Abstract
Cutaneous T-cell lymphoma (CTCL) is a heterogeneous non-Hodgkin’s lymphoma that may variably involve the skin, lymph nodes, and peripheral blood. Malignant burden ranges from cutaneous patches and plaques with little evidence of blood involvement to erythroderma often in association with frank leukemia, as in Sézary syndrome. Toward a better understanding of the pathogenesis of this CD4+ T-cell malignancy, we conducted a high-resolution genomic analysis combining DNA (23 samples) and mRNA (12 samples) data of peripheral blood isolates from CTCL patients across a spectrum of stages. Strikingly, even patients with limited involvement, e.g., normal CD4 counts, contained significant copy-number alterations. Defining genomic characteristics of CTCL blood involvement included gains on 8q and 17q, and deletions on 17p and chromosome 10. A consensus analysis of 108 leukemic CTCL samples demonstrated global similarities among patients with varied blood involvement, narrowing 38 of 62 loci. Toward an annotated framework for in vitro testing, we also characterized genomic alterations in five CTCL cell lines (HH, HUT78, PNO, SeAx, and Sez4), revealing intact core features of leukemic CTCL. Together, these studies produce the most comprehensive view of the leukemic CTCL genome to date, with implications for pathogenesis, molecular classification, and potential future therapeutic developments.
INTRODUCTION
Cutaneous T-cell lymphoma (CTCL) is a clinically heterogeneous malignancy of CD4+ skin-homing T cells (Berger and Edelson, 2003; Willemze and Meijer, 2006). Early stages of CTCL principally involve the skin; however, the risk of peripheral blood involvement increases with advancing stage (Schechter et al., 1987). Even patients with limited disease (stage IA, <10% body surface area involvement) may show evidence of an expanding clone in the blood as evidenced by flow cytometric T-cell subset analyses. Clonal expansion of malignant cells within the peripheral blood is also associated with a loss of the residual T-cell receptor diversity (Yawalkar et al., 2003), increased T-regulatory activity (Berger et al., 2005), and diminished CD8 counts (Klemke et al., 2005; Vonderheid et al., 2006). Responses to immune modifying treatments such as photopheresis (Girardi et al., 2003) and IFN-α (Olsen, 2003) have strongly suggested the immunogenicity of CTCL; yet, our understanding of the pathogenesis of this malignancy is still limited. Moreover, stage III and IV CTCL, particularly in those with depressed CD8 compartments, is commonly lethal, with survival not improved by conventional systemic therapies (Girardi et al., 2003; Olsen, 2003; Klemke et al., 2005; Vonderheid et al., 2006).
In recognition that cancer is fundamentally a genetic disease (Weber, 2002; Vogelstein and Kinzler, 2004), knowledge of the critical mutations underlying tumor initiation and progression, along with a growing array of targeted therapeutics, offer tremendous potential in improving the diagnosis and treatment of cancer (Demetri et al., 2002; Kantarjian et al., 2002; Paez et al., 2004). Early cytogenetic studies in CTCL suggested potential patterns in DNA copy-number mutations (Mao et al., 2002, 2003) and, importantly, some of these alterations, such as gain of 8q and loss of 6q and 13q, are associated with shorter survival (Fischer et al., 2004). Multiple levels of cellular dysregulation, restricted sample size, low-resolution data, and limited clinical information, however, have encumbered the discernment of the primary biological pathways in this malignancy. With accumulating genomic data and increased microarray resolution, additional candidate regions have been identified (Vermeer et al., 2008; Caprini et al., 2009; Laharanne et al., 2010), and now more recent advances in microarray technologies, coupled with improved computational algorithms, offer the unique opportunity to characterize the CTCL genome to the subgene level and study various modes of mutations.
Validation of genomic findings, however, is also limited by the lack of a genomically annotated in vitro system. Despite the challenges of ex vivo culture of CTCL cells in the absence of other cell interactions, a limited number of established CTCL cell lines do exist; yet, how representative these cell lines are of freshly derived patient samples is unclear. Established cell lines originally isolated from Sézary syndrome patients may require additional growth factors and are numerous passages from the initial patient, yet with a detailed knowledge of their genomes, our ability to perturb and assess CTCL biology could be greatly enhanced. Ultimately by characterizing the genomic landscape in leukemic CTCL, at both the DNA and RNA level and in the context of genetically characterized cell lines, we have begun to pinpoint potential targets and offer a framework in which to further study the mechanisms underlying transformation and pathogenesis in this malignancy.
RESULTS
Copy-number analysis identifies regions containing putative oncogenes and tumor suppressors
During malignant progression, acquired genetic alterations that confer a survival advantage would be expected to be selected for and altered at a higher rate. Therefore, identifying statistical outliers should determine regions of the genome with genes of potential biological significance. Our genomic DNA-based analysis sought to determine these “driver” copy-number mutations using an algorithm termed GISTIC (Genomic Identification of Significant Targets in Cancer; Beroukhim et al., 2007), which has proven successful in identifying both known and new oncogenes and tumor suppressors in multiple other cancers (Beroukhim et al., 2007, 2010; Weir et al., 2007; Lin et al., 2008). GISTIC takes into account both the frequency of a DNA gain or loss and the magnitude of this gain or loss at each probe. By comparing this calculated score to the background null hypothesis score, a significance value at each probe throughout the genome is determined. Significantly altered regions of the genome can then be further investigated for candidate oncogenes and tumor suppressors.
