Abstract
Thiol oxidation to disulfides and the reverse reaction, i.e., disulfide reduction to free thiols, are under the control of catalysts in vivo. Enzymatically assisted thiol-disulfide chemistry is required for the biogenesis of all energy-transducing membrane systems. However, until recently, this had only been demonstrated for the bacterial plasma membrane. Long considered to be vacant, the thylakoid lumen has now moved to the forefront of photosynthesis research with the realization that its proteome is far more complicated than initially anticipated. Several lumenal proteins are known to be disulfide bonded in Arabidopsis, highlighting the importance of sulfhydryl oxidation in the thylakoid lumen. While disulfide reduction in the plastid stroma is known to activate several enzymatic activities, it appears that it is the reverse reaction, i.e., thiol oxidation that is required for the activity of several lumen-resident proteins. This paradigm for redox regulation in the thylakoid lumen has opened a new frontier for research in the field of photosynthesis. Of particular significance in this context is the discovery of trans-thylakoid redox pathways controlling disulfide bond formation and reduction, which are required for photosynthesis.
Keywords: thylakoid lumen, photosynthesis, disulfide, thioredoxin
Thiol-disulfide chemistry refers to the oxidation of thiols (sulfhydryls) into disulfides and also the reversible reaction, i.e., the reduction of the disulfides to free thiols. In vivo, both chemical reactions are catalyzed by dedicated enzymes in cellular compartments where disulfide bonds and reduced thiols in protein targets need to be maintained at all times. Thiol-disulfide chemistry has been best studied in bacteria where it is required for the biogenesis of the periplasmic compartment (Denoncin and Collet, 2013). It has now become apparent that catalyzed thiol-disulfide reactions also operate in the mitochondrial intermembrane space (IMS) and the thylakoid lumen, which are topologically equivalent to the bacterial periplasm. In mitochondria, the Mia40/Erv1 proteins were discovered to be key enzymes of a disulfide relay system driving the import of cysteine-rich proteins into the IMS by an oxidative folding mechanism (Herrmann and Riemer, 2012). The thylakoid lumen has long been viewed as a vacant compartment, with the exception of the oxygen evolving complex (OEC) and some of the redox carriers (e.g., cytochrome f, Rieske, and plastocyanin-or its substitute cytochrome c6) involved in electron transfer during the light reactions of photosynthesis. This dogma has now been revised with the realization that numerous molecules reside in the thylakoid lumen in addition to the previously known photosynthetic proteins. Proteomics studies revealed that the lumenal proteome (~80–200 proteins) is far more complicated than initially anticipated and includes proteases, chaperones, isomerases, redox enzymes, and other proteins of unknown activities (Kieselbach and Schröder, 2003). It is possible that the proteins revealed by proteomics regulate photosynthesis or other yet-to-be discovered processes unrelated to photosynthesis. A recent study demonstrated that lumenal PPD5 is not involved in photosynthetic electron transfer reactions, but rather controls the synthesis of strigonolactone, a plant hormone regulating axillary bud formation (Roose et al., 2011). There is mounting evidence that several lumenal proteins, including components of the photosynthetic chain, contain one or several disulfide bonds (Kieselbach, 2013; Table 1). However the functional importance of the disulfide(s) has only been examined for a small number of these proteins. The molecular mass of disulfide bonded proteins in the thylakoid lumen range from 10 to 55 kDa and the disulfide bond forming cysteines do not appear to occur in a motif. This is at contrast with most of the disulfide bonded molecules in the mitochondrial IMS which are small proteins (6–18 kDa) containing two disulfide bonds in either a C(X)3C or a C(X)9C motif (Sideris and Tokatlidis, 2010). Interestingly, while disulfide bond reduction in stroma-localized targets serves to activate several enzymes (Meyer et al., 2009), it appears that it is the reverse reaction, i.e., disulfide bond formation, that is required for the activities of several lumen-resident proteins (Buchanan and Luan, 2005).
Table 1.
Lumenal disulfide bonded proteins in Arabidopsis.
