DA.ACI(Cia25) (2) (Cia10 and Cia25 congenics, respectively) congenic rats have a DA/Hsd background and ACI alleles at Cia10 on chromosomes 2 and Cia25 on chromosome 12 (Fig. 1A). All congenics were homozygous for the respective resistant alleles.
Fig. 1.
Congenic intervals containing arthritis severity quantitative trait loci (QTL) and their effect on arthritis severity. A: Cia3d and Cia5a congenics have a DA/Arb background and F344 alleles only on segments of chromosomes 4 and 10, respectively. Cia25 and Cia10 congenics have a DA/Hsd background and ACI alleles only on segments of chromosomes 12 and 2, respectively. B: at day 21 postarthritis induction DA/Arb and DA/Hsd rats developed similarly severe arthritis, while all 4 congenic strains were protected and had a significantly milder form of arthritis. **P ≤0.001 for each congenic vs. DA, Mann-Whitney.
All experiments involving animals were conducted with 8–12 wk old male rats under a protocol approved by the FIMR Institutional Animal Care and Use Committee.
PIA
Male DA and congenic rats were anesthetized and injected intradermally with 150 μl of pristane (2,6,10,14-tetramethylpentadecane; MP Bio, Solon, OH) divided into two injection sites at the base of the tail (day 0) (6, 44). Arthritis severity was assessed according to a previously described 80-point scoring system on day 21 postinduction of arthritis (6), followed by euthanasia and synovial tissue collection from the ankle joints.
RNA Preparation and Microarray Experiments
Synovial tissue total RNA was extracted with the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, including a DNase treatment step. RNA was quantified and assessed for purity with a NanoDrop spectrophotometer (Rockland, DE). RNA integrity was verified on a BioAnalyzer 2100 (Agilent, Palo Alto, CA).
Microarray
All reagents and procedures were previously optimized for use with the Illumina Whole-Genome Expression platform. Total RNA (200 ng) was amplified and biotinylated with the TotalPrep labeling kit (Ambion, Austin, TX). Samples were hybridized to RatRef-12 Expression BeadChips (Illumina, San Diego, CA), which contain 22,522 probes covering 21,922 rat genes selected primarily from the National Center for Biotechnology Information (NCBI) RefSeq database (release 16). Hybridization was carried out in Illumina IntelliHyb chambers, followed by washing and staining with Cy3-streptavidin. Each BeadChip was scanned on a high-resolution Illumina BeadArray reader with a two-channel 0.8 μm resolution confocal laser scanner.
Microarray Data Analyses
Fluorescence intensities were extracted with GenomeStudio 2.0 (Illumina). To correct for systematic biases of nonbiological origin, we normalized background-subtracted intensities of all probes using the cubic spline algorithm followed by log2-transformation. Probes consistently detected in all arrays (detection P ≤ 0.05 for all IDs) were used in quantitative comparative analyses. A ≥2-fold difference in intensity between groups and a t-test P ≤ 0.01 were considered significantly differentially expressed. Probes detected in >80% of the samples in one strain and in <20% of the samples of another were considered differentially detected and were compared with the Fisher's exact test. Probes differentially expressed or differentially detected between DA and each congenic strain were also compared between individual congenic strains to determine differences in expression and detection uniquely regulated by each QTL. Differentially expressed and detected probes were then used for pathway detection analyses with IPA 2013 (Ingenuity Systems, Redwood City, CA), plus searches in online public databases such as Genecards, Oncomine, BioGPS, Ensembl, and PubMed-based literature search. We excluded 3,580 probes from the analysis because they targeted pseudogenes or genes that had been withdrawn from NCBI/Rat Genome Database.
Quantitative PCR Expression Studies
Differences in the expression of selected genes were validated with quantitative (q)PCR. The qPCR conditions used have been described elsewhere (23). Briefly, total RNA (200 ng) from each sample was reverse transcribed using Superscript III (Invitrogen). Primers and qPCR probes were designed to target the same exons as the corresponding Illumina RatRef-12 Expression BeadChip probes and have been previously reported (5, 16). We used Universal ProbeLibrary (Roche, Indianapolis, IN) and Taqman (ABI, Applied Biosystems, Foster City, CA) probes labeled with FAM at the 5′-end and TAMRA at 3′-end. Reactions were prepared in duplicates with Absolute Blue qPCR Mix (Thermo Fisher, Waltham, MA), run on a LightCycler 480 thermocycler (Roche) using Relative Quantitative Software (Roche). Ct (threshold cycle) values were adjusted for GAPDH in each sample (ΔCt). Fold differences were calculated with the 2−ΔΔCt method (29). The same RNA samples used in the microarray experiments were used for qPCR validation.
Statistics
Differences in arthritis severity scores were calculated with the Mann-Whitney Test and ANOVA on ranks (nonnormally distributed data). Two-tailed t-test was used to compare log2-transformed microarray fluorescent intensities and Ct values, and a nominal cutoff P value of ≤0.01. Enrichment for differentially expressed genes within a chromosomal location was calculated using the χ2-test.
RESULTS
Rats Congenic for Cia3d, Cia5a, Cia25, and Cia10 Develop a Significantly Milder Form of Arthritis
Male DA rats developed severe arthritis by day 21 post-PIA, while males congenic for the Cia3d, Cia5a, Cia10, and Cia25 arthritis severity QTL were protected and had a significantly milder form of arthritis (median ASI, DA = 28.5, congenics = 6.0; P ≤ 0.001, Mann-Whitney; Fig. 1B). DA/Arb and DA/Hsd rats had similarly severe arthritis (median ASI; DA/Arb = 26.5, DA/Hsd = 29.0). Compared with DA, each individual congenic strain had significantly less severe arthritis (median ASI, DA = 28.5, Cia3d = 2.5, Cia5a = 5.5, Cia25 = 7.5, Cia10 = 12.5; P ≤0.001 for each congenic vs. DA; Mann-Whitney). Cia10 congenics were slightly less protected than Cia3d, Cia5a, and Cia25 congenics (P = 0.02, Mann-Whitney).
Differences in Gene Expression Between DA and Congenic Rats Reveal the Synovial Transcriptomes of Severe and Mild Forms of Arthritis
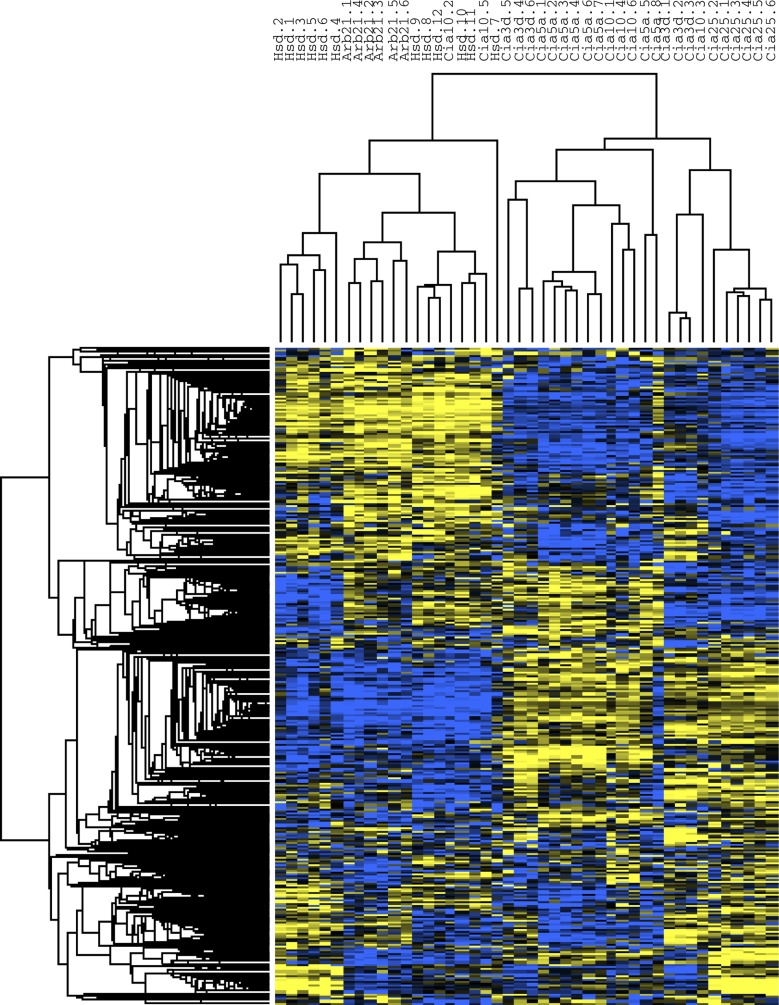
Of the 6,785 transcripts, 691, accounting for 685 of the 6,791 genes (∼10%) expressed in all synovial samples, were differentially expressed between DA (n = 18) and all four congenic strains (n = 26); 340 genes had increased expression in DA, and 345 genes were increased in the congenic strains (Supplemental Table S1, Fig. 2).1 DA rats (DA/Arb and DA/Hsd combined) clustered together, while 24 of the 26 congenics clustered in a separate group (Fig. 3).
