Abstract
Background
Intraductal papillary mucinous neoplasms (IPMNs) are the most common cystic precursor lesions of invasive pancreatic cancer. The recent identification of activating GNAS mutations at codon 201 in IPMNs is a promising target for early detection and therapy. The purpose of this study was to explore clinicopathological correlates of GNAS mutational status in resected IPMNs.
Methods
Clinical and pathologic characteristics were retrieved on 54 patients in whom GNAS codon 201 mutational status was previously reported (“historical group”, Wu et al. Sci Transl Med 3:92ra66, 2011). In addition, a separate cohort of 32 patients (validation group) was included. After microdissection and DNA extraction, GNAS status was determined in the validation group by pyrosequencing.
Results
GNAS activating mutations were found in 64 % of the 32 IPMNs included in the validation group, compared with a previously reported prevalence of 57 % in the historical group. Overall, 52 of 86 (61 %) of IPMNs demonstrated GNAS mutations in the two studies combined. Analysis of both groups confirmed that demographic characteristics, tumor location, ductal system involvement, focality, size, grade of dysplasia, presence of an associated cancer, and overall survival were not correlated with GNAS mutational status. Stratified by histological subtype, 100 % of intestinal type IPMNs demonstrated GNAS mutations compared to 51 % of gastric IPMN, 71 % of pancreatobiliary IPMNs, and 0 % of oncocytic IPMNs.
Conclusions
GNAS activating mutations can be reliably detected in IPMNs by pyrosequencing. In terms of clinicopathological parameters, only histological subtype was correlated with mutational frequency, with the intestinal phenotype always associated with GNAS mutations.
Intraductal papillary mucinous neoplasm (IPMN) is a cystic neoplasm of the pancreas characterized by intraductal papillary proliferation of neoplastic mucinous cells.1,2 IPMNs arise in the main pancreatic duct or its side branches (or both), can be unifocal or multifocal, and can involve the pancreatic head, body–tail, or, less frequently, the entire gland.1,2 Varying subtypes of differentiation can be seen in the neoplastic cells of IPMNs including gastric, intestinal, pancreatobiliary, and oncocytic subtypes.3 Among pancreatic cystic neoplasms, IPMNs are recognized precursor lesions of pancreatic adenocarcinoma, while others such as serous cystadenomas are benign and unless symptomatic generally do not require treatment.4,5 The preoperative differentiation of IPMNs from other pancreatic cysts can be challenging, and in spite of meticulous assessment, changes to the preoperative diagnosis can be as high as 30 % upon subsequent pathological appraisal.6,7 Even when an IPMN is correctly diagnosed, it is currently not possible to empirically predict its risk of developing an associated cancer; nor is it possible to assess the grade of dysplasia for that lesion unless resection and pathological examination are performed.
In a study by Wu et al.8 a novel next-generation sequencing technology was applied to unravel molecular pathways important for the pathogenesis of IPMNs. Unexpectedly, activating GNAS mutations at codon 201 were identified in approximately two-thirds of these neoplasms. Of note, no GNAS mutations were identified in other pancreatic cystic neoplasms; nor were they identified in conventional pancreatic ductal adenocarcinoma. This suggests that GNAS mutations are specific for the IPMN phenotype.8,9 In this study, we assessed the prevalence of GNAS-activating mutations in an additional set of surgically resected IPMNs using pyrosequencing and correlated GNAS mutational status with clinicopathological characteristics. Our results indicate that activating GNAS mutations are associated with the intestinal subtype morphology and are likely to be an early driver mutation in a significant subset of IPMNs.
MATERIALS AND METHODS
The present study was approved by the institutional review board of Johns Hopkins Medical Institutions. Previously published GNAS mutational status of patients included in the study by Wu et al.8 was retrieved. For all patients included in the above-mentioned study (historical group), clinicopathological characteristics were available. In addition, we reviewed the prospectively maintained Johns Hopkins Department of Surgery clinical database and identified a separate cohort of patients who underwent pancreatic resection at the Johns Hopkins Hospital (validation group). In both cohorts of patients, case selection was further restricted to patients surgically resected in or after the year 1996, when the World Health Organization diagnostic criteria for IPMN were implemented at our institution. For all the patients included in the study, a minimum follow-up of 2 years was available. Patients with cysts in the pancreatic head, uncinate process, or neck underwent pancreaticoduodenectomy. Distal pancreatectomy and splenectomy was performed for neoplasms located in the pancreatic body–tail. In case of main duct IPMNs diffusely involving the duct of Wirsung, a total pancreatectomy was performed. IPMN can be multifocal, and independent clones have been demonstrated in at least a subset of branch duct IPMNs.10 To limit confounding factors related to the analysis of multiple cysts within the same surgical specimen, patients who underwent total pancreatectomy for multifocal branch duct IPMN were excluded. After GNAS mutational status was assessed in the validation group, the whole cohort of patients was dichotomized on the basis of the presence or absence of mutated GNAS in their surgically resected IPMNs. Clinicopathological correlations and survival analysis of GNAS mutational status in IPMNs were performed.