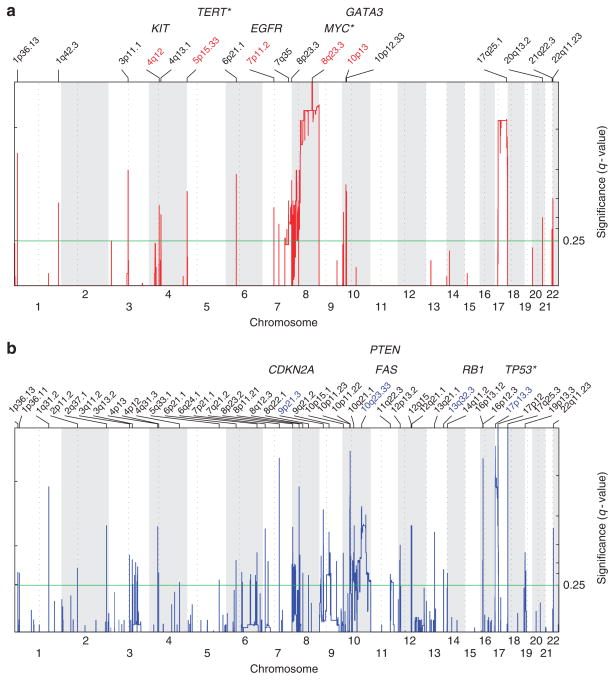
The CTCL landscape for amplifications based on GISTIC analysis of our samples is shown in Figure 1a, with significance per probe plotted across the genome. We identified 17 regions of amplifications in the CTCL genome seen in red (Figure 1a and Supplementary Table S1a online) and 40 regions of deletion in blue (Figure 1b and Supplementary Table S1b online). The most significant amplifications involved large regions of gain on 8q and 17q, in addition to many focal amplifications. Overall, there were many more regions of deletion than amplification, with the most significant deletions on 17p and 10. These regions of chromosomal amplification and loss are shown in Supplementary Table S1 online. We also identified focal amplifications of 4q12 including KIT and 7p11.2 including EGFR (Figure 1a, Supplementary Table S1a online). Although these amplifications were only found in a subset of samples, 4 of 23 (17%) and 3 of 23 (13%), respectively, half of those with 4q12 amplifications, and all of those with 7p11.2 amplifications, were noted to have nonresponsive or worsened skin disease after 1 year of treatment (Table 1 and Figure 3a). These amplifications could not be validated by PCR because of insufficient patient sample; however, given the potential clinical relevance and available targeted therapeutics, further exploration in vitro and in larger cohorts may be warranted. Moreover, VEGFA (vascular endothelial growth factor receptor) is another oncogene for which targeted therapy is available, and is the closest well-validated cancer gene to the focal amplification on 6p21.1 in 3 of 23 (13%) patients (Supplementary Table S1a online and Figure 3a). Thus, starting from the whole genome, the GISTIC algorithm narrowed and prioritized a list of candidate genes for further study.
Figure 1. Significant DNA copy-number alterations in leukemic cutaneous T-cell lymphoma (CTCL).
Statistically significant (a) amplifications and (b) deletions pinpointed by GISTIC (Genomic Identification of Significant Targets in Cancer) aggregate analysis of 23 CTCL patient samples. Chromosomal location is across the bottom with labeled cytobands corresponding to the center of the region and cancer-related genes from Beroukhim et al. (2010), or known CTCL genes labeled above the region. *A gene adjacent to the peak region. Significance reported as false discovery rate–corrected q-value.
Table 1.
Clinical data from the 24 CTCL patients with blood involvement
| Patient sample | Gender | Age | Status at 1 year | Initial skin stage/BSA | Skin score at 1 year | Treatment at sample collection | Evidence for blood involvement | CD4/ CD8 ratio | Population isolated | Gene expression data | SNP data |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 84 | Stable | T4; >50% | Improved | ECP | PCR for TCR-β/γ (+), CD45RO elevated | 1.7 | CD4+ | — | Yes |
| 2 | Female | 85 | Stable | T4; >50% | Improved | ECP, Targretin | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+CD26− elevated | 1.6 | CD4+CD7−CD26− | — | Yes |
| 3 | Male | 86 | Stable | T4; >50% | Improved | ECP, Targretin | PCR for TCR-β/γ (+), Vα | 2.9 | CD4+ | Yes | Yes |
| 4 | Female | 50 | Stable | T2; >10% | Improved | ECP, IFN-α | PCR for TCR-β/γ (+) | 38 | CD4+CD7− | Yes | Yes |
| 5 | Male | 66 | Deceased | T4; >50% | Nonresponsive | ECP | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+CD26− elevated | 99 | CD4+CD7−CD26− | Yes | Yes |
| 6 | Male | 52 | Improved | T3; >50% | Improved | ECP, Targretin | ? | 2.3 | CD4+ | — | Yes |
| 7 | Female | 76 | Improved | T3; <10% | Improved | ECP | PCR for TCR-β/γ (+) | 0.79 | CD4+ | Yes | Yes |
| 8 | Female | 83 | Improved | T2; 10–50% | Improved | ECP | PCR for TCR-β/γ (+), Vβ8a | 0.28 | CD4+ | — | Yes |
| 9 | Male | 53 | Stable | T4; 100% | Nonresponsive | ECP | PCR for TCR-β/γ (+), CD45RO elevated | 1.2 | CD4+ | Yes | — |
| 10 | Female | 86 | Worsened | T2; 10–50% | Worsened | ECP, Ontak | PCR for TCR-β/γ (+), CD4+CD26− elevated, Vβ2 | 19.7 | CD4+CD26− | Yes | Yes |
| 11 | Female | 72 | Worsened | T4; >50% | Nonresponsive | ECP, Targretin, IFN-α | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+CD26− elevated | 3.5 | CD4+CD7−CD26− | Yes | Yes |
