Abstract
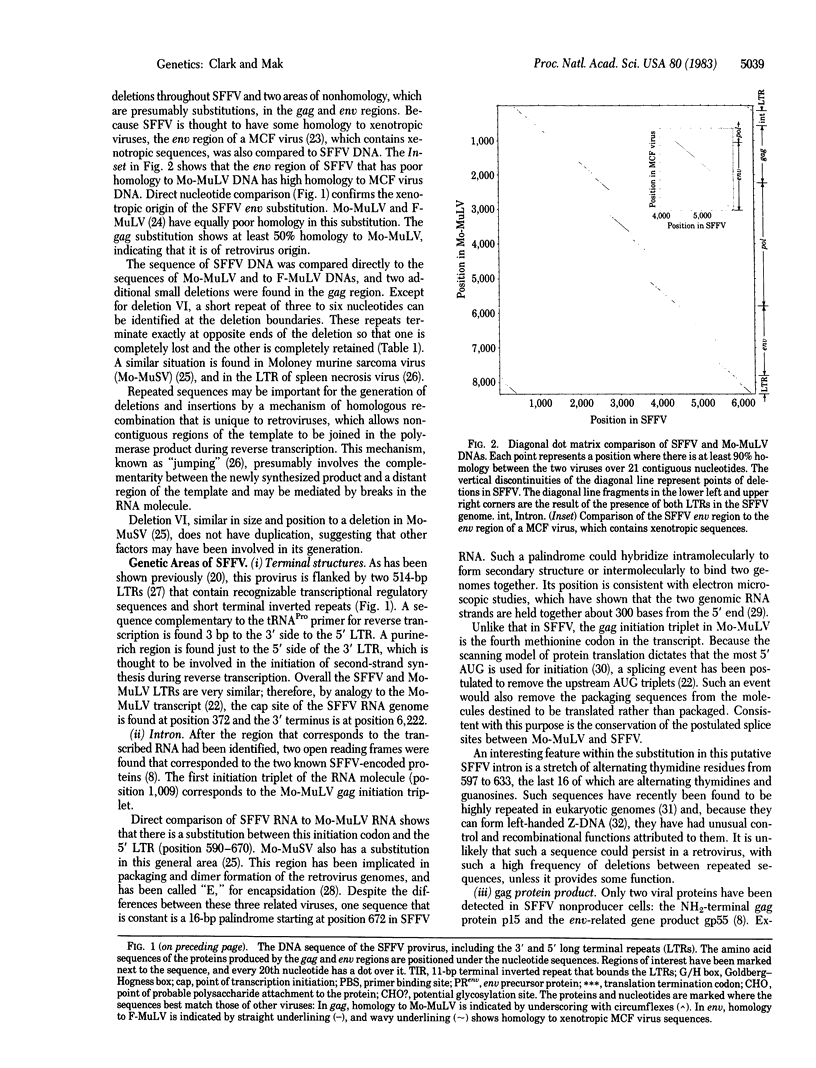
The Friend spleen focus-forming provirus is 6,296 base pairs (bp) in length. Compared to Moloney murine leukemia virus, it has undergone five major deletions, three substitutions, and a number of minor alterations. Otherwise, these viruses are about 90% homologous. A 16-bp palindrome is found in the region thought to be involved in packaging and dimerization of the RNA genome. Premature termination of translation of the gag polyprotein is attributed to a 13-bp deletion in the p12 region. A substitution of xenotropic env sequences was identified in the 5' region of the env gene; 150 nucleotides 3' to this substitution, a deletion of 585 bp removes the site where the normal env precursor protein is cleaved to form gp70 and p15(E), resulting in a fusion protein of Mr 44,725. Due to these changes, the env product gp55 is expected to have a substantially different conformation on the cell surface compared to either a xenotropic or ecotropic gp70 protein, and may be responsible for the rapid erythroleukemic potential of spleen focus-forming virus.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AXELRAD A. A., STEEVES R. A. ASSAY FOR FRIEND LEUKEMIA VIRUS: RAPID QUANTITATIVE METHOD BASED ON ENUMERATION OF MACROSCOPIC SPLEEN FOCI IN MICE. Virology. 1964 Nov;24:513–518. doi: 10.1016/0042-6822(64)90199-0. [DOI] [PubMed] [Google Scholar]
- Bender W., Chien Y. H., Chattopadhyay S., Vogt P. K., Gardner M. B., Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978 Mar;25(3):888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein A., Gamble C., Penrose D., Mak T. W. Presence and expression of Friend erythroleukemia virus-related sequences in normal and leukemic mouse tissues. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4455–4459. doi: 10.1073/pnas.76.9.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein A., Mak T. W., Stephenson J. R. The Friend virus genome: evidence for the stable association of MuLV sequences and sequences involved in erythroleukemic transformation. Cell. 1977 Sep;12(1):287–294. doi: 10.1016/0092-8674(77)90206-9. [DOI] [PubMed] [Google Scholar]
- Bosselman R. A., Van Griensven L. J., Vogt M., Verma I. M. Genome organization of retroviruses IX. Analysis of the genomes of Friend spleen focus-forming (F-SFFV) and helper murine leukemia viruses by heteroduplex-formation. Virology. 1980 Apr 15;102(1):234–239. doi: 10.1016/0042-6822(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Bosselman R. A., van Straaten F., Van Beveren C., Verma I. M., Vogt M. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J Virol. 1982 Oct;44(1):19–31. doi: 10.1128/jvi.44.1.19-31.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Chen R. Complete amino acid sequence and glycosylation sites of glycoprotein gp71A of Friend murine leukemia virus. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5788–5792. doi: 10.1073/pnas.79.19.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. P., Mak T. W. Nucleotide sequences of the murine retrovirus Friend SFFVp long terminal repeats: identification of a structure with extensive dyad symmetry 5' to the TATA box. Nucleic Acids Res. 1982 May 25;10(10):3315–3330. doi: 10.1093/nar/10.10.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R. J., Hettrick K. L., Greenberger J. S., Bennett M. Extended self-renewal capacity of pluripotent hemopoietic stem cells: association with persistent Friend spleen focus-forming virus. Cell. 1982 Dec;31(3 Pt 2):731–738. doi: 10.1016/0092-8674(82)90327-0. [DOI] [PubMed] [Google Scholar]
- Edwards S. A., Fan H. gag-Related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol. 1979 May;30(2):551–563. doi: 10.1128/jvi.30.2.551-563.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEND C. Cell-free transmission in adult Swiss mice of a disease having the character of a leukemia. J Exp Med. 1957 Apr 1;105(4):307–318. doi: 10.1084/jem.105.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S. C., Ivatt R. J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Ikawa Y., Kobayashi Y., Obinata M., Harada F., Hino S., Yoshikura H. RNA sequences and proteins specific to Friend strain of spleen focus-forming virus. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):875–885. doi: 10.1101/sqb.1980.044.01.094. [DOI] [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Yorifuji T., Tsuruta S. Rapid emergence of mink cell focus-forming (MCF) virus in various mice infected with NB-tropic friend virus. Virology. 