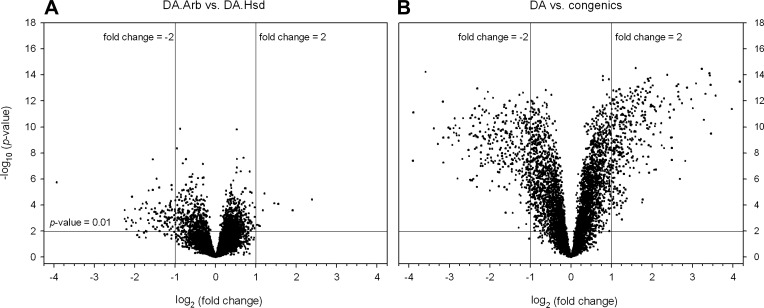
Fig. 2.
Volcano plot distribution of the microarray data. A: volcano plot distribution of the DA/Arb vs. DA/Hsd log-transformed data revealing a high degree of similarity with only a small number of differentially expressed genes, most of which had low fold-differences. B: combined DA (DA/Arb+DA/Hsd) vs. the combined congenics data reveals high number of differentially expressed genes including >2-fold differences.
Fig. 3.
Cluster analysis heat map. DA/Arb and DA/Hsd cluster together, while most of the congenic and subcogenic protected rats clustered as a separate group. The heat map shows in graphical form the significant differences between DA and congenics. Yellow, increased expression; blue, decreased expression.
This group of genes was differentially expressed in synovial tissues of the protected congenics strains compared with DA, independent of the specific congenic interval. Therefore, these observations suggest that these genes represent a central core group of mediators of arthritis severity independently of the genetic background. These genes are of potential great interest for further studies and for drug targeting.
Among these central core genes are those with the highest expression in DA compared with the combined congenics such Tnn (24.59-fold), Ccnb2 (18-fold), Slpi (15.67-fold), Cxcl1 (11-fold), Ccl7 (9.7-fold), Lbp (7.84-fold), and Mmp3 (11.88-fold) (Table 1). This central core of genes also includes those with the highest expression in congenics compared with DA such as Scd1 (14.93-fold), Akr1c19 (14.83-fold), Nnat (12.04-fold), Ces1d (10.41-fold), and Cidea (7.11-fold) (Table 2). Several genes within these central core genes such as Tnn, Lbp, Emb, Steap1, Mpz, Sult1a1, Slc2a4, Slc36a2, and Selenbp1 have a previously unknown involvement in arthritis and might become the focus of future studies.
Table 1.
Genes with the highest expression in DA relative to congenics
| Entrez | Symbol | Gene Name | Fold | t-Test P Value |
|---|---|---|---|---|
| 304913 | Tnn | tenascin N | 24.59 | 4.82 × 10−16 |
| 363088 | Ccnb2 | cyclin B2 | 18.00 | 3.49 × 10−14 |
| 84386 | Slpi | secretory leukocyte peptidase inhibitor | 15.67 | 4.44 × 10−12 |
| 171045 | Mmp3 | matrix metallopeptidase 3 | 11.88 | 4.13 × 10−13 |
| 81503 | Cxcl1 | [chemokine (C-X-C motif) ligand 1] | 11.00 | 3.35 × 10−10 |
| 498709 | Cks2 | CDC28 protein kinase regulatory subunit 2 | 10.93 | 6.01 × 10−14 |
| 308761 | Prc1 | protein regulator of cytokinesis 1 | 10.78 | 1.15 × 10−14 |
| 282836 | Cthrc1 | collagen triple helix repeat containing 1 | 10.70 | 7.87 × 10−15 |
| 363856 | Clec4m | C-type lectin domain family 4, member M | 9.99 | 3.03 × 10−13 |
| 287561 | Ccl7 | chemokine (C-C motif) ligand 7 | 9.71 | 2.97 × 10−11 |
| 304951 | Nuf2 | [NUF2, NDC80 kinetochore complex component] | 9.38 | 3.58 × 10−15 |
| 298845 | Emilin1 | elastin microfibril interfacer 1 | 8.51 | 4.78 × 10−14 |
| 297738 | Steap1 | six transmembrane epithelial antigen of the prostate 1 | 8.06 | 3.8 × 10−12 |
| 29469 | Lbp | lipopolysaccharide binding protein | 7.84 | 7.78 × 10−13 |
| 114122 | Vcan | versican | 7.31 | 1.01 × 10−13 |
| 29527 | Ptgs2 | prostaglandin-endoperoxide synthase 2 | 7.06 | 4.22 × 10−08 |
| 89824 | Chi3l1 | chitinase 3-like 1 | 6.68 | 1.56 × 10−12 |
| 81687 | Mmp9 | matrix metallopeptidase 9 | 6.45 | 1.01 × 10−06 |
| 24232 | C3 | complement component 3 | 6.41 | 8.15 × 10−13 |
| 64515 | Cdc20 | cell division cycle 20 homolog (S. cerevisiae) | 6.28 | 2.08 × 10−13 |
| 114511 | Emb | embigin | 6.28 | 4.70 × 10−13 |
| 64456 | Spon1 | spondin 1, extracellular matrix protein | 6.28 | 5.49 × 10−14 |
| 299112 | Pole2 | polymerase (DNA directed), epsilon 2 (p59 subunit) | 6.11 | 7.52 × 10−14 |
| 25060 | Hk3 | hexokinase 3 (white cell) | 5.82 | 4.78 × 10−12 |
| 25694 | Has2 | hyaluronan synthase 2 | 5.62 | 3.56 × 10−08 |
| 24770 | Ccl2 | chemokine (C-C motif) ligand 2 | 5.62 | 4.27 × 10−09 |
| 24451 | Hmox1 | heme oxygenase (decycling) 1 | 5.62 | 6.58 × 10−10 |
| 24494 | Il1b | interleukin 1 beta | 5.54 | 6.75 × 10−10 |
| 502902 | Clec7a | C-type lectin domain family 7, member A | 5.46 | 1.94 × 10−12 |
| 687856 | LOC687856 | similar to myeloid cell surface antigen CD33 precursor (Siglec-3) | 5.43 | 1.99 × 10−11 |
| 301229 | Trem1 | triggering receptor expressed on myeloid cells 1 | 5.35 | 7.26 × 10−12 |
| 294286 | Kifc1 | kinesin family member C1 | 5.21 | 1.04 × 10−14 |
Table 2.
Genes with the highest expression in congenics relative to DA
| Entrez | Symbol | Gene Name | Fold | t-Test P Value |
|---|---|---|---|---|
| 246074 | Scd1 | stearoyl-coenzyme A desaturase 1 | 14.93 | 4.01 × 10−08 |
| 307096 | Akr1c19 | aldo-keto reductase family 1, member C19 | 14.83 | 7.94 × 10−12 |
| 94270 | Nnat | neuronatin | 12.04 | 6.03 × 10−15 |
| 113902 | Ces1d | carboxylesterase 1D | 10.41 | 1.34 × 10−10 |
| 84356 | Abcd2 | ATP-binding cassette, subfamily D (ALD), member 2 | 9.71 | 2.13 × 10−09 |
| 25742 | S100b | S100 calcium binding protein B | 8.94 | 1.17 × 10−12 |
| 24564 | Mpz | myelin protein zero | 8.88 | 5.58 × 10−08 |
| 689147 | LOC689147 | hypothetical protein LOC689147 | 8.88 | 6.71 × 10−10 |
| 362282 | Pck1 | phosphoenolpyruvate carboxykinase 1 (soluble) | 8.11 | 6.76 × 10−10 |
| 25360 | Tshr | thyroid stimulating hormone receptor | 8.11 | 1.15 × 10−10 |
| 24212 | Atp1a2 | ATPase, Na+/K+-transporting, alpha 2 polypeptide | 7.89 | 1.44 × 10−10 |
| 25357 | Thrsp | thyroid hormone responsive | 7.62 | 2.5 × 10−09 |
| 83717 | Omd | osteomodulin | 7.46 | 1.31 × 10−10 |
| 296184 | Ankrd5 | ankyrin repeat domain 5 | 7.41 | 2.82 × 10−10 |
| 94199 | Dpep1 | dipeptidase 1 (renal) | 7.31 | 2.33 × 10−11 |
| 288271 | Mrap | melanocortin 2 receptor accessory protein | 7.21 | 8.04 × 10−11 |
| 291541 | Cidea | cell death-inducing DFFA-like effector a | 7.11 | 3.27 × 10−10 |
| 246235 | Slc36a2 | solute carrier family 36 (proton/amino acid symporter), member 2 | 7.06 | 4.69 × 10−10 |
| 25629 | Plin1 | perilipin 1 | 6.82 | 1.18 × 10−09 |
| 81670 | Gpt | glutamic-pyruvate transaminase (alanine aminotransferase) | 6.77 | 4.69 × 10−10 |
| 25139 | Slc2a4 | solute carrier family 2 (facilitated glucose transporter), member 4 | 6.73 | 1.27 × 10−10 |
| 24179 | Agt | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 6.41 | 2.93 × 10−08 |
| 83783 | Sult1a1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 | 6.32 | 2.22 × 10−11 |
| 64471 | Plekhb1 | [pleckstrin homology domain containing, family B member 1] | 5.78 | 6 × 10−09 |
| 114518 | Slc7a10 | [solute carrier family 7, member 10] | 5.70 | 1.85 × 10−08 |
| 246253 | Adipoq | adiponectin, C1Q and collagen domain containing | 5.66 | 3.88 × 10−10 |
| 259246 | LOC259246 | alpha-2u globulin PGCL1 | 5.62 | 1.18 × 10−06 |
| 81523 | Myoc | myocilin | 5.58 | 2.1 × 10−11 |
| 298107 | Mup5 | major urinary protein 5 | 5.54 | 1.34 × 10−06 |
| 140927 | Selenbp1 | selenium binding protein 1 | 5.50 | 3.3 × 10−10 |
| 252900 | Dgat2 | diacylglycerol O-acyltransferase 2 | 5.43 | 1.33 × 10−11 |
| 259244 | LOC259244 | alpha-2u globulin PGCL3 | 5.43 | 1.28 × 10−06 |
| 29533 | Pxmp2 | peroxisomal membrane protein 2 | 5.31 | 8.81 × 10−11 |
| 362527 | Mup4 | major urinary protein 4 | 5.31 | 1 × 10−06 |
| 364395 | C1qtnf9 | C1q and tumor necrosis factor related protein 9 | 5.28 | 5.26 × 10−10 |
| 25330 | Lipe | lipase, hormone sensitive | 5.17 | 3.87 × 10−12 |
| 312495 | Cyp26b1 | cytochrome P450, family 26, subfamily b, polypeptide 1 | 5.17 | 5.02 × 10−09 |
| 310625 | Rhbg | Rh family, B glycoprotein | 5.13 | 1.91 × 10−11 |
| 291018 | Ecm2 | extracellular matrix protein 2, female organ and adipocyte specific | 5.13 | 4.36 × 10−10 |
| 365345 | Cyb5r2 | cytochrome b5 reductase 2 | 5.03 | 5.44 × 10−10 |
| 287167 | LOC287167 | globin, alpha | 5.03 | 3.9 × 10−09 |
The genes expressed in increased levels in DA suggest a likely proinflammatory, proproliferative, and invasive role and potential targets for antagonistic therapies. The genes expressed in increased levels in the congenics suggest protective pathways such as those regulated by nuclear receptors and increased anabolic metabolism and potential targets for agonistic and pathway activating agents. Genes expressed in increased levels in DA include several key arthritis mediators such as cytokines [IL-1β, IL-18, lymphotoxin-β, lipopolysaccharide-induced TNF factor (Litaf)], chemokines and their receptors (Ccl2, Ccl7, Cxcl1, Ccr5), and class II MHC molecules (HLA-DMA, -DMB, -DQA1, -DRA, -DRB1) (Table 3). DA also had increased levels of enzymes implicated in the formation of reactive oxygen species (subunits of NADPH oxidase and superoxide dismutase), as well as damage-associated and pattern recognition receptors (DAMPs and PRRs, respectively) (Table 3). Additionally, DA synovial tissues expressed increased levels of adhesion molecules, proteases, collagens, and other components of the extracellular matrix (ECM), enzymes involved in the synthesis of ECM components, and numerous genes involved in cell cycle regulation and cell proliferation (Table 3).