| 12 | Male | 62 | Improved | T4; >50% | Improved | ECP, Targretin | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+CD26− elevated, previous Vβ5a | 2.8 | CD4+CD7−CD26− | Yes | Yes |
| 13 | Male | 31 | Improved | T4; >50% | Improved | ECP | PCR for TCR-β/γ (+), previous Vα12.1 | 1.6 | CD4+ | — | Yes |
| 14 | Male | 76 | Stable | T2; 10–50% | Worsened | ECP | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD26− elevated | 18.5 | CD4+CD26− | — | Yes |
| 15 | Female | 56 | Improved | T4; >50% | Improved | ECP | PCR for TCR-β/γ (+), CD45RO elevated, Vβ5a | 1.4 | CD4+ | — | Yes |
| 16 | Male | 84 | Worsened | T4; >50% | Worsened | ECP, Targretin | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+CD26− elevated, Vβ5a | 4.9 | CD4+CD7−CD26− | — | Yes |
| 17 | Female | 80 | Improved | T4; >50% | Improved | ECP | PCR for TCR-β/γ (−), previous CD4/CD8 ratio of 11.6 | 3.3 | CD4+ | Yes | Yes |
| 18 | Female | 88 | Deceased | T4; >50% | Improved | ECP, Targretin | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD26− elevated, Vβ5c | 3.6 | CD4+CD26− | Yes | Yes |
| 19 | Male | 54 | Stable | T4; >50% | Improved | ECP, Targretin, IFN-α | PCR for TCR-β/γ (+) | 3.1 | CD4+ | Yes | Yes |
| 20 | Male | 65 | Stable | T4; >50% | Improved | ECP, Targretin | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+, CD26− elevated, Vβ3 | 3.4 | CD4+CD7−CD26− | Yes | Yes |
| 21 | Male | 75 | Deceased | T4; >50% | Nonresponsive | ECP, MTX, IFNα | PCR for TCR-β/γ (+), CD45RO elevated, CD4+CD7− elevated, CD4+CD26− elevated | 7.5 | CD4+CD7−CD26− | — | Yes |
| 22 | Male | 63 | Deceased | T4; >50% | Improved | ECP | PCR for TCR-β/γ (−), Vβ5a | 7.6 | CD4+ | — | Yes |
| 24 | Male | 45 | Improved | T2; 10–50% | Improved | ECP, Targretin, IFN-α | PCR for TCR-β/γ (−), previous Vα12.1 | 3.1 | CD4+ | — | Yes |
| 25 | Female | 37 | No data | T4; >50% | Nonresponsive | ECP | PCR for TCR-β/γ (−), previous Vβ20 | 1.2 | CD4+ | — | Yes |
Abbreviations: BSA, body surface area; CTCL, cutaneous T-cell lymphoma; ECP, extracorporeal photopheresis; MTX, methotrexate; SNP, single-nucleotide polymorphism.
Please see Materials and Methods for information regarding collection of data.
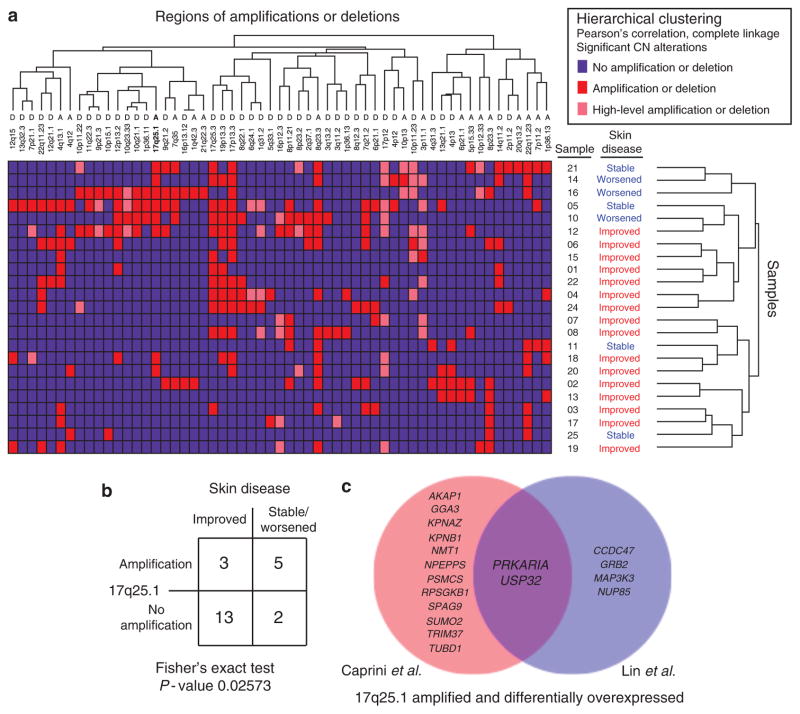
Figure 3. 17q25.1 amplification candidate targets.
(a) Unsupervised analysis of significant copy-number (CN) mutations in cutaneous T-cell lymphoma (CTCL) as defined by GISTIC (Genomic Identification of Significant Targets in Cancer) analysis was performed with hierarchical clustering. Each patient’s skin disease severity after 1 year of treatment is annotated next to the sample number. (b) Fisher’s exact test showed 17q25.1 amplification to be more common in patients with stable/worsened skin disease after 1 year of treatment; however, this was not significant after Bonferroni multiple-hypothesis correction. (c) Using Comparative Marker Selection, differential expression analysis of samples with 17q25.1 amplification versus no amplification reveals candidate targets that are amplified and overexpressed in the data set of Caprini et al. (2009; shown in red), our data set (shown in blue), and the overlap (shown in purple).
Consensus analysis of the leukemic CTCL genome
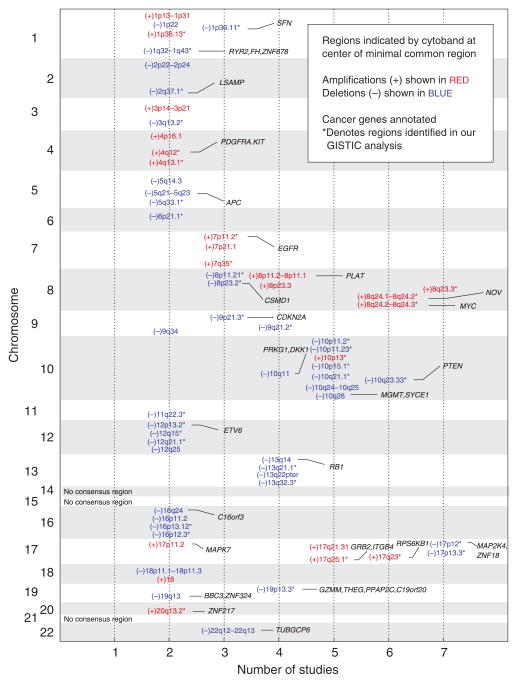
The clinical heterogeneity and relative rarity of CTCL have hindered the genomic characterization of CTCL. To address some of these concerns, we performed a meta-analysis of published data focusing on patients with Sézary syndrome, integrating regions of amplification and deletion from 108 samples comprising 7 studies (Supplementary Table S2 online), including our own, to create a consensus view of the CTCL genome (Figure 2). In total, we determined 21 regions of amplification and 42 regions of deletion. Notably, all of our samples had significant copy-number alterations and many patterns in the consensus analysis mirrored our GISTIC analysis (Figure 1 and Supplementary Table S1 online), thus corroborating our data set and analysis. A majority of minimal common regions were narrowed and defined by our study (38 of 63 regions), and there were no instances where a region was found in all of the studies except ours.