| protein | Gene locus | Classification | Function |
|---|---|---|---|
| PsbO1 (2 cys, 1 SS) | AT5G66570 | PSII subunit | Oxygen evolution |
| PsbO2 (2 cys, 1 SS) | AT3G50820 | PSII subunit | Oxygen evolution |
| PSI-N (4 cys, 2 SS) | AT5G64040 | PSI subunit | Regulation of PSI activity? |
| Rieske (2 cys, 1 SS) | AT4G03280 | Cytochrome b6f subunit | Electron transfer in b6f complex |
| Cyt c6A (2 cys, 1 SS) | AT5G45040 | c-type cytochrome | Unknown |
| STT7/STN7 (2 cys, 1 SS) | AT1G68830 | Kinase | State transition |
| FKBP13 (4 cys, 2 SS) | AT5G45680 | Peptidyl-prolyl cis-trans isomerase | Unknown |
| FKBP20-2 (2 cys, 1 SS) | AT3G60370 | Peptidyl-prolyl cis-trans isomerase | PSII supercomplex assembly |
| PrxQ (2 cys, 1 SS) | AT3G26060 | Peroxiredoxin | Unknown |
| VDE (13 cys, ≥4 SS) | AT1G08550 | Violaxanthin de-epoxidase | Photoprotection |
| PPD6 (2 cys, 1SS) | AT3G56650 | PsbP domain protein | Unknown |
| TL15 (2 cys, 1 SS) | AT2G44920 | Pentapeptide repeat | Unknown |
| TL17 (4 cys, 2 SS) | AT5G53490 | Pentapeptide repeat | Unknown |
| TL20.3 (4 cys, ≥1 SS) | AT1G12250 | Pentapeptide repeat | Unknown |
| TL29 (2 cys, 1 SS) | AT4G09010 | Ascorbate peroxidase | Unknown |
| CtD1 (5 cys, ≥1 SS) | AT4G17740 | Protease | Processing of D1 subunit |
| CtD1-like (4 cys, ≥1 SS) | AT5G46390 | Protease | Unknown |
| Deg5 (2 cys, 1 SS) | AT4G18370 | Protease | Degradation of lumenal proteins? |
The number of lumenal cysteines (cys) that can form an intramolecular disulfide and the number of disulfides (SS) are indicated in parenthesis. The number of disulfide is experimentally determined. Bolded font indicates that the presence and the number of the disulfide has been confirmed. “≥n SS” indicates that the protein contain at least n disulfide(s). Experimental evidence for the presence of disulfides comes from a combination of thiol trapping/labeling experiments, structure determination or the finding that the enzymatic activity is sensitive to disulfide reducing agents such as DTT or abolished by site-directed mutagenesis of conserved cysteine residues. Several single cysteine containing lumenal proteins (not indicated here) were recovered in thiol trapping experiments, suggesting that intermolecular disulfides can be formed. See references (Hall et al., 2010; Hall, 2012; Kieselbach, 2013).
In bacteria, the requirement for disulfide bond reduction in the periplasmic space was established mainly through studies of cytochrome c maturation. Cytochromes c are metalloproteins with one or several hemes covalently linked to a CXXCH motif on the protein (Thony-Meyer, 1997; Hamel et al., 2009). The need for a disulfide-reducing activity in the context of bacterial cytochrome c assembly seemed obvious because the periplasmic space is also the compartment where cysteine-containing proteins are oxidized by dedicated catalysts (Denoncin and Collet, 2013). The current view is that the CXXCH motif is oxidized after translocation of apocytochromes c to the periplasmic space and then reduced by a reducing pathway to provide free sulfhydryls for ligation with heme (Bonnard et al., 2010; Sanders et al., 2010). This question had received little attention in the context of plastid cytochromes c because thiol-disulfide chemistry as a catalyzed process was not believed to take place in the thylakoid lumen, the compartment where heme attachment to apocytochromes c occurs. The discovery of trans-thylakoid redox pathways controlling disulfide bond formation and reduction in Chlamydomonas and Arabidopsis has now changed this perception.
TWO DISULFIDE-REDUCING PATHWAYS OPERATE IN THE LUMEN
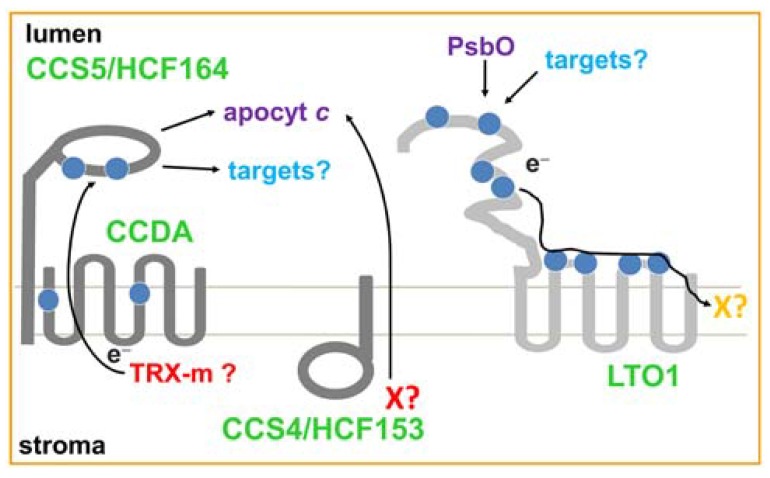
A central component of the bacterial disulfide-reducing pathways is the thiol-disulfide oxido-reductase of the DsbD family (Cho and Collet, 2013). In bacteria, members of this family (CcdA, DsbD, ScsB) are cytoplasmic membrane proteins conveying reducing power from the cytosol to the active sites of several target molecules in the periplasm. Reducing power is transferred across the cytoplasmic membrane through sequential thiol-disulfide exchanges (Cho and Collet, 2013). One target required for cytochrome c assembly is the membrane-bound, periplasm-facing, thioredoxin-like protein (CcmG/ResA/CcsX). CcmG/ResA/CcsX is postulated to reduce a disulfide in the CXXCH heme-binding site of apocytochrome c prior to heme ligation to the cysteines (Bonnard et al., 2010; Sanders et al., 2010). In the plastid lumen, the involvement of a disulfide-reducing pathway was first suspected based on the presence of CCDA, an ortholog of bacterial CcdA from the DsbD family, at the thylakoid membrane (Page et