Table 3.
Gene functional categories expressed in increased levels in synovial tissues from DA or in the combined congenic rats
| Category | Genes |
|---|---|
| DA | |
| Cytokines and related genes | Il1β, Il6*, Il18, Ltb, Il13ra2, Il17a*, Il17ra, Il4ra, Litaf, Jak2 |
| Chemokines and receptors | Ccl2, Ccl7, Cxcl1, Cxcl13, Ccr5 |
| Class II MHC | CD74, HLA-DMA, HLA-DMB, HLA-DQA1, HLA-DRA, HLA-DRB1 |
| Phagocytosis | Fcer1 g, Fcgr1a, Fcgr3a, Fcrls |
| Oxidative metabolism | Ncf1, Ncf4, Sod2 |
| DAMPs and PRRs | Cd14, Cd68, Cd163, Tlr2, Clec7a, Clec4a1, Clec4a3, Clec10a, Mrc1, Pycard |
| Adhesion molecules | CD44, Clec4m, Itgam, Itgb2, Itgb7, Lman1, Selp |
| Proteases | Adamts1, Mmp3, Mmp9, Mmp14, Plau |
| ECM components | Col1a1, Col3a1, Col5a1, Col5a2, Col11a1, Col16a1, Emilin1, Tnn, Vcan |
| ECM synthesis | Has1, Has2, Lox, Loxl3, Pcolce, Plod2 |
| Cell cycle | Ccnb2, Cks2, Cdc20, Cenpk, Cdca8, Chek2, Fos |
| Congenic strains | |
| Detoxification | Adh1, Cyb561, Cyb5r2, Cyb5r3, Cyp2j3, Cyp26b1, Cyp27a1, Mt3, Phyh, Tst |
| Lipid metabolism | Adipoq, Dgat1, Dgat2, Lipe, Lpin1, Lpl, Mgll, Plin1, Scd1 |
| Oxidative metabolism | Cat, Gstk1, Gstm1, Gstm2, Gstm7, Gstp1, Gstt1, Gstt2 |
Results from quantitative (q)PCR. DAMPs, damage-associated molecular patterns; PRRs, pattern recognition receptors; ECM, extracellular matrix.
Genes expressed in increased levels in the congenics include those involved in detoxification and xenobiotic processes (cytochrome P450; Cyp), genes involved in the synthesis of vitamin D (Cyp27a1), genes involved in lipid metabolism (Dgat1), and inhibitors of oxidation (Table 3).
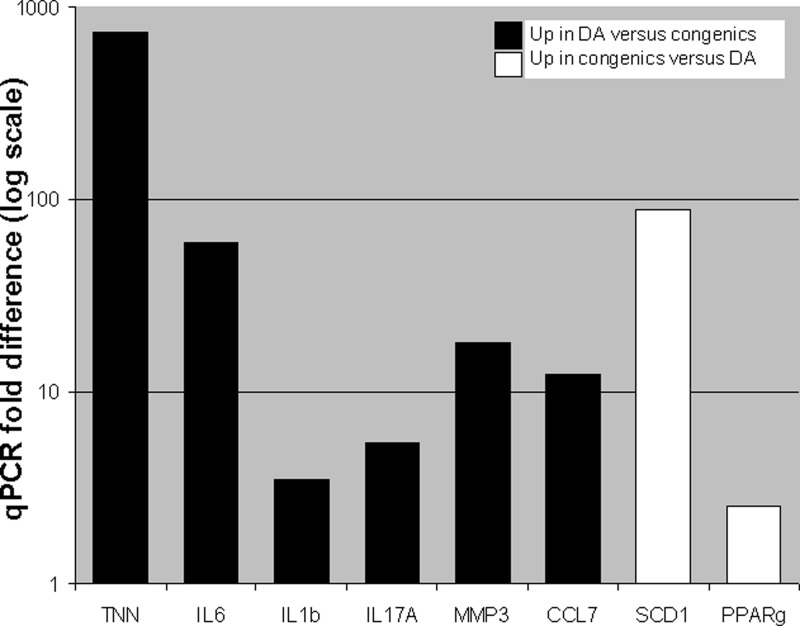
The expression levels of selected genes were confirmed and validated by qPCR (Fig. 4). Furthermore, since the Illumina rat microarray does not contain IL-17A and, in our experience, has inconsistent detection of IL-6, we also quantified these two key cytokines with qPCR and found them to be significantly increased in DA and reciprocally decreased in congenic with fold differences of 5.4 and 59, respectively (Fig. 4).
Fig. 4.
Quantitative (q)PCR confirmation and analyses of selected genes. qPCR confirmed the microarray increased expression of IL-1β, Tnn, Mmp3, and Ccl7 in DA compared with congenics. We used qPCR to analyze IL-17A and IL-6 since the first was not represented in the microarray and the second had variable detection. Both IL-17A and IL-6 were had significantly increased expression in DA and reciprocally reduced in congenics. Additionally, we use qPCR to confirm the increased expression of Scd1 and Pparg in congenics compared with DA. All P values ≤ 4 × 10−8, t-test.
Biological functions promoting inflammation and accumulation of immune cells were expressed in significantly increased levels or activated in DA rats compared with the congenic strains (Fig. 5, Supplemental Table S2). Processes involved in leukocyte proliferation and migration were prominently overrepresented among genes with increased expression in DA, as were functions related to activation of lymphocytes and myeloid cells, phagocytosis, and angiogenesis (Supplemental Table S2).
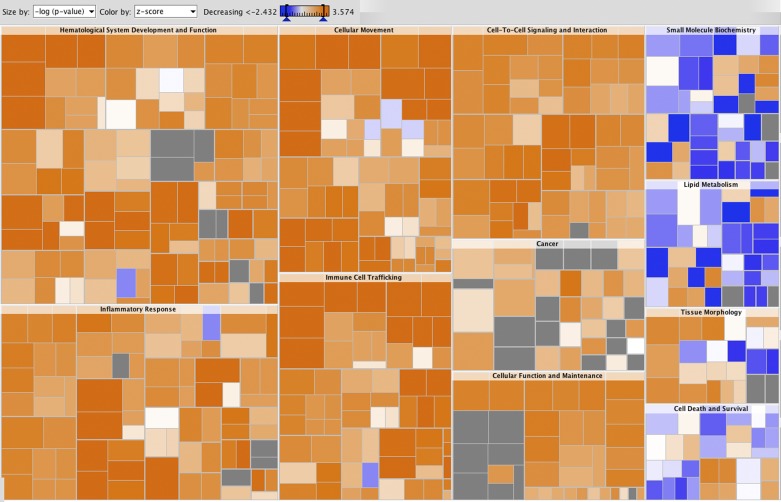
Fig. 5.