1981 Sep;113(2):644–655. doi: 10.1016/0042-6822(81)90193-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Linemeyer D. L., Menke J. G., Ruscetti S. K., Evans L. H., Scolnick E. M. Envelope gene sequences which encode the gp52 protein of spleen focus-forming virus are required for the induction of erythroid cell proliferation. J Virol. 1982 Jul;43(1):223–233. doi: 10.1128/jvi.43.1.223-233.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M. E., Reynolds F. H., Jr, Van de Ven W. J., Stephenson J. R., Mak T. W., Bernstein A. Anemia- and polycythemia-inducing isolates of Friend spleen focus-forming virus. Biological and molecular evidence for two distinct viral genomes. J Exp Med. 1980 Jun 1;151(6):1477–1492. doi: 10.1084/jem.151.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager D. L., Mak T. W., Bernstein A. Quantitative colony method for tumorigenic cells transformed by two distinct strains of Friend leukemia virus. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1703–1707. doi: 10.1073/pnas.78.3.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak T. W., Axelrad A. A., Bernstein A. Fv-2 locus controls expression of Friend spleen focus-forming virus-specific sequences in normal and infected mice. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5809–5812. doi: 10.1073/pnas.76.11.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak T. W., Gamble C. L., MacDonald M. E., Bernstein A. Host control of sequences specific to Friend erythroleukemia virus in normal and leukemic mice. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):893–899. doi: 10.1101/sqb.1980.044.01.096. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risser R. Friend erythroleukemia antigen. A viral antigen specified by spleen focus-forming virus and differentiation antigen controlled by the Fv-2 locus. J Exp Med. 1979 May 1;149(5):1152–1167. doi: 10.1084/jem.149.5.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti S., Davis L., Feild J., Oliff A. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J Exp Med. 1981 Sep 1;154(3):907–920. doi: 10.1084/jem.154.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti S., Troxler D., Linemeyer D., Scolnick E. Three laboratory strains of spleen focus-forming virus: comparison of their genomes and translational products. J Virol. 1980 Jan;33(1):140–151. doi: 10.1128/jvi.33.1.140-151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. Spontaneous variation and synthesis in the U3 region of the long terminal repeat of an avian retrovirus. J Virol. 1982 Jan;41(1):163–171. doi: 10.1128/jvi.41.1.163-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Green N., Liu F. T., Niman H. L., Lerner R. A. Chemical synthesis of a polypeptide predicted from nucleotide sequence allows detection of a new retroviral gene product. Nature. 1980 Oct 30;287(5785):801–805. doi: 10.1038/287801a0. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Axelrad A. A. Fv-2 locus controls the proportion of erythropoietic progenitor cells (BFU-E) synthesizing DNA in normal mice. Cell. 1980 Jan;19(1):225–236. doi: 10.1016/0092-8674(80)90404-3. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Function of the retrovirus long terminal repeat. Cell. 1982 Jan;28(1):3–5. doi: 10.1016/0092-8674(82)90367-1. [DOI] [PubMed] [Google Scholar]
- Temin H. M. The DNA provirus hypothesis. Science. 1976 Jun 11;192(4244):1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- Troxler D. H., Lowy D., Howk R., Young H., Scolnick E. M. Friend strain of spleen focus-forming virus is a recombinant between ecotropic murine type C virus and the env gene region of xenotropic type C virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4671–4675. doi: 10.1073/pnas.74.10.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Vorlíckovă M., Kypr J., Stokrová S., Sponar J. A Z-like form of poly(dA-dC).poly(dG-dT) in solution? Nucleic Acids Res. 1982 Feb 11;10(3):1071–1080. doi: 10.1093/nar/10.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5' long terminal repeat and the start of the gag gene. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Gamble C. L., Clark S. P., Joyner A., Shibuya T., MacDonald M. E., Mager D., Bernstein A., Mak T. W. Clonal analysis of early and late stages of erythroleukemia induced by molecular clones of integrated spleen focus-forming virus. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6893–6897. doi: 10.1073/pnas.78.11.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]