al., 2004; Motohashi and Hisabori, 2010; Figure 1). Another component in the plastid is HCF164 (High Chlorophyll Fluorescence), a membrane-anchored, lumen-facing, thioredoxin-like protein with similarity to bacterial CcmG/ResA/CcsX (Lennartz et al., 2001; Figure 1). The disulfide-reducing activity of HCF164 was inferred from the fact that a recombinant form of its lumenal domain displays disulfide reductase activity in vitro (Lennartz et al., 2001; Motohashi and Hisabori, 2006). Loss of function of CCDA or HCF164 in Arabidopsis produces a photosynthetic-deficient phenotype due to a defect in cytochrome b6f complex assembly (Lennartz et al., 2001; Page et al., 2004). However, the biochemical activity and site of action of CCDA and HCF164 in the assembly process remained unknown until the characterization of the Chlamydomonas ccs4 and ccs5 mutants (Xie et al., 1998). The ccs4 and ccs5 mutants (ccs for cytochrome c synthesis), which are partially photosynthetic deficient, display a block in the conversion of apo to holoform of lumen resident cytochromes c, namely membrane-bound cytochrome f and soluble cytochrome c6. The ccs mutants are deficient for cytochrome b6f assembly, which is dependent on cytochrome f, the thylakoid membrane-bound cytochrome c. This assembly defect is at the step of heme attachment to apocytochromes c, a chemical reaction taking place in the thylakoid lumen (Howe and Merchant, 1992, 1993, 1994; Xie et al., 1998). The Chlamydomonas CCS5 gene was cloned and shown to encode a thioredoxin-like protein with similarity to Arabidopsis HCF164 (Gabilly et al., 2010; Figure 1). Arabidopsis HCF164 can complement the cytochrome c assembly defect when expressed from the plastid genome of the Chlamydomonas ccs5 mutant, demonstrating that HCF164 and CCS5 are functionally equivalent. This also suggests that the defect in cytochrome b6f assembly in the Arabidopsis hcf164 mutant is caused by a block in the conversion of apo to holocytochrome f. The role for CCS5 as an apocytochrome c disulfide reductase in cytochrome c assembly was formulated based on the following findings: (a) the ccs5-null mutant can be chemically rescued in vivo by application of exogenous thiols (such as DTT); (b) CCS5 interacts with plastid apocytochromes c in yeast two-hybrid assays; and c) a recombinant form of CCS5 is able to reduce a disulfide bonded CXXCH in apocytochrome c in vitro (Gabilly et al., 2010). The proposed model is that the thiol-disulfide oxido-reductase CCDA and the thioredoxin-like protein CCS5/HCF164 define a trans-thylakoid pathway for the delivery of reductants from the stroma to the lumen (Lennartz et al., 2001; Page et al., 2004; Motohashi and Hisabori, 2006; Gabilly et al., 2010; Motohashi and Hisabori, 2010; Gabilly et al., 2011). This pathway is required for cytochrome b6f assembly but it is conceivable that it also acts as the transducer of reducing power to regulate the thiol-dependent activity of other lumenal targets (Motohashi and Hisabori, 2006). For instance, in vitro and in organello experiments showed that the reduction of the two disulfides in PSI-N (Table 1), a subunit of photosystem I (PSI), is dependent upon HCF164. The source of reducing power on the stromal side is not known but in organello experiments support the role of the thioredoxin Trx-m as a possible electron donor to CCDA and HCF164 (Motohashi and Hisabori, 2010).
FIGURE 1.
FIGURE 1. Trans-thylakoid thiol metabolizing pathways. The disulfide-reducing (CCDA, CCS5/HCF164 and CCS4/HCF153) and thiol-oxidizing pathways (LTO1) are represented in dark and light grey, respectively. Cysteines are represented as blue balls. Electron (e-) routes are indicated by arrows. The topology of CCS4/HCF164 is hypothetical but the positive-inside rule predicts a stromal localization for the C-terminal domain of the protein. CCS5/HCF164 is a membrane anchored lumen-facing thioredoxin-like protein. LTO1 contains a VKOR-like membrane domain and a lumen-facing thioredoxin-like domain. Stromal reductants for the disulfide reducing pathways are indicated in red. The electron acceptor for the thiol-oxidizing pathway is indicated in orange. Thioredoxin-m (Trx-m) is the possible reductant for the CCDA-CCS5/HCF164 pathway. The CCS4/HCF153 pathway might be involved in the transport of a reductant such as glutathione or ascorbate (X?). The final electron acceptor of the LTO1-dependent pathway is not known and is postulated to be a phylloquinone (X?). CCDA and LTO1 topology was deduced using PhoA/LacZ topological reporters (Page et al., 2004; Feng et al., 2011; Karamoko et al., 2011). The thylakoid membrane is shown in light grey.