Histopathology
Two experts in pancreatic pathology (RHH and AM) confirmed all histological diagnoses using recent guidelines.11 On the basis of the maximum cytoarchitectural atypia, IPMNs were classified as low grade, intermediate grade, or high grade. IPMNs were classified as gastric, intestinal, pancreatobiliary, or oncocytic subtypes with respect to the predominant phenotypic features of the neoplastic epithelium.12 IPMNs were further classified as noninvasive or as having an associated invasive adenocarcinoma.
Laser-Capture Microdissection
Archival formalin-fixed, paraffin-embedded tissue blocks of selected IPMNs and matched normal pancreatic tissues were collected. For laser-capture microdissection, 10–30 sections (10 μm) were embedded onto UV-treated PALM membrane slides (Carl Zeiss MicroImaging, Inc., Thornwood, NY). After deparaffinization in xylene/ethanol and hematoxylin and eosin staining, small neoplasms or those with substantial inflammatory component were subjected to laser-capture microdissection on a PALM Micro Beam System (Carl Zeiss MicroImaging, Inc., Thornwood, NY). All other lesions were microdissected with a sterile needle under a stereomicroscope (SMZ1500, Nikon, Tokyo, Japan). Approximately 5,000–10,000 cells were collected from each lesion, with an estimated neoplastic cellularity >80 %. In addition, matched nonneoplastic pancreatic tissue was manually macrodissected. Subsequently, DNA was extracted using the QIAamp DNA Micro Kit (Qiagen Inc., Valencia, CA) per the manufacturer's instructions.
GNAS Pyrosequencing
On the basis of previously published findings, GNAS mutational analysis was restricted to the codon 201 hot spot contained within exon 8 because no mutations outside of this codon have been described in IPMNs.13,14 Pyrosequencing was performed as published.15,16 Briefly, DNA extracted from microdissected tissue was PCR amplified on GeneAmp9700 Thermal Cycler (Applied Biosystem) such that each reaction contained: 1× buffer (Qiagen), 1.5 mmol/L MgCl2, 0.2 mmol/L of each dNTP, 5 pmol of forward primer (5′-CCAGACCTTTGCTTTAG ATTGG-3′), 5 pmol of reverse primer (5′-biotin-TCCACC TGGAACTTGGTCTC-3′), 0.8 U of HotStart TaqDNA polymerase (Qiagen), 10 ng of DNA, and dH2O for a 25 μl final volume. Cycling conditions were as follows: 95 °C for 15 min; 38 cycles of 95 °C for 20 s; 53 °C for 30 s; 72 °C for 20 s; and a final extension at 72 °C for 5 min. The amplicons were sequenced with the PyroMark Q24 (Qiagen) with PyroMark Gold reagents (Qiagen) containing 0.3 μmol/L sequencing primer (5′-TTTGTTTCAGGACCTGCTTCGC-3′) and annealing buffer. The nucleotide triphosphate dispensation order for codon 201 was TGCTAGTGTCTGACTCTGATCT.