Figure 2. Consensus analysis of the leukemic cutaneous T-cell lymphoma (CTCL) genome.
Analyzing seven CTCL genomic studies (Mao et al., 2002, 2003; Fischer et al., 2004; Vermeer et al., 2008; Caprini et al., 2009; Laharanne et al., 2010), including our own, common regions of amplification (shown in red) and deletion (shown in blue) were defined by at least two studies and the minimal common region was determined by at least one study. The first column indicates the chromosome number. The second column indicates the number of studies including the minimal common region. Cancer-related genes from Beroukhim et al. (2010) are shown in the final column.
Annotating these regions, we identified known oncogenes and tumor suppressors in addition to candidates that, to our knowledge, have not been previously reported. The most significant gain was a broad region on 8q, including MYC. This finding supports results from Vermeer et al. (2008) and Laharanne et al. (2010) suggesting the primary role of MYC in oncogenesis. The second most significant region of gain encompasses a large region on 17q that has yet to be fully studied in CTCL. STAT3 (signal transducer and activator of transcription 3) has previously been suggested as a candidate of gain at this locus (Vermeer et al., 2008); however, in our analysis, STAT3 is not at the peak, implying that there may be other gene targets of the amplification and drivers of oncogenesis on 17q. Although our analysis identified genes involved in many cancers, potential CTCL-specific oncogenes were also considered. A focal amplification of 10p13 including GATA3 (GATA binding protein 3), a transcription factor that promotes T-helper 2 cytokine skewing (Skapenko et al., 2004; Ho et al., 2009), was also identified. The importance of GATA3 in CTCL was previously proposed based on a supervised gene expression analysis (Kari et al., 2003). Its amplification in malignant cells may thus represent one mechanism of its activation and consequent CTCL-mediated suppression of cell-mediated immunity via T-helper 2 cytokine production. Deletions were most commonly found on chromosome 17p with TP53 nearby, chromosome 10 including PTEN and FAS, 13q including RB1, and 9p21.3 including CDKN2A, reaffirming previous findings (Vermeer et al., 2008; Caprini et al., 2009; Laharanne et al., 2010).
17q amplification candidate targets
Ultimately, one of the goals of cancer genomics is tying patient genotype to clinical phenotype to better understand pathogenesis and identify opportunities to therapeutically intervene. After defining the most significant copy-number alterations in CTCL, we used unsupervised pattern recognition algorithms to determine whether any clinical phenotypes were linked to copy-number alterations (Figure 3a). Using hierarchical clustering, we found 17q25.1 amplification to be more common in patients who did not appear to improve and had stable/worsened skin disease after 1 year of treatment (Figure 3b, Fisher’s exact test, P-value 0.026), although this was not significant when using a Bonferroni correction for multiple hypotheses. Seeking to identify candidate targets of amplification at this locus, we integrated gene expression data with our copy-number analysis (Figure 3c). Comparing samples with and without 17q25.1 gain, we identified genes that were both amplified and differentially overexpressed. This analysis was additionally performed in an independent matched data set of CTCL samples with blood involvement (Caprini et al., 2009). Two genes from our consensus analysis (Figure 2) were identified: RPS6KB1 from the analysis of the data set of Caprini et al. (2009), and GRB2 from our data set. RPS6KB1 is involved in the phosphatidylinositol 3-kinase/mammalian target of rapamycin pathways and its amplification and overexpression is believed to affect prognosis in breast cancer (Heinonen et al., 2008). GRB2 is involved in mitogen-activated protein kinase signaling and therapeutic targets are already under development (Bocanegra et al., 2010). Finally, overlapping genes in common included PRKAR1A and USP32 (Figure 3c). It is noteworthy that PRKAR1A regulates cAMP levels and has been found to induce 3- to 4-fold increased expression of IL-2 when it is overexpressed (Elliott et al., 2004). Moreover, we found that IL-2 receptor-β expression, the receptor subunit that binds IL-2 and has been specifically linked to proto-oncogene induction (Manoukian et al., 2009), was concordantly highest in patients with PRKAR1A amplifications in comparison with those without amplifications. Thus, we consider that PRKAR1A amplification may represent one mechanism of IL-2 pathway activation, potentially enhancing malignant T-cell proliferation through upregulation of JAK/STAT, phosphatidylinositol 3-kinase/AKT, and mitogen-activated protein kinase pathways (Elliott et al., 2004; Manoukian et al., 2009).