The Chlamydomonas CCS4 gene encodes a small protein with limited similarity to Arabidopsis HCF153, a thylakoid membrane anchored protein required for the assembly of the cytochrome b6f complex (Lennartz et al., 2006; Gabilly et al., 2011; Figure 1). The CCS4 protein contains an N-terminal hydrophobic stretch that could serve as a membrane anchor and a C-terminal domain rich in charged residues. On the basis of the positive-inside rule that governs the topology of bacterial and thylakoid membrane proteins (von Heijne, 1989; Gavel et al., 1991), the C-terminal domain of CCS4 is predicted to be exposed to the stromal side of the thylakoid membrane. Surprisingly, CCS4 does not display any motif or residue (such as cysteines) suggesting a role in thiol-based redox chemistry. Yet, the thiol-dependent photosynthetic rescue of the ccs4 mutant and the suppression of the ccs4 phenotype by ectopic expression of CCDA, the thiol/disulfide oxido-reductase of the DsbD family, at the thylakoid membrane, confirms the activity of CCS4 in a disulfide-reducing pathway for cytochrome c assembly. Moreover, the CCDA-dependent suppression of the ccs4 mutant confirms the placement role of CCDA in plastid cytochrome c maturation. Indeed, earlier studies in Arabidopsis supported, but did not establish, the requirement of plastid CCDA in the conversion of apo- to holocytochromes c (Page et al., 1997; Motohashi and Hisabori, 2010). Interestingly, the ccs4-null ccs5-null double mutant displays a synthetic phenotype characterized by a complete loss of b6f assembly, an indication that CCS4 and CCS5 are redundant. This functional redundancy suggests that CCS4 might control a different disulfide-reducing pathway than the CCS5/HCF164 dependent one (Figure 1). The CCDA-dependent suppression of the ccs4 mutant can be explained by a compensatory effect due to enhanced expression of the thiol-disulfide oxido-reductase CCDA. At the present time, the activity of CCS4 in such a pathway remains elusive. While the CCDA dependent pathway relies on the transfer of reductants through sequential thiol-disulfide exchanges, it is conceivable that CCS4 controls the transport across the thylakoid membrane of a molecule acting as a reducing agent (Figure 1). In bacteria, operation of such redundant routes for export of reductant to the periplasm have been postulated (Pittman et al., 2005). The nature of the exported reductant can only be speculated upon but glutathione or ascorbate is an obvious candidate. While the presence of glutathione in the thylakoid lumen remains to be established, ascorbate is known to function in this compartment as an alternative electron donor to PSII and a co-factor for violaxanthin de-epoxidase (VDE) dependent photoprotection (Tóth et al., 2013).
DISCOVERY OF A DISULFIDE-FORMING ENZYME IN THE THYLAKOID LUMEN
In the periplasmic space of most proteobacteria, the thiol-oxidizing pathway consists of a disulfide bond catalyzing system defined by soluble DsbA and membrane-bound DsbB (Denoncin and Collet, 2013). DsbA catalyzes disulfide bridge formation in cysteine-containing substrates that are translocated across the membrane into the periplasmic space. DsbB operates by recycling reduced DsbA to its oxidized form with transfer of electrons to quinones, which are membrane-soluble redox carriers in the respiratory chain. The fact that no DsbA or DsbB-like enzymes can be detected in the genomes of photosynthetic eukaryotes or Cyanobacteria, the presumed ancestors of the chloroplast, reinforced the view that disulfide bond formation did not take place in the thylakoid lumen. However, the operation of catalyzed disulfide bond formation in the lumen is supported by the finding that bacterial alkaline phosphatase (PhoA), an enzyme requiring two disulfide bonds for activity, and basic pancreatic trypsin inhibitor BPTI (aprotinin), a molecule containing three disulfide bonds, are active when targeted to this compartment in tobacco (Sone et al., 1997; Bally et al., 2008; Tissot et al., 2008). A novel class of disulfide-forming enzymes with similarity to VKOR (Vitamin K epoxide Oxidoreductase) was recently recognized in some bacterial phyla