Statistical Analysis
Patient characteristics were compared between those who had an IPMN that harbored a codon 201 GNAS mutation and those who had an IPMN that did not by Wilcoxon rank sum tests for continuous measures and Fisher's exact test for categorical measures. Pairwise comparisons between various clinical and pathological characteristics and their respective GNAS mutation frequency were performed by Fisher's exact test. Overall survival was calculated as the date of surgery to the date of death or to the date last known alive and estimated using the Kaplan–Meier method. Hazard ratios estimated from Cox proportional hazards models were used to compare overall survival between patients with and without a codon 201 GNAS mutation while controlling for whether the patient had invasive cancer, age at surgery, and histological subtype. Analyses were completed by R software, version 2.15.1.13
RESULTS
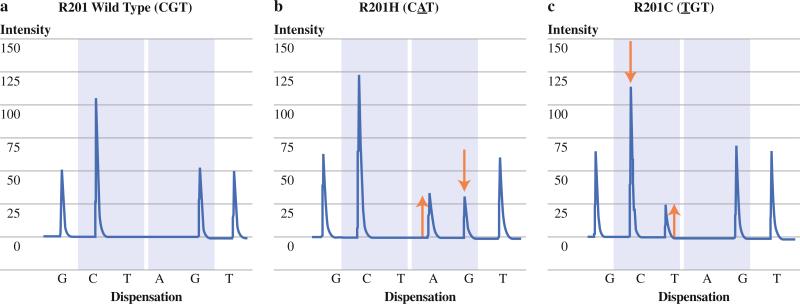
On the bases of the time frame considered for this study (1996–2009) and the exclusion of total pancreatectomy specimens for multifocal disease, 54 IPMNs in the historical group were suitable for inclusion.8 Within this cohort, 31 IPMNs (57 %) harbored a codon 201 GNAS mutation, as assessed by a sensitive PCR/ligation assay.8 An additional cohort of 32 separate patients with IPMNs resected at Hopkins and not previously reported met the aforementioned inclusion criteria (validation group). A total of 21 IPMNs within this cohort (64 %) harbored GNAS-activating mutations (Fig. 1). After combining the two groups, a total of 86 IPMNs were suitable for clinicopathological analysis, of which 52 (61 %) harbored GNAS codon 201 mutations and 34 (39 %) were GNAS wild type.
FIG. 1.
Pyrograms of GNAS mutational status. a Wild type GNAS codon 201 is retained. GNAS codon 201 exhibits missense mutations b CGT to CAT, resulting in amino acid substitution of an arginine with histidine (R201H) and c CGT to TGT, resulting in amino acidic substitution of an arginine with cysteine (R201C)
Two independent statistical correlations were performed. First, the prevalence of any given clinical or pathological trait was compared in the GNAS mutant versus GNAS wild type subsets of IPMNs for statistically significant differences (Table 1). Second, we performed a series of pairwise comparisons between various clinical and pathological characteristics for significant differences in their respective GNAS mutation frequency in order to determine whether the proportion of mutated samples was different between samples belonging to a specific category (Table 2). We found no association with respect to age, gender, history of smoking, alcohol use, and the presence or absence of GNAS mutations (Table 1). Within the mutant GNAS group, 10 IPMNs (29 %) had low-grade dysplasia, while 10 IPMNs (19 %) had low-grade dysplasia within the GNAS wild type group (P = 0.305). Similarly, no statistical difference in the prevalence of either intermediate-grade or high-grade dysplasia was observed between GNAS mutant IPMNs and GNAS wild type IPMNs: 7 patients (21 %) vs. 12 patients (23 %) (P > 0.99), and 17 patients (50 %) vs. 30 patients (58 %) (P = 0.514), respectively. Further, by pairwise comparison of different grades of dysplasia with respect to their mutational status (Table 2), the prevalence of GNAS mutations in IPMNs with low-grade (LG) dysplasia was similar to that of IPMNs with intermediate-dysplasia (IG) or high-grade (HG) dysplasia. Ten of 20 LG (50 %) vs. 12 of 19 IG (63 %) (P = 0.523); and 10 of 20 LG (50 %) vs. 30 of 47 HG (64 %) (P = 0.415). Similarly, no difference was observed when comparing IG IPMNs (12 of 19, 63 %) with HG IPMNs (30 of 47, 64 %), (P > 0.99).
TABLE 1.