Genomic comparison of leukemic CTCL cell lines and leukemic CTCL patient samples
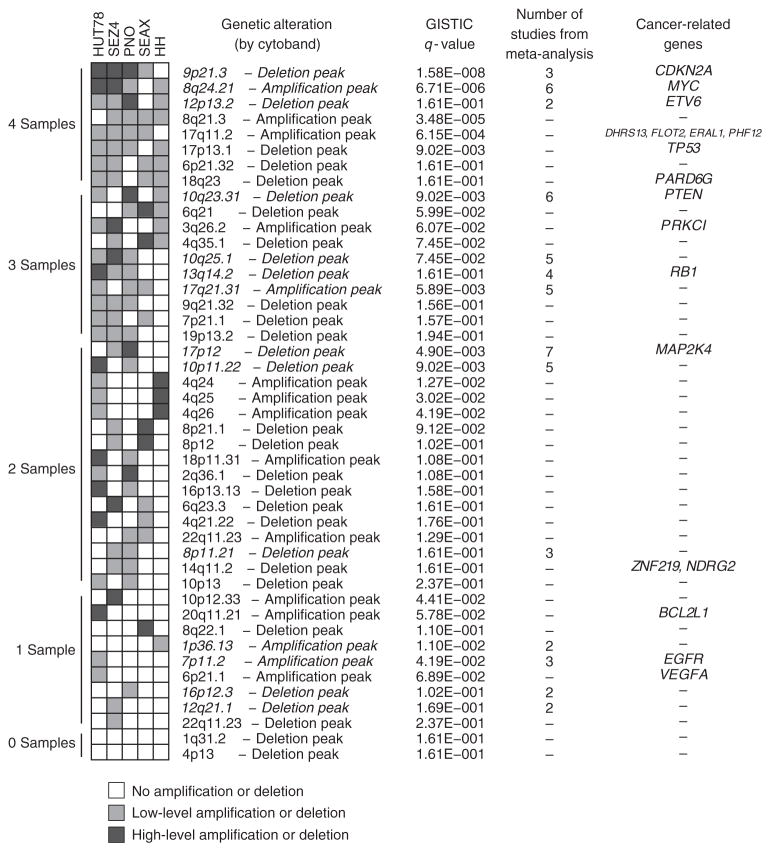
Given the challenges of isolating malignant cells in CTCL, and the difficulty in culturing cells from patients, many investigators have used established CTCL lines for in vitro studies. A major concern of cell line models is the potential dissimilarity between cell lines, which have undergone multiple passages and sometimes require additional growth factors, and primary patient samples. To address this question, we generated DNA copy-number and mRNA expression data for five established cell lines derived from the peripheral blood of Sézary syndrome patients: HH, HUT78, PNO, SeAx, and Sez4 (Supplementary Table S3 online). In Figure 4, we display all significant copy-number alterations in these cell lines defined by GISTIC analysis (details of regions in Supplementary Table S4 online) and compare them with genetic alterations from our 23 patient samples and meta-analysis. Overall, there were many similarities, and the core features of leukemic CTCL, gain of 8q and 17q, and loss of chromosome 10 and 17p were maintained. Among the most significant genetic alterations in common between primary leukemic CTCL samples and the representative cell lines were the following: deletion of 9p21.3 wherein lies CDKN2A (cyclin-dependent kinase inhibitor 2A), and amplification of 8q24.21 wherein lies MYC. Other recurrent regions in common with meta-analysis results included amplification of 17q21.31 and deletion of 10q23.31, 10q25.1, and 13q14.2. There were, nevertheless, also some notable differences. Alterations seen in HH and HUT78 cell lines, which were not significantly observed in primary samples, were gain of a region on 4q24–q26 and loss of 4q35.1 and 6q21 (Figure 4, Supplementary Tables S1 and S4 online). In addition, on a relative scale, chromosome 17p loss and 17q gain were present, but less pronounced, in the cell lines. Thus, although there are several additional alterations in some of the cell lines, our studies revealed that these established cell lines do demonstrate core genomic features of primary leukemic CTCL patients and identified those specific differences.
Figure 4. Genomic alterations in leukemic cutaneous T-cell lymphoma (CTCL) cell lines.
A heatmap view shows the presence of amplifications and deletions in five cell lines derived from Sézary syndrome patients. The heatmap is sorted by the number of cell lines affected, ranging from 0 to 4. Genetic alterations and q-values are derived from a combined GISTIC (Genomic Identification of Significant Targets in Cancer) run of 5 cell lines and 23 leukemic CTCL patient samples. Genetic alterations are listed by the cytoband in the middle of each region and regions in italics were found in the meta-analysis.
DISCUSSION
CTCL continues to offer unique opportunities to study T-cell biology and cancer. Advances in genomic technology along with improved modeling and an increasing number of genomic studies are steadily expanding our knowledge of the mutations underlying both malignant T-cell transformation and the immunogenic aspects of CTCL. Our high-resolution analysis and meta-analysis of patients with leukemic disease helps discern some of the common DNA copy-number alterations characteristic of CTCL, and we will continue to learn more as we study the mutations of patients with variable skin (e.g., patch, plaque, tumor, erythrodermic) and blood involvement, and those ranging from the earliest to the most advanced stages of disease.
Despite their promise, genomic studies in CTCL must navigate many challenges. Foremost are limitations presented by sample size and data quality, which are affected by both the purity and yield of malignant cell isolation. These hurdles, largely due to the low incidence and heterogeneity of disease and technical options available, continue to restrain the potential contribution of genomics to our understanding of CTCL biology. Our desire to understand key genes in pathogenesis led us to incorporate a spectrum of CTCL patients with variable blood involvement, predominantly including those with earlier stages of disease. Focusing on patients with early stages of disease may have allowed us to concentrate on early genetic events and enriches for “driver” mutations, as opposed to “passenger” events that occur as a result of genomic instability.
Despite this potential benefit, characterizing early leukemic disease makes concerns about malignant cell isolation particularly critical to consider. Ultimately, we utilized loss of the immunophenotypic markers CD7 and/or CD27 to enrich for the malignant cells in the population we isolated. The loss of CD7 expression has often been correlated with expansion of clonal T cells in CTCL, and the dominant clone in CTCL patients has been observed to have a CD4+CD7− phenotype (Rappl et al., 2001). Although this phenotype can also be found in reactive T-cell populations, it may nevertheless be the most common major T-cell antigen lost, with 46 to 76% of leukemic CTCL patients showing decreased CD7 expression (Klemke et al., 2008). Similarly, loss of CD26 has also been used as a marker for leukemic CTCL, being found in clonal T-cell populations and estimated to occur in 59.3% of Sézary syndrome patients (Klemke et al., 2008). Notably, CD4+CD26− populations have also been found to significantly correlate with the percentage of Sézary cells within the lymphoid population and, in particular, have been found to be expanded in patients with CD4/CD8 ratios <10 (Bernengo et al., 2001). Neither of these markers are ideal, as there exist normal cells expressing these immunophenotypes; however, in the setting where a CD7− or CD26− population was expanded outside of normal ranges, we effectively used these markers to more specifically select and enrich for the dominant malignant clone.