lacking the typical DsbAB components (including Cyanobacteria) and in all photosynthetic eukaryotes (Dutton et al., 2008; Singh et al., 2008; Grossman et al., 2010). VKOR is well studied for its involvement in the reduction of vitamin K, a phylloquinone required as a co-factor for the γ-carboxylation of clotting factors in blood (Tie and Stafford, 2008). Recent work shows that the enzymatic activity of VKOR is also linked to oxidative folding of proteins in the ER lumen (Rishavy et al., 2011; Schulman et al., 2011). A first indication that the thylakoid lumen houses a thiol-oxidizing pathway came from the identification of a VKOR-like protein in Cyanobacteria (Singh et al., 2008). In vitro reconstitution of disulfide bond formation with the purified cyanobacterial enzyme demonstrated the sulfhydryl oxidase activity of the protein (Li et al., 2010). However, the localization of the protein at the thylakoid membrane and its relevant targets of action in the lumen were not documented (Singh et al., 2008). The identity of the thiol-oxidizing catalyst in the thylakoid lumen of plastids has now emerged through the discovery of LTO1 (Lumen Thiol Oxidoreductase 1), a thylakoid membrane protein containing a VKOR domain fused to a thioredoxin-like moiety (Figure 1; Furt et al., 2010; Feng et al., 2011; Karamoko et al., 2011). Topological studies using bacterial topological reporters established a lumenal location for the LTO1 domains carrying the redox motifs and conserved cysteines (Figure 1). Previous studies with bacterial VKOR-like proteins have demonstrated that the thioredoxin-like domain carries a DsbA-like activity while the VKOR-like central domain is functionally equivalent to DsbB (Singh et al., 2008; Dutton et al., 2010; Wang et al., 2011). In Arabidopsis, loss of LTO1 function is associated with a severe phototrophic growth defect (Karamoko et al., 2011; Lu et al., 2013). Measurements of the photosynthetic activity indicate that lto1 mutants display a limitation in the electron flow from Photosystem II (PSII). In accord with these measurements, lto1 mutants show a severe depletion of several of the structural subunits of PSII (including subunits of the OEC) but no change in the accumulation of the cytochrome b6f complex or PSI and no defect in the activity of ATP synthase. In a yeast two-hybrid assay, the lumen-facing thioredoxin-like domain of LTO1 was shown to interact with PsbO, a lumenal PSII subunit in the OEC known to be disulfide bonded (Table 1). In vitro, the thioredoxin-like domain of LTO1 is able to introduce a disulfide bond in the PsbO target when recombinant forms of the molecules are used. Because the redox state of the sulfhydryls in PsbO was shown to be a determinant for the stability of this subunit and also for PSII accumulation (Burnap et al., 1994; Nikitina et al., 2008; Hall et al., 2010), it is likely that loss of disulfide bond formation in PsbO in the lto1 mutants accounts for the PSII assembly defect. It is not known if the ability of LTO1 to form a disulfide bond in PsbO is linked to the import of this protein into the lumen, similarly to the Mia40-dependent pathway in mitochondria (Herrmann and Riemer, 2012). In organello import experiments showed that PsbO translocates at a different site than PsbP, another subunit of the PSII OEC (Hashimoto et al., 1997). It is plausible that this translocation step is assisted by LTO1 but this awaits experimental testing.
The final electron acceptor of the LTO1-dependent disulfide bond forming pathway is currently unknown (Figure 1). It is likely to be a phylloquinone based on the fact that the Arabidopsis protein reduces phylloquinone in an in vitro enzymatic assay (Furt et al., 2010). The role of phylloquinone as a structural cofactor tightly bound to PSI is well documented (Brettel, 1997). However, the occurrence of a pool of phylloquinone that is not bound to PSI suggests phylloquinone might participate in redox processes in addition to the known electron transfer reactions through PSI (Gross et al., 2006; Lohmann et al., 2006). This pool may act as an electron acceptor for the LTO1-dependent disulfide bond forming pathway in vivo.