Heat map of biological functions activated in DA and in the congenic strains. Gene clusters suggesting biological functions to be activated in DA are marked in shades of orange and those to be activated in the congenics are marked in shades of blue. The square size in the figure is proportion to the −log(P value), and the colors represent z-scores. Color intensity refers to the z-score for enrichment of each functional subcategory. The highest z-scores were in the “cell movement” and “immune cell trafficking” (neutrophils, myeloid cells, phagocytes) and “inflammatory response.” The subcategory with the lowest z-score was “metabolism of triacylglycerol.”
In addition to the 691 genes that were differentially expressed, 124 genes were differentially detected. Specifically, 85 genes were detected in 80% or more of the DA samples but in only 20% or less of the congenic samples (Supplemental Table S3). Conversely, 39 genes were detected in 80% or more of the congenic samples but in 20% or less of the DA samples (Supplemental Table S3). Genes preferentially detected in DA were particularly enriched for proteins important for cell cycle progression (29/85 in DA, 1/39 in congenics) and inflammatory/immune responses (14/85 in DA, 0/39 in congenics). Prominent in the latter group were CD2, CD8b, Ciita, Cxcl5, Cx3cr1, Itk, and Zap70. Conversely, genes preferentially detected in the congenic strains were enriched for energy/lipid metabolism (6/39 in the congenics and 0/85 in DA).
Upstream Regulators of Gene Expression
The Ingenuity software has a function that looks for pathways, processes, and genes that might have increased activity based on expression levels of their downstream targets. Therefore, this analysis predicted increased or decreased activity of 1,161 upstream regulators with P ≤ 0.01. Many of those regulators overlap, like PPARG and rosiglitazone, and several genes were included in more than one upstream regulator group of genes.
DA.
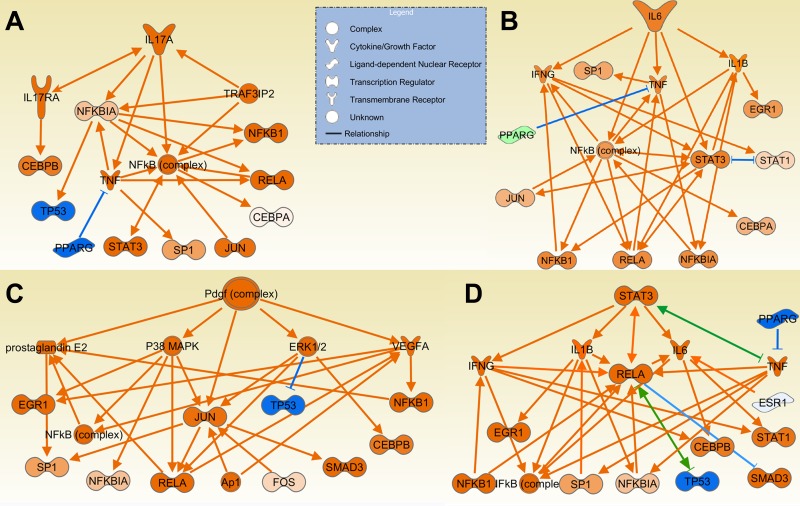
The top 20 upstream regulators predicted to be activated in DA and reciprocally inhibited in congenics include cytokines implicated in arthritis pathogenesis such as IL-1β, IL-6 (Fig. 6A), IFN-γ, PDGF (Fig. 6C), TGF-β, and TNF-α (Table 4, Supplemental Table S4). Stat3 (Fig. 6D), a key gene required for cytokine receptor signal transduction and Th17 differentiation, was also among the top 20 upstream genes in DA (Table 4, Supplemental Table S4). Other inflammatory and arthritis mediators predicted to have increased activity were BCR, C5, CXCL10, IL-2, IL-17 (Fig. 6B), IL-18, LEP, NOS, OSM, TNFSF11 (RANKL), TNFSF12 (TWEAK).
Fig. 6.
Upstream regulators with increased activity in DA and reduced activity in the congenics. IL-6 (A), IL-17A (B), PDGF (C), and STAT3 (D) were among the upstream regulators with the highest z-scores and P values, suggesting increased activity in DA and reduced activity in congenics. Orange lines with arrows indicate effect that increases expression or activity, while blue lines with vertical dash indicate an effect that decreases expression or activity. Green line and a combination of arrow plus dash indicate mixed activity or activity that, based on the literature varies, according to experimental conditions.
Table 4.
Selected predicted upstream regulators for DA and congenic rats
| Activated in | Upstream Regulator | Molecule Type | z-Score | P Value of Overlap |
|---|---|---|---|---|
| DA | TNF | cytokine | 6.83 | 3.39 × 10−33 |
| LPS | chemical drug | 6.07 | 1.33 × 10−24 | |
| IFNG | cytokine | 5.43 | 4.09 × 10−17 | |
| TGFB1 | growth factor | 4.86 | 4.51 × 10−23 | |
| IL1B | cytokine | 4.85 | 4.42 × 10−13 | |
| MYD88 | cell signaling | 4.46 | 4.85 × 10−08 | |
| IL6 | cytokine | 4.26 | 1.74 × 10−14 | |
| Poly rI:rC RNA | chemical reagent | 3.81 | 3.49 × 10−09 | |
| STAT3 | transcription regulator | 3.68 | 9.33 × 10−21 | |
| RELA | transcription regulator | 3.67 | 3.38 × 10−09 | |
| TNFSF12 | cytokine | 3.61 | 2.68 × 10−07 | |
| LEP | growth factor | 3.53 | 4.85 × 10−09 | |
| ERK1/2 | cell signaling | 3.52 | 8.04 × 10−07 | |
| TLR4 | transmembrane receptor | 3.50 | 2.82 × 10−06 | |
| Jnk | cell signaling | 3.46 | 2.20 × 10−07 | |
| BCR (complex) | complex | 3.39 | 2.25 × 10−04 | |
| NFkB (complex) | complex | 3.36 | 1.20 × 10−09 | |
| IL17A | cytokine | 2.66 | 1.09 × 10−07 | |
| cigarette smoke | chemical toxicant | 2.99 | 5.11 × 10−05 | |
| carrageenan | chemical - nonmammalian | 2.58 | 3.58 × 10−05 | |
| Congenics | PPARG | ligand-dependent nuclear receptor | 4.15 | 2.13 × 10−16 |
| PPARA | ligand-dependent nuclear receptor | 2.38 | 1.51 × 10−18 | |
| PPARD | ligand-dependent nuclear receptor | 2.87 | 9.02 × 10−05 | |
| GW3965 | LXR agonist | 3.20 | 7.78 × 10−07 | |
| NR1H3/LXRA | ligand-dependent nuclear receptor | 2.69 | 1.10 × 10−04 | |
| L-triiodothyronine/THR | chemical - endogenous mammalian | 2.65 | 2.00 × 10−09 | |
| dexamethasone/GR | chemical drug | 3.69 | 5.80 × 10−23 | |
| SB203580 | p38 MAPK inhibitor | 3.27 | 2.23 × 10−16 | |
| U0126 | ERK inhibitor | 3.38 | 3.09 × 10−06 | |
| PD98059 | chemical - kinase inhibitor | 2.90 | 6.07 × 10−17 | |
| TP53 | transcription regulator | 2.14 | 1.65 × 10−16 | |
| let-7 | microRNA | 2.98 | 3.70 × 10−02 | |
| diphenyleneiodonium | chemical drug | 3.79 | 1.08 × 10−11 | |
| N-acetyl-L-cysteine | chemical drug | 2.41 | 5.92 × 10−09 | |
| APOE | transporter | 2.54 | 1.80 × 10−12 | |
| resveratrol | chemical drug | 2.84 | 8.09 × 10−08 | |
| valsartan | chemical drug | 2.40 | 5.94 × 10−10 |
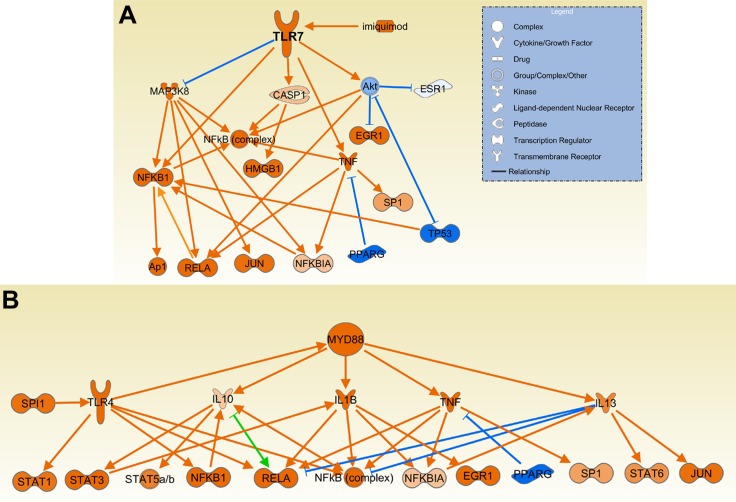
Erk, Jnk, Mapk, NfkB, p38, RelA, and Smad3 were among the cell signaling mediators upstream regulators predicted to be activated in DA. Increased activation via Toll-like receptors was also predicted, with LPS (overall top second upstream gene/pathway prediction), MyD88, poly I:C, TLR2, TL3, TLR4, TLR7 (imiquimod), and TLR9 among the top predicted activators in DA vs. congenics (Table 4, Supplemental Table S4, and Fig. 7).
Fig. 7.