The potential ramifications of reduced malignant cell purity affect all leukemic CTCL genomic studies and the resulting data must be viewed in light of isolation methodology limitations and the patient population. Gene expression values can be biased as a direct result of cell population contaminants. Moreover, gene expression was also likely affected by the different therapies each patient received; however, the impact of these treatments could not be identified in global clustering. DNA copy-number changes are theoretically more robust, but there can be attenuation of absolute signal from normal cell admixture. Notably, all of our samples had copy-number alterations, even those isolations from patients with relatively small changes in flow cytometric parameters. Nevertheless, had they not (e.g., due to isolation of the incorrect population of cells or inclusion of a patient with undetectable malignancy), our copy-number algorithm was designed to remove samples without any significant copy-number alterations from inclusion in further analysis. Additional concerns about the data purity and quality are also partially alleviated through the meta-analysis, which enabled not only increased sample size but also allowed us to place our study in the context of other published data.
Our consensus analysis included six other genomic studies that focused on Sézary syndrome patients; i.e., those with more advanced leukemic disease. The results of this analysis and the similarities of alterations on a genome-wide level between our data and these other data sets corroborate our own analysis. Indeed, it appears that our samples captured a large proportion of the genomic diversity in leukemic CTCL with some patterns characteristic of malignancy observed even in our patients with early levels of blood involvement. These loci of recurrent gains and losses, observed in both Sézary syndrome patients and our patients, who are skewed toward earlier stages of disease, thus likely contain genes perturbed at an early stage that have a role in pathogenesis. In addition, including data and analysis from 108 patients enabled us to determine recurrent commonalities in DNA copy-number alterations resulting in one of the more comprehensive descriptions of the leukemic CTCL genome. At the same time, we recognize that this view reflects an evolving picture that we hope will be built upon and refined by future studies to further our understanding of the genomic landscape in CTCL.
Although the genomic landscapes are similar from a global view between our patients with early leukemic disease and other studies of patients with more advanced disease, we also make direct comparisons at individual loci. By generating our own high-resolution data set and then using an algorithm that identified driver genetic mutations, we narrowed and verified significant regions of gain and loss in leukemic CTCL. Our analysis confirmed findings from previous studies such as the significance of 8q amplification including MYC, chromosome 10 loss including PTEN and FAS, 9p21.3 deletion including CDKN2A, 13q deletion including RB1, and 17p loss including TP53. In addition, we began to narrow regions with 38 of the 63 regions in the meta-analysis refined by our analysis. In particular, we focused on 17q and, through integration of gene expression data, identified genes of interest that, to our knowledge, have not been previously reported.
We also notably discovered copy-number amplifications within or adjacent to KIT, EGFR, and VEGFA. Cancer is increasingly classified and additionally defined by key mutations. The discovery of these amplifications obligates further study as to whether a subset of CTCL patients may benefit from targeted therapies, in addition to any potential roles these genes and their products may have in cutaneous biology. Clinical data offer the possibility of correlating genotype to clinical phenotype to help delineate important biology. Unfortunately, our analyses were limited by our sample number and no significant correlations could be made.
In our cluster analysis of significant copy-number alterations, we found 17q25.1 amplification to be more common in patients with stable/progressive skin disease 1 year after treatment than those who did not improve; however, the association was not significant after multiple hypothesis correction. Additional confounders include the multiple treatment options that these patients received as seen in Table 1. Nevertheless, 17q25.1 was the second most significant region of amplification, making it an extreme statistical outlier. This, together with the fact that a total of ~44% samples from our consensus analysis included a 17q25 gain, suggests that there are genes at or near this locus driving oncogenesis. Using orthogonal genomic expression data focusing on samples that are amplified and differentially over-expressed in two data sets, PRKAR1A was nominated from our list of candidates. In our cell line data, PRKAR1A had among the highest mean expression of all genes on chromosome 17 (ranked 11th of 1235 genes; data not shown) and indeed the entire genome (98.7th percentile of expression; data not shown). Given the importance of the IL-2 pathway in CTCL and the availability of denileukin difitox, PRKAR1A represents an interesting target meriting in vitro validation and further study. It is important to note, however, that broad gain of 17q suggests multiple oncogenic targets and therefore PRKAR1A is likely not the sole focus of amplification. Indeed, GRB2 and RPS6KB1 represent other candidates in addition to STAT3. Thus, although the potential role of PRKAR1A in activating the IL-2 pathway is conceivable, more importantly, our analysis provides a framework with which to prioritize future functional work. Hopefully, future studies with full clinical data and larger numbers will enable informative clinical correlations.
Looking toward the potential development of biomarkers and targeted therapeutics, functional validation of genomic hypotheses will require robust, genetically characterized in vitro and in vivo models. We hope to have aided this effort through our profiling of five established cell lines from Sézary syndrome patients. Although these lines contain some commonalities with primary leukemic patients, there are also additional genetic alterations not observed in our meta-analysis of 108 patient samples. Whether these genetic alterations represent a small proportion of true CTCL biology or whether they reflect new mutations from selection under culture is unclear. Thus, these cell lines should not be used blindly. Individual experiments will hopefully benefit from our genomic annotation, and together they may serve as an initial framework in which to embark upon functional validation of genomic hypotheses and potential pharmacogenomic endeavors. Finally, as more studies are published and sample size grows, the ability to perform meta-analyses and correlate genotypes to clinical phenotypes will reveal additional avenues and insights into the interface of cancer biology and immunology in this malignancy.