OUTLOOK
It is conceivable that catalyzed thiol-oxidation in the lumen extends to other disulfide-bond containing targets in addition to PsbO (Table 1). In vitro experiments suggest that the thioredoxin-like domain of LTO1 is also able to catalyze the formation of the two disulfide bonds in FKBP13 (Lu et al., 2013), a peptidyl-prolyl cis-trans isomerase whose activity is dependent upon sulfhydryl oxidation (Gopalan et al., 2004; Table 1). It is not known if additional enzymes with sulfhydryl oxidase activity besides LTO1 also operate in the thylakoid lumen. In bacteria, the disulfide-reducing pathway is also required to maintain the reduction state of periplasmic oxido-reductases that shuffle disulfide bonds that are incorrectly formed (Kadokura and Beckwith, 2010; Depuydt et al., 2011). The need for disulfide bond isomerization is critical for proteins containing more than two cysteines such as VDE, an enzyme involved in photoprotection whose activity depends on sulfhydryl oxidation (Kanervo et al., 2005; Table 1). Interestingly, the activity of recombinant VDE was initially reported to be low, presumably because of improper protein folding due to incorrect disulfide linkages (Bugos and Yamamoto, 1996; Hieber et al., 2002). Expression in the cytosol of a bacterial strain engineered for disulfide bond formation and isomerization resulted in a high level of VDE activity for the purified enzyme, an indication that isomerization of disulfide bond in the lumen is also likely to be required for yielding an active enzyme (Saga et al., 2010). It is possible that LTO1 exhibits this activity based on the finding that its thioredoxin-like domain is active as a disulfide-bond isomerase in vitro (Lu et al., 2013). Another thylakoid membrane protein displaying disulfide-bond isomerase activity in vitro is LQY1, an enzyme required for the repair and re-assembly of photodamaged PSII (Lu, 2011). However, it is not known if the active site of LQY1 faces the lumenal or stromal side of the thylakoid membrane. Further experimental work is required to establish the molecular identity of the missing redox components defining the thiol-metabolizing pathways, identify their relevant targets of action and understand how they control photosynthesis or other processes in the thylakoid lumen.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by a National Science Foundation grant (MCB-0920062) to Patrice P. Hamel. We thank Dr. Lamb and Dr. Ruiz for critical reading of the manuscript.
REFERENCES
- Bally J., Paget E., Droux M., Job C., Job D., Dubald M. (2008). Both the stroma and thylakoid lumen of tobacco chloroplasts are competent for the formation of disulphide bonds in recombinant proteins. Plant Biotechnol. J. 6 46–61 10.1111/j.1467-7652.2007.00298.x [DOI] [PubMed] [Google Scholar]
- Bonnard G., Corvest V., Meyer E. H., Hamel P. P. (2010). Redox processes controlling the biogenesis of c-type cytochromes. Antioxid. Redox Signal. 13 1385–1401 10.1089/ars.2010.3161 [DOI] [PubMed] [Google Scholar]
- Brettel K. (1997). Electron transfer and arrangement of the redox cofactors in photosystem I. Biochim. Biophys. Acta 1318 322–373 10.1016/S0005-2728(96)00112-0 [DOI] [Google Scholar]
- Buchanan B. B., Luan S. (2005). Redox regulation in the chloroplast thylakoid lumen: a new frontier in photosynthesis research. J. Exp. Bot. 56 1439–1447 10.1093/jxb/eri158 [DOI] [PubMed] [Google Scholar]
- Bugos R. C., Yamamoto H. Y. (1996). Molecular cloning of violaxanthin de-epoxidase from romaine lettuce and expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 93 6320–6325 10.1073/pnas.93.13.6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap R. L., Qian M., Shen J. R., Inoue Y., Sherman L. A. (1994). Role of disulfide linkage and putative intermolecular binding residues in the stability and binding of the extrinsic manganese-stabilizing protein to the photosystem II reaction center. Biochemistry 33 13712–13718 10.1021/bi00250a023 [DOI] [PubMed] [Google Scholar]
- Cho S. H., Collet J. F. (2013). Many roles of the bacterial envelope reducing pathways. Antioxid. Redox Signal. 18 1690–1698 10.1089/ars.2012.4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoncin K., Collet J. F. (2013). Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid. Redox Signal. 19 63–71 10.1089/ars.2012.4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt M., Messens J., Collet J.-F. (2011). How proteins form disulfide bonds. Antioxid. Redox Signal. 15 49–66 10.1089/ars.2010.3575 [DOI] [PubMed] [Google Scholar]
- Dutton R. J., Boyd D., Berkmen M., Beckwith J. (2008). Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. USA 105 11933–11938 10.1073/pnas.0804621105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton R. J., Wayman A., Wei J.-R., Rubin E. J., Beckwith J., Boyd D. (2010). Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc. Natl. Acad. Sci. USA 107 297–301 10.1073/pnas.0912952107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W. K., Wang L., Lu Y., Wang X. Y. (2011). A protein oxidase catalysing disulfide bond formation is localized to the chloroplast thylakoids. FEBS J. 278 3419–3430 10.1111/j.1742-4658.2011.08265.x [DOI] [PubMed] [Google Scholar]