Upstream regulators MyD88 and TLR7 had a predicted increased activity in DA. MyD88 (A), a key mediator of signaling through most of the TLRs, and several TLRs such as TLR7 (B) and their ligands were among the upstream regulators with the highest z-scores and P values, suggesting increased activity in DA and reduced activity in congenics. Orange lines with arrows indicate effect that increases expression or activity, while blue lines with vertical dash indicate an effect that decreases expression or activity. Green line and a combination of arrow plus dash indicate mixed activity, or activity that, based on the literature, varies according to experimental conditions.
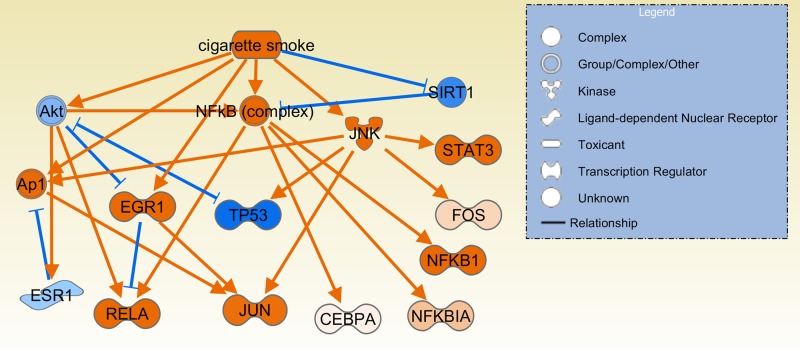
Interestingly, cigarette smoking, a well-known environmental risk factor for RA (10, 31, 43), was identified as an upstream regulator/process (z-score = 3, P =0.00011; Table 4, Fig. 8). These observations could provide new insight into processes central to arthritis pathogenesis that are enhanced or activated by smoking.
Fig. 8.
Cigarette smoking as a predicted upstream regulator in arthritis. Cigarette smoking was one of the environmental risk factors associated with rheumatoid arthritis that was predicted to be an upstream regulator with increased activity in DA and reduced activity in congenics. These observations provide new clues to previously unknown molecular pathogenic processes upregulated or activated by smoking. Orange lines with arrows indicate effect that increases expression or activity, while blue lines with vertical dash indicate an effect that decreases expression or activity.
Additionally, activated upstream regulators included chemicals not previously associated with RA such as trinitrobenzenesulfonic acid, carrageenan [an additive commonly used by the food industry that is capable of inducing inflammation (28) and arthritis in rodents (14)], and 17-alpha-ethinylestradiol.
MicroRNAs such as Let-7 and the inflammation regulatory mir-21 were also predicted to have reduced activity in DA compared with congenics.
Congenics.
Gene expression in the congenic strains suggested increased nuclear receptor (NR) activation (and conversely reduced activity in DA), with seven NRs and NR agonists (PPARA, PPARD, PPARG, dexamethasone/GR, l-triiodothyronine/THR, GW3965/LXR, 1,25-dihydroxy vitamin D3/VDR) among the top upstream regulators predicted to be activated (Table 4, Supplemental Table S4, and Fig. 9A).
Fig. 9.
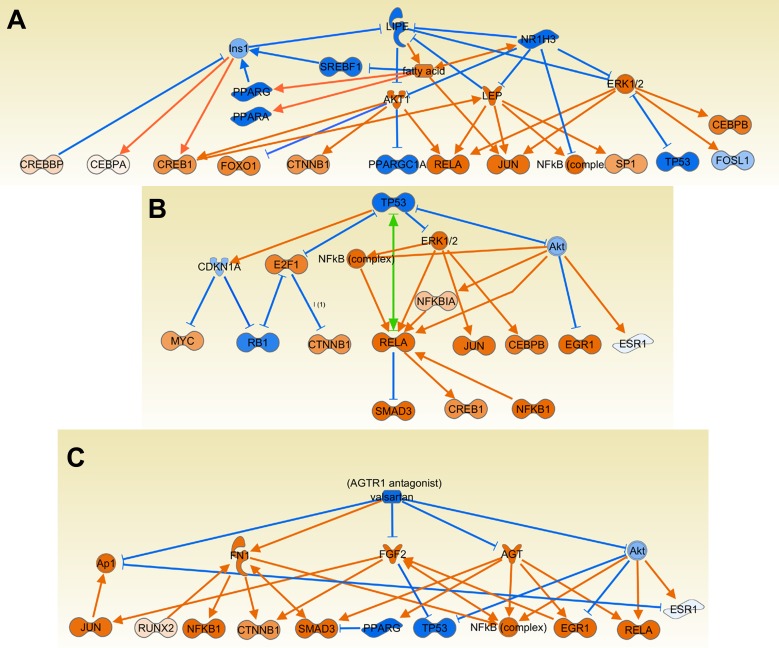
Predicted upstream regulators inhibited in DA rats but activated in congenics. LIPE and LXR pathway (A), p53 (TP53) (B), and Agtr2 (valsartan) (C) blockade were considered as activated in congenics but inhibited in DA, suggesting that increased LXR and p53 expression and activity and reduced Agtr2 activity in congenics are important to maintaining an inflammation-free synovial tissue. Orange lines with arrows indicate effect that increases expression or activity, while blue lines with vertical dash indicate an effect that decreases expression or activity. Green line and a combination of arrow plus dash indicate mixed activity or activity that, based on the literature, varies according to experimental conditions.
TP53 (p53) was increased in congenics as opposed to reduced or inactivated in DA (Table 4, Supplemental Table S4, and Fig. 9B). That observation is interesting in light of previous descriptions of p53 inactivating somatic mutations in RA synovial tissues and further supports a potential role for p53 in disease pathogenesis.
MEK1 and p38 activities were reduced in the congenics as suggested by the increased upstream regulators PD98059 and SB203580, which are MEK1 and p38 inhibitors, respectively. Also, reduced NADPH oxidase activity and reduced oxidative stress was suggested by the increased upstream regulator diphenyleneiodonium and N-acetyl-l-cysteine. Angiotensin II receptor activity was also inhibited in congenics as suggested by the upstream regulator angiotensin II receptor inhibitor valsartan (Table 4, Supplemental Table S4, and Fig. 9C).
QTL-specific Gene Expression
We looked for genes with differential expression restricted to a single congenic strain compared with DA. These genes have the potential to reveal pathways controlled by the arthritis regulatory gene(s) located within the QTL interval. Among the 6,785 genes detected in all samples, 298 genes were differentially expressed between one congenic strain and DA. These congenic-specific genes were also significantly different from the remaining congenic strains, making them unique for a single strain.
Expression differences between congenic strains was classified into two major patterns: 1) unique to a given QTL compared with both DA and all other congenics and 2) congenic-specific detection (or lack of detection) compared with other strains (Tables 5 and 6). The specific genes are shown in Table 6. We focused on genes expressed with unique or opposite patterns. We also looked for a greater-than-expected number of differentially expressed genes located within their respective QTL interval but found that only in Cia25 congenics and Cia25 interval genes.
Table 5.
Expression patterns of genes differentially expressed between each congenic and the remaining congenics combined
| Strain | Differentially Expressed | Differentially Detected |
|---|---|---|
| Cia3d | 29 | 22 |
| Cia5a | 58 | 62 |
| Cia25 | 90 | 58 |
| Cia10 | 0 | 4 |
Differentially Expressed, unique and differentially expressed genes compared with all congenics and DA combined. Differentially Detected, number of genes detected only in one congenic strain and not in the others.
Table 6.