MATERIALS AND METHODS
Patient selection and clinical data collection
Genomic data (DNA data from 23 patients, mRNA data from 12 patients, and 11 patients with both) was generated from the peripheral blood of 24 patients at the Yale Cutaneous Lymphoma Center clinic over a 3-month period in 2008. All patients were treated with extracorporeal photopheresis on a monthly basis. Although the primary focus of study was on patients with early blood involvement, several patients with advanced disease were also included (exclusion and inclusion criteria and rationale for patient selection are detailed in Supplementary Information online). Some patients additionally received therapies such as bexarotene, methotrexate, IFN-α, or vorinostat (Table 1). Briefly, criteria for study inclusion included at least 10% body surface involvement (T2) skin disease, as well as demonstration of peripheral blood involvement by one or more of the following: (1) positive PCR for TCR-β/γ in blood, or (2) expansion of CD4+CD7−, CD4+CD26−, or CD45RO populations outside of normal ranges, or (3) Vα or β family expansion outside of normal ranges. The majority of patients showed limited blood involvement (e.g., CD4/CD8 ratios <10), although patients 5, 10, and 14 fulfilled a diagnosis of Sézary syndrome. Written informed consent was obtained in accordance with the Declaration of Helsinki Principles and approval was obtained from the Yale School of Medicine Human Investigation Committee. Clinical information was gathered at the time of sample collection and 1 year later (Table 1 and Supplementary Table S5 online).
Isolation of enriched malignant cell populations
In the final cycle of leukapheresis before 8-methoxypsoralen exposure, lymphocyte-enriched blood was drawn from each patient, and whole blood was drawn from age- and sex-matched controls. All samples were processed similarly. Blood was layered on Isolymph and processed as described by the manufacturer (CTL Scientific Supply, Deer Park, NY), and detailed in Supplementary Information online. Using flow cytometric parameters, expanded populations representing ≥20% of the peripheral blood were identified and sorted for CD4+CD7−, CD4+CD26−, or CD4+CD7−CD26− populations. Otherwise, the CD4− population was isolated. Negative selection followed by positive selection was performed using the Magnetic-bead Antibody Cell Sorting LS column and magnet following the manufacturer’s instruction (Miltenyi Biotech, Auburn, CA), and detailed in the Supplementary Information online. FACS was performed after isolation to confirm purity ≥70% (mean 96%, median 99% CD4+).
Cell lines and culture conditions
HUT78, HH, SeAx, Sez4, and PNO were kindly provided by collaborators and were originally derived from the peripheral blood of Sézary syndrome patients (please see Supplementary Table S3 online for details). Each was maintained in culture at 37 °C, 5% CO2, in specific media described in Supplementary Table S3 online.
Genomic data generation
From the freshly isolated cells and cell lines, RNA was extracted using Qiagen RNeasy kit and stored in RNAlater (Qiagen, Valencia, CA), and DNA was extracted using Puregene (Gentra, Valencia, CA) and stored in TE buffer. After standard quality control measurements and quantification, 500 ng of genomic DNA was input for human SNP 6.0 arrays and 300 ng of total RNA was input for human Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA) at the Keck Microarray Core facility, according to the manufacturer’s specifications. Details about data access, assays, and quality controls are in Supplementary Information online.
Copy-number analysis
Genomic data were processed using algorithms written in MATLAB and packages in GenePattern (Reich et al., 2006). Affymetrix. CEL files were converted to .SNP files by an adaption of SNP File Creator for SNP 6.0. GISTIC preprocessing and normalization were done as described previously (Beroukhim et al., 2007; Lin et al., 2008). GISTIC parameters are further detailed in Supplementary Information online.
Consensus analysis
Studies used in the consensus analysis focused on CTCL patients with blood involvement, specifically Sézary syndrome (Supplementary Table S2 online). Lower-resolution studies with <10 patients included all available data. Higher-resolution studies with >10 patients only included regions identified by each study’s respective authors. Common regions were defined by at least two studies and minimal common regions were defined by at least one study. Cancer-related genes were annotated from genes identified in Beroukhim et al., 2010.
Cluster analysis
Significant regions of amplifications and deletions defined by GISTIC were analyzed using hierarchical clustering (Pearson’s correlation, complete linkage; Figure 3a, detailed in Supplementary Information online). 17q25.1 was identified through visual analysis of clusters and with Fisher’s exact test (Figure 3b).
Gene expression integration with DNA copy number
Gene expression was integrated with copy number through Comparative Marker Selection (parameters in Supplementary Information online). Samples with an amplification in the region were compared with those without an amplification to identify differentially expressed genes (Figure 3c). GSE17601 (Caprini et al., 2009) was downloaded with GEOImporter and annotated using GeneCruiser.
Cell line analysis
Cell line SNP array data were processed as described above in copy-number analysis section. A heatmap of 28 samples (5 cell lines and 23 patient samples) was generated from the all_lesions file as part of standard GISTIC output. Cancer-related genes were annotated from genes identified in Beroukhim et al. (2010).
Supplementary Material
Acknowledgments
This study was supported by (1) Argall and Anna Hull Cancer Research Award from the Yale Cancer Center, (2) Skin Cancer Foundation Research grant, (3) American Skin Association Medical Student Award, (4) Etta S. Chidsey Award in Cancer Research, (5) Yale School of Medicine Student Fellowship, and (6) Affymetrix to WML as part of their Collaborators in Cancer Research Program. Affymetrix had no scientific involvement in this study. We also thank Martine Bagot, Armand Bensussan, and Anne Marie-Cardine of INSERM; Eric Vonderheid and Alain Rook of University of Pennsylvania; Emmanuel Contassot and Lars French of University Hospital of Zurich; and Francine Foss of Yale University for their generosity in providing CTCL cell lines.