- Furt F., Oostende C., Widhalm J. R., Dale M. A., Wertz J., Basset G. J. (2010). A bimodular oxidoreductase mediates the specific reduction of phylloquinone (vitamin K) in chloroplasts. Plant J. 64 38–46 10.1111/j.1365-313X.2010.04305.x [DOI] [PubMed] [Google Scholar]
- Gabilly S. T., Dreyfuss B. W., Karamoko M., Corvest V., Kropat J., Page M. D., et al. (2010). CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J. Biol. Chem. 285 29738–29749 10.1074/jbc.M109.099069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabilly S. T., Kropat J., Karamoko M., Page M. D., Nakamoto S. S., Merchant S. S., et al. (2011). A novel component of the disulfide-reducing pathway required for cytochrome c assembly in plastids. Genetics 187 793–802 10.1534/genetics.110.125369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y., Steppuhn J., Herrmann R, Von Heijne G. (1991). The ‘positive-inside rule’ applies to thylakoid membrane proteins. FEBS Lett. 282 41–46 10.1016/0014-5793(91)80440-E [DOI] [PubMed] [Google Scholar]
- Gopalan G., He Z., Balmer Y., Romano P., Gupta R., Héroux A., et al. (2004). Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen. Proc. Natl. Acad. Sci. USA 101 13945–13950 10.1073/pnas.0405240101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J., Cho W. K., Lezhneva L., Falk J., Krupinska K., Shinozaki K., et al. (2006). A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J. Biol. Chem. 281 17189–17196 10.1074/jbc.M601754200 [DOI] [PubMed] [Google Scholar]
- Grossman A. R., Karpowicz S. J., Heinnickel M., Dewez D., Hamel B., Dent R., et al. (2010). Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106 3–17 10.1007/s11120-010-9555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. (2012). The Chloroplast Lumen – New Insights into Thiol Redox Regulation and Functions of Lumenal Proteins. Ph.D. thesis, Umeå University, Sweden. [Google Scholar]
- Hall M., Mata-Cabana A., Akerlund H. E., Florencio F. J., Schroder W. P., Lindahl M., et al. (2010). Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 10 987–1001 10.1002/pmic.200900654. [DOI] [PubMed] [Google Scholar]
- Hamel P., Corvest V., Giege P., Bonnard G. (2009). Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim. Biophys. Acta 1793 125–138 10.1016/j.bbamcr.2008.06.017 [DOI] [PubMed] [Google Scholar]
- Hashimoto A., Ettinger W. F., Yamamoto Y., Theg S. M. (1997). Assembly of newly imported oxygen-evolving complex subunits in isolated chloroplasts: sites of assembly and mechanism of binding. Plant Cell 9 441–452 10.1105/tpc.9.3.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. M., Riemer J. (2012). Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J. Biol. Chem. 287 4426–4433 10.1074/jbc.R111.270678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieber D., Bugos R., Verhoeven A., Yamamoto H. (2002). Overexpression of violaxanthin de-epoxidase: properties of C-terminal deletions on activity and pH-dependent lipid binding. Planta 214 476–483 10.1007/s00425-001-0704-2 [DOI] [PubMed] [Google Scholar]
- Howe G., Merchant S. (1992). The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J. 11 2789–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G., Merchant S. (1993). Maturation of thylakoid lumen proteins proceeds post-translationally through an intermediate in vivo. Proc. Natl. Acad. Sci. USA 90 1862–1866 10.1073/pnas.90.5.1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G., Merchant S. (1994). Role of heme in the biosynthesis of cytochrome c6. J. Biol. Chem. 269 5824–5832 [PubMed] [Google Scholar]
- Kadokura H., Beckwith J. (2010). Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid. Redox Signal. 13 1231–1246 10.1089/ars.2010.3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanervo E., Suorsa M., Aro E.-M. (2005). Functional flexibility and acclimation of the thylakoid membrane. Photochem Photobiol. Sci. 4 1072–1080 10.1039/b507866k [DOI] [PubMed] [Google Scholar]
- Karamoko M., Cline S., Redding K., Ruiz N., Hamel P. P. (2011). Lumen thiol oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23 4462–4475 10.1105/tpc.111.089680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach T. (2013). Oxidative folding in chloroplasts. Antioxid. Redox Signal. 19 72–82 10.1089/ars.2012.4582 [DOI] [PubMed] [Google Scholar]
- Kieselbach T, Schröder W. (2003). The proteome of the chloroplast lumen of higher plants. Photosyn. Res. 78 249–264 10.1023/B:PRES.0000006913.86689.f1 [DOI] [PubMed] [Google Scholar]
- Lennartz K., Bossmann S., Westhoff P., Bechtold N., Meierhoff K. (2006). HCF153, a novel nuclear-encoded factor necessary during a post-translational step in biogenesis of the cytochrome bf complex. Plant J. 45 101–112 10.1111/j.1365-313X.2005.02605.x [DOI] [PubMed] [Google Scholar]
- Lennartz K., Plücken H., Seidler A., Westhoff P., Bechtold N., Meierhoff K. (2001). HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13 2539–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Schulman S., Dutton R. J., Boyd D., Beckwith J., Rapoport T. A. (2010). Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 463 507–512 10.1038/nature08720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann A., Schottler M. A., Brehelin C., Kessler F., Bock R., Cahoon E. B., et al. (2006). Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 281 40461–40472 10.1074/jbc.M609412200 [DOI] [PubMed] [Google Scholar]