Genes differentially expressed and detected in only one congenic strain compared with the others
| Increased |
Decreased |
|||
|---|---|---|---|---|
| Strain | Expression | Detection | Expression | Detection |
| Cia3d | Adipor2, Ccdc61, Cpt1c, Fam193b, Hsf4, Khk, Pik3r6, Rock2, Scnn1b, Vgll4, Zfp579 | Ankrd55, Col4a3, Fam5b, Frs3, Gfra4, Gjb1, Gp9, Heatr8, Hoxa9l, Prdm9, RGD1566314, Scn8a, Toag1, Ybx2, Zfp651 | Eef1b2l, Eif2 s1, Fam114a1l1, Fbln1, Hsp90aa1, LOC 689408, Npm1, Pabpc1, Pah, Pdia3, Ppih, Prss35, Ptma, RGD1562725, RGD1563124, RGD1565900, Spred1, Wdfy1 | A2m, Cysltr1, LOC290595, Nipsnap3b, Pabpc1l2b, Ptn, Zfp367 |
| Cia5a | Abhd5, Abi1, Adamts5, Aldh9a1, Angpt2, Arhgap29, Cald1, Chmp7, Crim1, Ddx21, Dock9, Dstnl1, Eef1a1, Eif1a, Eif4ebp2, Epas1, Fam43a, Fez2, Ggta1, Itga1, LOC498453, LOC688922, Mapre1, Mcam, MGC105560, Msn, Mtmr6, Nckap1, Pik3r1, Pls3, Ppp3cb, RGD1308874, RGD1561778, RGD1564400, RGD1565496, Sept7, Slc31a2, Tek, Tmem158, Tmem87a, Tspan4, Ube2q2, Wdr45l, Yy1, Zdhhc20, Zfp235, Zfp36l2, Zmpste24, Zmynd11 | Aanat, Acss3, Aif1l, Ankrd13c, Atg13, Atp13a3, Bmi1, Bsdc1, Ccdc79, Cdh5, Ctu1, Ddrgk1, D oc2b, Enpp4, Fbxw11, Foxn3, Gab1, Gabpb2, Ifit2, Kitlg, LOC497978, LOC679565, LOC682571, Mki67ip, Neil2, Nek7, Nxt2, Pex10, Pik3r3, Plekhm2, Ppp1r15b, Prok1, Prr14l, Prrg1, Rassf3, RGD1309104, RGD1561481, Rmnd5a, Stag2, Tbx18, Tmem182, Tril, Twsg1, V gll3, Xxylt1 | Atxn7l2, Bmp1, Cercam, Ctrl, Fbf1, Nebl, Peli3, Pla2 g2d, RGD1308106, RGD1309310, Stat6, Trerf1, Tut1 | C5, AceII, Lhx9, Tns3, Ccnt1, Olr150, Rtp3, Phactr1, Fdxr, Tchp, Lrp4, LOC299458, RGD1307525, RGD1560663, RGD1564313, Stc2, Hapln3, Col7a 1, LOC499530, Mcf2, Mef2d, Chrna7, Magi1 |
| Cia25 | Anp32a, C1 galt1c1, Ccdc84, Cep57, Cnn1, Cpeb2, Csrp1, Eif2ak4, Emp2, Gstp1, Gulp1, Ik, LOC259246, LOC301444, LOC361117, LOC498544, LOC501441, LOC503325, LOC684681, LOC684988, LOC689031, Marveld1, Mcfd2, Med31, Meox2, Mospd2, Mpdz, Myl12b, Nfib, Nme7, Nptn, Numa1, Pgrmc1, Pon2, Pura, RGD1305721, RGD1559566, RGD1560186, RGD1561310, RGD1562469, RGD1562905, RGD1563431, RGD1563551, RGD1564055, RGD1565370, RGD1565806, Rpl30, Rsf1, Shox2, Skiv2l2, Slc12a2, Slc44a1, Vamp2, Ywhag, Zfp672 | Mccc2, Atpif1, Nicn1, Hmgb1, Shbg, Rnf113a2, Krt1-5, Tcte4 | Actb, Agap3, Ahr, Akt1, Atf1, B3 gnt9, Cap1, Cpne3, Cpox, Cs, Ctnnb1, Dpy19l1, Hip1, LOC308990, LOC500959, Lrat, Nudcd1, Obfc2a, Ppp1r18, Rab7b, Rab8a, Rftn1, rnf141, Shc1, Slc35e3, Stat1, Tm6 sf1, Tmem165, Tpcn2, Tpm4, Ube2q2, Wbscr22, Wdr45l, Yy1 | Abcb8, Abi3, Acap1, Adcy10, Arid5a, Atp11a, Camkmt, Cbl, Ccdc99, Ccpg1, Cd4, Cdca7, Ch25 h, Cxcl10, Det1, Dmxl1, Dusp18, Elf1, G2e3, Gbp5, Gga2, Hvcn1, Hyal3, Il13ra1, Ireb2, Klhl6, Leng4, Madd, Map3k 14, Mcm3, Mfsd11, Mgrn1, Nadkd1, Olr211, Olr987, Pask, Ptprc, RGD1306151, RGD1307704, RGD1563701, Rpl5, Spi1, Swsap1, Tll1, Tmem173, Tsc1, Unc45b, Usp24, Vom2r30, Zbed4 |
| Cia10 | (no genes) | Aifm3, Pcnxl2, Tff3 | (no genes) | Srp54a |
The specific strain-specific differentially detected genes are listed below.
Cia3d congenics.
F344 alleles at Cia3d interval were uniquely associated with differences in the expression of 29 genes and detection of 22 genes (Tables 5 and 6).
Eleven genes were expressed in increased levels only in this congenic strain, including Adipor2 and Rock2, and 15 genes were uniquely detected in Cia3d congenics, including Col4a3 and Prdm9.
Eighteen genes were uniquely expressed in decreased levels in Cia3d, including nine genes related with translation, protein synthesis, or protein folding. Seven genes were detected in all DA and all other congenics but not by Cia3d congenics and included Cysltr1 and Ptn.
Cia5a congenics.
F344 alleles at Cia5a were uniquely associated with differences in the expression of 58 genes and unique detection of 62 genes (Tables 5 and 6). Genes with uniquely increased expression in Cia5a congenics included Adamts5, Itga1, Moesin, and Slc31a2.
Thirteen genes had decreased expression only in Cia5a, including Pla2g2d, Stat6, and Bmp1; 23 genes were detected in samples from the other strains but not from Cia5a and included C5, Ace2, and LOC299458 (Ig heavy chain variable gene segment). Overall, Cia5a-specific expression suggested a suppressive activity on actin organization, cell movement, and cell cycle.
Cia25 congenics.
The congenic strain with the highest number of QTL-specific differentially expressed and detected genes was Cia25. ACI alleles at Cia25 were uniquely associated with differences in the expression of 90 genes and detection of 58 genes (Tables 5 and 6).
Genes expressed in increased levels only in Cia25 included Ik (Ik is a suppressor of MHC class II expression), Nfib, Slc12a2, and Slc44a1.
Genes differentially expressed only in Cia25 congenics were also significantly enriched for genes mapping to the Cia25 interval (P < 0.001, χ2 with Yates correction). Specifically, three Cia25 interval genes (Orai2, Hip1, and Wbscr22) had decreased expression in Cia25 congenic rats but not in the other strains, while one Cia25 interval gene (Ywag) was increased only in the Cia25 congenics.
Cia10 congenics.
There were no genes with significant expression differences between Cia10 and the three congenics. However, three genes were uniquely detected (Aifm3, Pcnxl2, Tff3), and one was uniquely not detected (Srp54a) in Cia10 compared with the other congenics. The known function of these four genes does not provide a clear unified pathway for Cia10.
Upstream regulator analysis of individual congenics.
Few genes belonging to a single pathway were differentially expressed or preferentially detected in any single congenic strain. Therefore, analyses of upstream regulators were underpowered for reliable results. No significant congenic-specific upstream regulators were detected.
DA/Arb and DA/Hsd Rats Have Similar Synovial Gene Expression During Arthritis
Cia10 and Cia25 congenics were generated on a DA/Hsd background, while Cia3d and Cia5a were generated on a DA/Arb background. Both DA/Arb and DA/Hsd originated from the same strain, but different stocks of the same strain maintained at different locations over years may occasionally diverge genetically from each other due to incomplete inbreeding at the time of the stock formation, contamination during breeding, or subsequent spontaneous mutations. Pairwise comparison of the expression profiles of synovial samples collected 21 days postinduction of PIA from six DA/Arb and twelve DA/Hsd male rats identified 99 (1.46%) differentially expressed genes out of 6,791 genes reliably detected in all samples (Fig. 2, Supplemental Table S5). Ninety were increased in DA/Hsd, including Higd1a, Rpl30, Ccl19, and Rbb7, while nine were increased in DA/Arb including Mmp9, Timp2, Tnn, and Wisp. While these genes reached significantly different levels between the two DA strains, their expression was mostly in the same direction compared with congenics.
DISCUSSION
Little is known about the genetic regulation of disease severity or the synovial genes mediating articular damage. In the present study we analyzed the gene expression of over 23,000 genes in synovial tissues of four different congenic strains and DA from two different sources. These analyses revealed genes that have a consistently reduced expression and genes that have a consistently increased expression in the synovial tissues of the protected congenics.
These observations across different arthritis-protected congenic strains that share the DA genetic background but differ at a single locus suggest that these are core genes required for the development of or protection against arthritis, synovial hyperplasia, and joint damage. The identification and further functional characterization of these genes should provide new insight into disease pathogenesis and generate new targets for therapy.
Several of these core genes had reduced expression in synovial tissues from protected congenics compared with susceptible DA and included proinflammatory mediators such as IL-1β, IL-6, IL-17A, IL-18, and Mmp3. However, this group of core genes also included several genes not previously implicated in arthritis, such as Tnn (tenascin N), Ccl7, Slpi, and Prc1. We have recently reported increased expression of Tnn in synovial tissues from DA rats and reduced expression in Cia5a congenics (1), but the current report significantly expands the potential relevance of this gene to arthritis in general. Tnn is involved in osteoblast differentiation and angiogenesis (30, 32), and its expression can be induced by TNF-α (42) and IL-1β (Brenner M and Gulko PS, unpublished observations). It is expressed in synovial fibroblasts and is inducible in macrophages (Brenner M and Gulko PS, unpublished observations). Tnn is related to tenascin C, which is an endogenous activator of TLR4 and a key mediator of arthritis (34). Therefore, while Tnn's role in arthritis remains unknown, it is reasonable to speculate that it might be relevant and also a good soluble target for therapy.
Similarly, genes with increased expression in synovial tissues from protected congenics and reciprocally decreased expression in DA suggested a protective effect. These genes included the nuclear receptors Nr1d1 (Rev-erbα), Pparg, Rxrg, and Thrb and other genes such as Scd1, Nnat, Mpz, Mrap, Slc2a4. These genes could have a central homeostatic role in preserving an inflammation-free and nonhyperplastic synovial tissue. Strategies that stimulate some of these nuclear receptors have been successfully used to treat arthritis in rodents (38, 47), and the other new genes could also become good targets for futures studies and therapeutic intervention.