Abbreviations
- CTCL
cutaneous T-cell lymphoma
- GISTIC
Genomic Identification of Significant Targets in Cancer
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
References
- Berger CL, Edelson RL. Current concepts of the immunobiology and immunotherapy of cutaneous T cell lymphoma: insights gained through cross-talk between the clinic and the bench. Leuk Lymphoma. 2003;44:1697–703. doi: 10.1080/1042819031000104033. [DOI] [PubMed] [Google Scholar]
- Berger CL, Tigelaar R, Cohen J, et al. Cutaneous T-cell lymphoma: malignant proliferation of T-regulatory cells. Blood. 2005;105:1640–7. doi: 10.1182/blood-2004-06-2181. [DOI] [PubMed] [Google Scholar]
- Bernengo MG, Novelli M, Quaglino P, et al. The relevance of the CD4+ CD26− subset in the identification of circulating Sézary cells. Br J Dermatol. 2001;144:125–35. doi: 10.1046/j.1365-2133.2001.04014.x. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocanegra M, Bergamaschi A, Kim YH, et al. Focal amplification and oncogene dependency of GAB2 in breast cancer. Oncogene. 2010;29:774–9. doi: 10.1038/onc.2009.364. [DOI] [PubMed] [Google Scholar]
- Caprini E, Cristofoletti C, Arcelli D, et al. Identification of key regions and genes important in the pathogenesis of Sézary syndrome by combining genomic and expression microarrays. Cancer Res. 2009;69:8438–46. doi: 10.1158/0008-5472.CAN-09-2367. [DOI] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Shanks RA, Khan IU, et al. Down-regulation of IL-2 production in T lymphocytes by phosphorylated protein kinase A-RIIβ. J Immunol. 2004;172:7804–12. doi: 10.4049/jimmunol.172.12.7804. [DOI] [PubMed] [Google Scholar]
- Fischer TC, Gellrich S, Muche JM, et al. Genomic aberrations and survival in cutaneous T cell lymphomas. J Invest Dermatol. 2004;122:579–86. doi: 10.1111/j.0022-202X.2004.22301.x. [DOI] [PubMed] [Google Scholar]
- Girardi M, Knobler R, Edelson R. Selective immmunotherapy through extracorporeal photochemotherapy: yesterday, today, and tomorrow. Hematol Oncol Clin North Am. 2003;17:1391–403. doi: 10.1016/s0889-8588(03)00106-0. [DOI] [PubMed] [Google Scholar]
- Heinonen H, Nieminen A, Saarela M, et al. Deciphering downstream gene targets of PI3K/mTOR/p70S6K pathway in breast cancer. BMC Genomics. 2008;9:348. doi: 10.1186/1471-2164-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–35. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–52. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- Kari L, Loboda A, Nebozhyn M, et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med. 2003;197:1477–88. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke CD, Brade J, Weckesser S, et al. The diagnosis of Sézary syndrome on peripheral blood by flow cytometry requires the use of multiple markers. Br J Dermatol. 2008;159:871–80. doi: 10.1111/j.1365-2133.2008.08739.x. [DOI] [PubMed] [Google Scholar]
- Klemke CD, Mansmann U, Poenitz N, et al. Prognostic factors and prediction of prognosis by the CTCL Severity Index in mycosis fungoides and Sézary syndrome. Br J Dermatol. 2005;153:118–24. doi: 10.1111/j.1365-2133.2005.06676.x. [DOI] [PubMed] [Google Scholar]
- Laharanne E, Oumouhou N, Bonnet F, et al. Genome-wide analysis of cutaneous T-cell lymphomas identifies three clinically relevant classes. J Invest Dermatol. 2010;130:1707–18. doi: 10.1038/jid.2010.8. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–73. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukian G, Hagemeister F. Denileukin diftitox: a novel immunotoxin. Expert Opin Biol Ther. 2009;9:1445–51. doi: 10.1517/14712590903348135. [DOI] [PubMed] [Google Scholar]
- Mao X, Lillington D, Scarisbrick JJ, et al. Molecular cytogenetic analysis of cutaneous T-cell lymphomas: identification of common genetic alterations in Sézary syndrome and mycosis fungoides. Br J Dermatol. 2002;147:464–75. doi: 10.1046/j.1365-2133.2002.04966.x. [DOI] [PubMed] [Google Scholar]
- Mao X, Lillington DM, Czepulkowski B, et al. Molecular cytogenetic characterization of Sézary syndrome. Genes Chromosomes Cancer. 2003;36:250–60. doi: 10.1002/gcc.10152. [DOI] [PubMed] [Google Scholar]
- Olsen EA. Interferon in the treatment of cutaneous T-cell lymphoma. Dermatol Ther. 2003;16:311–21. doi: 10.1111/j.1396-0296.2003.01643.x. [DOI] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Rappl G, Muche JM, Abken H, et al. CD4(+)CD7(−) T cells compose the dominant T-cell clone in the peripheral blood of patients with Sézary syndrome. J Am Acad Dermatol. 2001;44:456–61. doi: 10.1067/mjd.2001.110900. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Schechter GP, Sausville EA, Fischmann AB, et al. Evaluation of circulating malignant cells provides prognostic information in cutaneous T cell lymphoma. Blood. 1987;69:841–9. [PubMed] [Google Scholar]
- Skapenko A, Leipe J, Niesner U, et al. GATA-3 in human T cell helper type 2 development. J Exp Med. 2004;199:423–8. doi: 10.1084/jem.20031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer MH, van Doorn R, Dijkman R, et al. Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Res. 2008;68:2689–98. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Vonderheid EC, Pena J, Nowell P. Sézary cell counts in erythrodermic cutaneous T-cell lymphoma: implications for prognosis and staging. Leuk Lymphoma. 2006;47:1841–56. doi: 10.1080/10428190600709655. [DOI] [PubMed] [Google Scholar]
- Weber BL. Cancer genomics. Cancer Cell. 2002;1:37–47. doi: 10.1016/s1535-6108(02)00026-0. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemze R, Meijer CJ. Classification of cutaneous T-cell lymphoma: from Alibert to WHO-EORTC. J Cutan Pathol. 2006;33(Suppl 1):18–26. doi: 10.1111/j.0303-6987.2006.00494.x. [DOI] [PubMed] [Google Scholar]
- Yawalkar N, Ferenczi K, Jones DA, et al. Profound loss of T-cell receptor repertoire complexity in cutaneous T-cell lymphoma. Blood. 2003;102:4059–66. doi: 10.1182/blood-2003-04-1044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.