- Lu Y. (2011). The occurrence of a thylakoid-localized small zinc finger protein in land plants. Plant Signal. Behav. 6 1881–1885 10.4161/psb.6.12.18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Wang H. R., Li H., Cui H. R., Feng Y. G., Wang X. Y. (2013). A chloroplast membrane protein LTO1/AtVKOR involving in redox regulation and ROS homeostasis. Plant Cell Rep. 32 1427–1440 10.1007/s00299-013-1455-9 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Buchanan B. B., Vignols F., Reichheld J.-P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43 335–367 10.1146/annurev-genet-102108-134201 [DOI] [PubMed] [Google Scholar]
- Motohashi K., Hisabori T. (2006). HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 281 35039–35047 10.1074/jbc.M605938200 [DOI] [PubMed] [Google Scholar]
- Motohashi K., Hisabori T. (2010). CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid. Redox Signal. 13 1169–1176 10.1089/ars.2010.3138 [DOI] [PubMed] [Google Scholar]
- Nikitina J., Shutova T., Melnik B., Chernyshov S., Marchenkov V., Semisotnov G., et al. (2008). Importance of a single disulfide bond for the PsbO protein of photosystem II: protein structure stability and soluble overexpression in Escherichia coli. Photosynth. Res. 98 391–403 10.1007/s11120-008-9327-9 [DOI] [PubMed] [Google Scholar]
- Page M. D., Saunders N. F., Ferguson S. J. (1997). Disruption of the Pseudomonas aeruginosa dipZ gene, encoding a putative protein-disulfide reductase, leads to partial pleiotropic deficiency in c-type cytochrome biogenesis. Microbiology 143 3111–3122 10.1099/00221287-143-10-3111 [DOI] [PubMed] [Google Scholar]
- Page M. L. D., Hamel P. P., Gabilly S. T., Zegzouti H., Perea J. V., Alonso J. M., et al. (2004). A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 279 32474–32482 10.1074/jbc.M404285200 [DOI] [PubMed] [Google Scholar]
- Pittman M. S., Robinson H. C., Poole R. K. (2005). A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 280 32254–32261 10.1074/jbc.M503075200 [DOI] [PubMed] [Google Scholar]
- Rishavy M. A., Usubalieva A., Hallgren K. W., Berkner K. L. (2011). Novel insight into the mechanism of the vitamin K oxidoreductase (VKOR). J. Biol. Chem. 286 7267–7278 10.1074/jbc.M110.172213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J. L., Frankel L. K., Bricker T. M. (2011). Developmental defects in mutants of the PsbP domain protein 5 in Arabidopsis thaliana. PLoS ONE 6:e28624. 10.1371/journal.pone.0028624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga G., Giorgetti A., Fufezan C., Giacometti G. M., Bassi R., Morosinotto T. (2010). Mutation analysis of violaxanthin de-epoxidase identifies substrate-binding sites and residues involved in catalysis. J. Biol. Chem. 285 23763–23770 10.1074/jbc.M110.115097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C., Turkarslan S., Lee D.-W., Daldal F. (2010). Cytochrome c~biogenesis: the Ccm system. TIM 18 266–274 10.1016/j.tim.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman S., Wang B., Li W., Rapoport T. A. (2011). Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. USA 107 15027–15032 10.1073/pnas.1009972107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris D. P., Tokatlidis K. (2010). Oxidative protein folding in the mitochondrial intermembrane space. Antioxid. Redox Signal. 13 1189–1204 10.1089/ars.2010.3157 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Bhattacharyya-Pakrasi M., Pakrasi H. B. (2008). Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J. Biol. Chem. 283 15762–15770 10.1074/jbc.M800982200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Kishigami S., Yoshihisa T., Ito K. (1997). Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272 6174–6178 10.1074/jbc.272.10.6174 [DOI] [PubMed] [Google Scholar]
- Thony-Meyer L. (1997). Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61 337–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J. K., Stafford D. W. (2008). Structure and function of vitamin K epoxide reductase. Vitam. Horm. 78 103–130 10.1016/S0083-6729(07)00006-4 [DOI] [PubMed] [Google Scholar]
- Tissot G., Canard H., Nadai M., Martone A., Botterman J., Dubald M. (2008). Translocation of aprotinin, a therapeutic protease inhibitor, into the thylakoid lumen of genetically engineered tobacco chloroplasts. Plant Biotechnol. J. 6 309–320 10.1111/j.1467-7652.2008.00321.x [DOI] [PubMed] [Google Scholar]
- Tóth S. Z., Schansker G., Garab G. (2013). The physiological roles and metabolism of ascorbate in chloroplasts. Physiol. Plant. 148 161–175 10.1111/ppl.12006 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1989). Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341 456–458 10.1038/341456a0 [DOI] [PubMed] [Google Scholar]
- Wang X., Dutton R. J., Beckwith J., Boyd D. (2011). Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase. Antioxid. Redox Signal. 14 1413–1420 10.1089/ars.2010.3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Culler D., Dreyfuss B. W., Kuras R., Wollman F. A., Girard-Bascou J., et al. (1998). Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: one chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]