Based on the differential expression of genes we were able to detect 1,161 candidate upstream regulators. These possible upstream regulators are upstream genes, pathways, chemicals, or drugs that induce similar gene expression signatures. Expected upstream regulators that were considered activated in DA include those known to be central to arthritis pathogenesis such as TNF-α, PDGF, IL-1β, IL-6, IL-17, and IL-18. STAT3 mediates cell responses to IL-6, OSM, and LIF; is critical for the development of Th17 T cells (15, 19); and was detected as an activated upstream regulator. Several activated upstream regulators were unexpected and included pattern recognition receptors and associated genes/molecules, such as LPS, poly I:C, MYD88, TLR2, TLR3, TLR4, TLR7 (imiquimod), and TLR9, to name a few. The high number of TLR-related upstream regulators is of great interest as more evidence accumulates supporting a central role for these innate immunity receptors, their signaling pathways (9, 18, 22, 34, 41), and their exogenous (35) and endogenous ligands (8, 34) in arthritis.
Cigarette smoking is a major risk factor for developing RA (10, 31, 43), but little is known about the mechanism mediating this effect. Our analyses detected cigarette smoking as one of the activated upstream regulators in DA compared with congenics (Supplemental Table S4). While our rats were not exposed to cigarette smoke or any other type of smoke, the genes differentially expressed in the synovial tissues from DA rats include those induced or suppressed by cigarette smoking. These observations provide new and interesting clues to the possible pathogenic mechanisms induced or enhanced by smoking.
Chemicals other than cigarettes were also identified as activated upstream regulators. One of the most intriguing was carrageenan, which is a common additive used by the food industry. It is known to induce inflammation when injected subcutaneously (28) and even arthritis following intra-articular injection (14). However, it has not been previously considered an environment of dietary risk factor for arthritis or inflammation. Our results raise the possibility that exposure to carrageenan might increase the expression of genes implicated in disease pathogenesis and joint damage.
The protected congenics had increased expression of nuclear receptors such as Pparg, Lxra, and others. In fact, seven different nuclear receptors or nuclear receptor ligands, including Gr and Vdr, were among the activated upstream regulators with the most significant P values and z-scores. We have previously shown that nuclear receptors may have a homeostatic role in keeping an inflammation-free synovial environment (5, 25, 27). The present observations expand our initial discoveries beyond the Cia25 locus and make them more likely to be broadly relevant to disease.
Genes involved in antioxidant and detoxification pathways were also expressed in increased levels in congenics, compared with DA. These included Adh1, Cyb561, Cyb5r2, Cyb5r3, Cyp2j3, which also mediates activation of vitamin D (the ligand for the anti-inflammatory nuclear receptor VDR) (50); Cyp26b1, regulator of the metabolism of the nuclear receptor agonist retinoic acid (21); Cyp27a1; Mt3; and Tst. Some of these genes, such as Cyp2j3 and Cyp26b1, also regulate the synthesis and inactivation of ligands to anti-inflammatory nuclear receptors (vitamin D and retinoic acid, respectively) (21, 50), again supporting a potentially positive feedback on anti-inflammatory processes around nuclear receptors and disease protection.
P53 (and its pathway) was identified as an activated upstream regulator in congenics, which was inhibited or suppressed in DA. Gene-inactivating somatic mutations on p53 have been previously described in synovial tissues and synovial cells from patients with RA (11), and p53 regulates joint damage in rodent models of arthritis (49). The inhibited upstream regulator activity of p53 detected in DA suggests a synovial tissue inactivation similar to that found in RA, favoring synovial tissue proliferation with cartilage and bone invasion.
Another interesting activated upstream regulator in DA that was inhibited in the protected congenics is Agtr2 (angiotensin II receptor, type 2). Agtr2 is expressed in RA synovial tissues (45), and Agtr2 blockers reduce disease severity and joint damage in rodent models of arthritis (39, 40). Our results support a broad role for Agtr2 in arthritis pathogenesis, which is not genetically restricted to any single specific arthritis severity locus and raise the idea that Agtr2 blockers might be beneficial in patients with RA.
In addition to characterizing the core nongenetically determined differentially expressed genes, we also looked at strain-specific differential expression and uniquely detected genes. There were several congenic-specific differentially expressed genes that point to specific processes regulated the arthritis gene(s) located within each QTL interval (Table 6). Cia3d congenics expressed uniquely lower levels of Ccl21 and complement factor H and increased levels of Adipor2 and Bmp4. Cia5a congenics had decreased expression of a secreted form of phospholipase 2 (Pla2g2d), Stat6, and Bmp1 and increased expression of Itga1, Adamts5, and moesin. Cia25 congenics expressed lower levels of Itga4, Ahr, and Stat2 and increased levels of IK cytokine (a suppressor of MHC class II expression). Cia10 did not have uniquely differentially expressed genes but had specific detection of Aifm3, Pcnxl2, and Tff3 and was the only strain that did not express Srp54a. These and other congenic-specific expressions differentiate the processes regulated by each congenic and should facilitate the QTL gene discovery effort.
We also confirmed the high degree of similarity between DA rats obtained from two sources and therefore were able to use them together as a single group for our analyses. This is relevant as it makes data generated with DA rats from these sources comparable and more reliable.
In conclusion, we describe a new core set of nongenetically restricted genes that differentiate arthritic DA from protected congenic rats. Our analyses reveal several known and previously unknown pro- and anti-inflammatory pathways implicated in disease pathogenesis and also suggest new potential environmental and dietary risk factors. These discoveries should facilitate future studies including gene discovery and target selection for therapy.
GRANTS
Funded by the National Institutes of Health Grants R01-AR-46213, R01-AR-052439, and R01-AI-54348 to P. S. Gulko.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.B., T.L., and P.S.G. conception and design of research; M.B. and P.S.G. performed experiments; M.B., T.L., and P.S.G. analyzed data; M.B., T.L., and P.S.G. interpreted results of experiments; M.B. and P.S.G. prepared figures; M.B., T.L., and P.S.G. edited and revised manuscript; M.B., T.L., and P.S.G. approved final version of manuscript; P.S.G. drafted manuscript.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Brenner M, Gulko PS. The arthritis severity locus Cia5a regulates the expression of inflammatory mediators including Syk pathway genes and proteases in pristane-induced arthritis. BMC Genomics 13: 710, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner M, Laragione T, Mello A, Gulko PS. Cia25 on rat chromosome 12 regulates severity of autoimmune arthritis induced with pristane and with collagen. Ann Rheum Dis 66: 952–957, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner M, Laragione T, Shah A, Mello A, Remmers EF, Wilder RL, Gulko PS. Identification of two new arthritis severity loci that regulate levels of autoantibodies, interleukin-1β, and joint damage in pristane- and collagen-induced arthritis. Arthritis Rheum 64: 1369–1378, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner M, Linge CP, Li W, Gulko PS. Increased synovial expression of nuclear receptors correlates with protection in pristane-induced arthritis: a possible novel genetically regulated homeostatic mechanism. Arthritis Rheum 63: 2918–2929, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner M, Meng H, Yarlett N, Griffiths M, Remmers E, Wilder R, Gulko P. The non-MHC quantitative trait locus Cia10 contains a major arthritis gene and regulates disease severity, pannus formation and joint damage. Arthritis Rheum 52: 322–332, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Brenner M, Meng HC, Yarlett NC, Joe B, Griffiths MM, Remmers EF, Wilder RL, Gulko PS. The non-MHC quantitative trait locus Cia5 contains three major arthritis genes that differentially regulate disease severity, pannus formation, and joint damage in collagen- and pristane-induced arthritis. J Immunol 174: 7894–7903, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain ND, Kim SJ, Vila OM, Volin MV, Volkov S, Pope RM, Arami S, Mandelin AM, 2nd, Shahrara S. Ligation of TLR7 by rheumatoid arthritis synovial fluid single strand RNA induces transcription of TNFalpha in monocytes. Ann Rheum Dis 72: 418–426, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SY, Shiau AL, Li YT, Lin YS, Lee CH, Wu CL, Wang CR. Suppression of collagen-induced arthritis by intra-articular lentiviral vector-mediated delivery of Toll-like receptor 7 short hairpin RNA gene. Gene Ther 19: 752–760, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Criswell LA, Saag KG, Mikuls TR, Cerhan JR, Merlino LA, Lum RF, Pfeiffer KA, Woehl B, Seldin MF. Smoking interacts with genetic risk factors in the development of rheumatoid arthritis among older Caucasian women. Ann Rheum Dis 65: 1163–1167, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firestein GS, Echeverri F, Yeo M, Zvaifler NJ, Green DR. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci USA 94: 10895–10900, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulko PS. Contribution of genetic studies in rodent models of autoimmune arthritis to understanding and treatment of rheumatoid arthritis. Genes Immun 8: 523–531, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Gulko PS, Kawahito Y, Remmers EF, Reese VR, Wang J, Dracheva SV, Ge L, Longman RE, Shepard JS, Cannon GW, Sawitzke AD, Wilder RL, Griffiths MM. Identification of a new non-major histocompatibility complex genetic locus on chromosome 2 that controls disease severity in collagen- induced arthritis in rats. Arthritis Rheum 41: 2122–2131, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Hansra P, Moran EL, Fornasier VL, Bogoch ER. Carrageenan-induced arthritis in the rat. Inflammation 24: 141–155, 2000 [DOI] [PubMed] [Google Scholar]
- 15.Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, Getnet D, Goldberg MV, Maris CH, Housseau F, Yu H, Pardoll DM, Drake CG. Cutting edge: an in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol 179: 4313–4317, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Jenkins E, Brenner M, Laragione T, Gulko PS. Synovial expression of Th17-related and cancer-associated genes is regulated by the arthritis severity locus Cia10. Genes Immun 13: 221–231, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joe B, Remmers EF, Dobbins DE, Salstrom JL, Furuya T, Dracheva S, Gulko PS, Cannon GW, Griffiths MM, Wilder RL. Genetic dissection of collagen-induced arthritis in Chromosome 10 quantitative trait locus speed congenic rats: evidence for more than one regulatory locus and sex influences. Immunogenetics 51: 930–944, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Joosten LA, Koenders MI, Smeets RL, Heuvelmans-Jacobs M, Helsen MM, Takeda K, Akira S, Lubberts E, van de Loo FA, van den Berg WB. Toll-like receptor 2 pathway drives streptococcal cell wall-induced joint inflammation: critical role of myeloid differentiation factor 88. J Immunol 171: 6145–6153, 2003 [DOI] [PubMed] [Google Scholar]
- 19.Ju JH, Heo YJ, Cho ML, Jhun JY, Park JS, Oh HJ, Moon SJ, Kwok SK, Park KS, Park SH, Kim HY. Modulation of STAT-3 in rheumatoid synovial T cells suppresses Th17 differentiation and increases the proportion of Treg cells. Arthritis Rheum 64: 3543–3452, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Kremer JM, Westhovens R, Leon M, Di Giorgio E, Alten R, Steinfeld S, Russell A, Dougados M, Emery P, Nuamah IF, Williams GR, Becker JC, Hagerty DT, Moreland LW. Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N Engl J Med 349: 1907–1915, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Krivospitskaya O, Elmabsout AA, Sundman E, Soderstrom LA, Ovchinnikova O, Gidlof AC, Scherbak N, Norata GD, Samnegard A, Torma H, Abdel-Halim SM, Jansson JH, Eriksson P, Sirsjo A, Olofsson PS. A CYP26B1 polymorphism enhances retinoic acid catabolism and may aggravate atherosclerosis. Mol Med 18: 712–718, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyburz D, Rethage J, Seibl R, Lauener R, Gay RE, Carson DA, Gay S. Bacterial peptidoglycans but not CpG oligodeoxynucleotides activate synovial fibroblasts by toll-like receptor signaling. Arthritis Rheum 48: 642–650, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Laragione T, Brenner M, Li W, Gulko PS. Cia5d regulates a new fibroblast-like synoviocyte invasion-associated gene expression signature. Arthritis Res Ther 10: R92, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laragione T, Brenner M, Mello A, Symons M, Gulko PS. The arthritis severity locus Cia5d is a novel genetic regulator of the invasive properties of synovial fibroblasts. Arthritis Rheum 58: 2296–2306, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laragione T, Gulko PS. Liver X receptor (LXR) regulates rheumatoid arthritis fibroblast-like synoviocyte invasiveness, matrix metalloproteinase 2 activation, interleukin-6 and CXCL10. Mol Med 18: 1009–1017, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laragione T, Gulko PS. mTOR regulates the invasive properties of synovial fibroblasts in rheumatoid arthritis. Mol Med 16: 352–358, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laragione T, Shah A, Gulko PS. The vitamin D receptor regulates rheumatoid arthritis synovial fibroblast invasion and morphology. Mol Med 18: 194–200, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laragione T, Yarlett NC, Brenner M, Mello A, Sherry B, Miller EJ, Metz CN, Gulko PS. The arthritis severity quantitative trait loci Cia4 and Cia6 regulate neutrophil migration into inflammatory sites and levels of TNF-alpha and nitric oxide. J Immunol 178: 2344–2351, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta delta C(T) method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 30.Martina E, Degen M, Ruegg C, Merlo A, Lino MM, Chiquet-Ehrismann R, Brellier F. Tenascin-W is a specific marker of glioma-associated blood vessels and stimulates angiogenesis in vitro. FASEB J 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattey DL, Hutchinson D, Dawes PT, Nixon NB, Clarke S, Fisher J, Brownfield A, Alldersea J, Fryer AA, Strange RC. Smoking and disease severity in rheumatoid arthritis: association with polymorphism at the glutathione S-transferase M1 locus. Arthritis Rheum 46: 640–646, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Meloty-Kapella CV, Degen M, Chiquet-Ehrismann R, Tucker RP. Effects of tenascin-W on osteoblasts in vitro. Cell Tiss Res 334: 445–455, 2008 [DOI] [PubMed] [Google Scholar]
- 33.Meng H, Griffiths M, Remmers E, Kawahito Y, Li W, Neisa R, Cannon R, Wilder R, Gulko P. Identification of two novel female-specific non-MHC loci regulating collagen-induced arthritis severity and chronicity, and evidence of epistasis. Arthritis Rheum 50: 2695–2705, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Midwood K, Sacre S, Piccinini AM, Inglis J, Trebaul A, Chan E, Drexler S, Sofat N, Kashiwagi M, Orend G, Brennan F, Foxwell B. Tenascin-C is an endogenous activator of Toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nat Med 15: 774–780, 2009 [DOI] [PubMed] [Google Scholar]
- 35.Miyata M, Kobayashi H, Sasajima T, Sato Y, Kasukawa R. Unmethylated oligo-DNA containing CpG motifs aggravates collagen-induced arthritis in mice. Arthritis Rheum 43: 2578–2582, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Navarro-Cano G, Del Rincon I, Pogosian S, Roldan JF, Escalante A. Association of mortality with disease severity in rheumatoid arthritis, independent of comorbidity. Arthritis Rheum 48: 2425–2433, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Nishimoto N, Yoshizaki K, Miyasaka N, Yamamoto K, Kawai S, Takeuchi T, Hashimoto J, Azuma J, Kishimoto T. Treatment of rheumatoid arthritis with humanized anti-interleukin-6 receptor antibody: a multicenter, double-blind, placebo-controlled trial. Arthritis Rheum 50: 1761–1769, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Park MC, Kwon YJ, Chung SJ, Park YB, Lee SK. Liver X receptor agonist prevents the evolution of collagen-induced arthritis in mice. Rheumatology (Oxford) 49: 882–890, 2010 [DOI] [PubMed] [Google Scholar]
- 39.Price A, Lockhart JC, Ferrell WR, Gsell W, McLean S, Sturrock RD. Angiotensin II type 1 receptor as a novel therapeutic target in rheumatoid arthritis: in vivo analyses in rodent models of arthritis and ex vivo analyses in human inflammatory synovitis. Arthritis Rheum 56: 441–447, 2007 [DOI] [PubMed] [Google Scholar]
- 40.Refaat R, Salama M, Abdel Meguid E, El Sarha A, Gowayed M. Evaluation of the effect of losartan and methotrexate combined therapy in adjuvant-induced arthritis in rats. Eur J Pharmacol 698: 421–428, 2013 [DOI] [PubMed] [Google Scholar]
- 41.Sacre SM, Andreakos E, Kiriakidis S, Amjadi P, Lundberg A, Giddins G, Feldmann M, Brennan F, Foxwell BM. The Toll-like receptor adaptor proteins MyD88 and Mal/TIRAP contribute to the inflammatory and destructive processes in a human model of rheumatoid arthritis. Am J Pathol 170: 518–525, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherberich A, Tucker RP, Degen M, Brown-Luedi M, Andres AC, Chiquet-Ehrismann R. Tenascin-W is found in malignant mammary tumors, promotes alpha8 integrin-dependent motility and requires p38MAPK activity for BMP-2 and TNF-alpha induced expression in vitro. Oncogene 24: 1525–1532, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Silman AJ, Newman J, MacGregor AJ. Cigarette smoking increases the risk of rheumatoid arthritis. Results from a nationwide study of disease-discordant twins. Arthritis Rheum 39: 732–735, 1996 [DOI] [PubMed] [Google Scholar]
- 44.Vingsbo C, Sahlstrand P, Brun JG, Jonsson R, Saxne T, Holmdahl R. Pristane-induced arthritis in rats: a new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non-major histocompatibility complex genes. Am J Pathol 149: 1675–1683, 1996 [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh DA, Suzuki T, Knock GA, Blake DR, Polak JM, Wharton J. AT1 receptor characteristics of angiotensin analogue binding in human synovium. Br J Pharmacol 112: 435–442, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinblatt ME, Kremer JM, Bankhurst AD, Bulpitt KJ, Fleischmann RM, Fox RI, Jackson CG, Lange M, Burge DJ. A trial of etanercept, a recombinant tumor necrosis factor receptor:Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N Engl J Med 340: 253–259, 1999 [DOI] [PubMed] [Google Scholar]
- 47.Wiesenberg I, Chiesi M, Missbach M, Spanka C, Pignat W, Carlberg C. Specific activation of the nuclear receptors PPARgamma and RORA by the antidiabetic thiazolidinedione BRL 49653 and the antiarthritic thiazolidinedione derivative CGP 52608. Mol Pharmacol 53: 1131–1138, 1998 [PubMed] [Google Scholar]
- 48.Wolfe F, Rasker JJ, Boers M, Wells GA, Michaud K. Minimal disease activity, remission, and the long-term outcomes of rheumatoid arthritis. Arthritis Rheum 57: 935–942, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Yamanishi Y, Boyle DL, Pinkoski MJ, Mahboubi A, Lin T, Han Z, Zvaifler NJ, Green DR, Firestein GS. Regulation of joint destruction and inflammation by p53 in collagen- induced arthritis. Am J Pathol 160: 123–130, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamasaki T, Izumi S, Ide H, Ohyama Y. Identification of a novel rat microsomal vitamin D3 25-hydroxylase. J Biol Chem 279: 22848–22856, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.