Characteristics of patients by GNAS mutation status
| Characteristic | GNAS wild type (n = 34) | GNAS mutant (n = 52) | P |
|---|---|---|---|
| Age, y, median (range) | 71 (48–85) | 71 (35–91) | 0.533 |
| Gender | 0.266 | ||
| Female | 17 (50 %) | 19 (37 %) | |
| Male | 17 (50 %) | 33 (63 %) | |
| Smoking history | 0.235 | ||
| Never | 25 (76 %) | 31 (62 %) | |
| Ever | 8 (24 %) | 19 (38 %) | |
| Unknown | 1 | 2 | |
| Alcohol use | >0.99 | ||
| None | 7 (44 %) | 11 (42 %) | |
| Some | 9 (56 %) | 15 (58 %) | |
| Unknown | 18 | 26 | |
| Dysplasia | |||
| Low grade | 19 (29 %) | 10 (19 %) | 0.305 |
| Intermediate grade | 7 (21 %) | 12 (23 %) | >0.99 |
| High grade | 17 (50 %) | 30 (58 %) | 0.514 |
| Focality | >0.99 | ||
| Unifocal | 23 (79 %) | 37 (77 %) | |
| Multifocal | 6 (21 %) | 11 (23 %) | |
| Unknown | 5 | 4 | |
| Duct | 0.948 | ||
| Main | 12 (38 %) | 18 (41 %) | |
| Mixed | 4 (12 %) | 5 (11 %) | |
| Branch | 16 (50 %) | 21 (48 %) | |
| Unknown | 2 | 8 | |
| Cyst size | >0.99 | ||
| <3 cm | 17 (53 %) | 28 (55 %) | |
| ≥3 cm | 15 (47 %) | 23 (45 %) | |
| Unknown | 2 | 1 | |
| Size, cm, median (range) | 2.2 (0.5–8) | 2.5 (0.6–8) | 0.35 |
| Invasive cancer | 0.809 | ||
| No | 25 (74 %) | 36 (69 %) | |
| Yes | 9 (26 %) | 16 (31 %) | |
| Site | |||
| Pancreatobiliary | 2 (7 %) | 5 (11 %) | 0.694 |
| Gastric | 26 (87 %) | 27 (61 %) | 0.02 |
| Intestinal | 0 | 12 (27 %) | 0.001 |
| Oncocytic | 2 (7 %) | 0 | 0.161 |
| Location | 0.493 | ||
| Distal | 21 (62 %) | 36 (69 %) | |
| Head–neck–uncinate process | 13 (38 %) | 16 (31 %) |
TABLE 2.
Pairwise comparison of mutant GNAS samples by grade of dysplasia, epithelial differentiation, and duct involvement
| Dysplasia grade | n | Mutant, n (%) |
|---|---|---|
| Low-grade dysplasia | 20 | 10 (50) |
| Intermediate-grade dysplasia | 19 | 12 (63) |
| P | 0.523 | |
| Intermediate-grade dysplasia | 19 | 12 (63) |
| High-grade dysplasia | 47 | 30 (64) |
| P | >0.99 | |
| Low-grade dysplasia | 20 | 10 (50) |
| High-grade dysplasia | 47 | 30 (64) |
| P | 0.415 | |
| Gastric | 53 | 27 (51) |
| Pancreatobiliary | 7 | 5 (71) |
| P | 0.432 | |
| Intestinal | 12 | 12 (100) |
| Pancreatobiliary | 7 | 5 (71) |
| P | 0.123 | |
| Oncocytic | 2 | 0 (0) |
| Pancreatobiliary | 7 | 5 (71) |
| P | 0.167 | |
| Intestinal | 12 | 12 (100) |
| Gastric | 53 | 27 (51) |
| P | 0.001 | |
| Oncocytic | 2 | 0 (0) |
| Gastric | 53 | 27 (51) |
| P | 0.491 | |
| Oncocytic | 2 | 0 (0) |
| Intestinal | 12 | 12 (100) |
| P | 0.011 | |
| Mixed duct | 9 | 5 (56) |
| Main duct | 30 | 18 (60) |
| P | >0.99 | |
| Branch duct | 37 | 21 (57) |
| Main duct | 30 | 18 (60) |
| P | 0.809 | |
| Branch duct | 37 | 21 (57) |
| Mixed duct | 9 | 5 (56) |
| P | >0.99 |
P values are for testing whether the proportion of mutated samples is different between patient groups
IPMNs with mutant GNAS and the IPMNs with wild type GNAS had a similar distribution among the main duct, mixed duct, and branch duct types (38, 12 and 50 % vs. 41, 11 and 48 %, respectively; P = 0.948) (Table 1). Nor were any differences observed in pairwise comparisons of the abovementioned categories; thus, 5 out of 9 (56 %) mixed duct IPMNs harbored GNAS mutations, compared with 18 out of 30 (60 %) of main duct IPMNs (P > 0.99) (Table 2). The prevalence of GNAS mutations in branch duct IPMNs (21 of 37, 57 %) was not different from the prevalence observed in main duct IPMNs (18 of 30, 60 %) or with that observed in mixed duct IPMNs (5 of 9, 56 %) (P = 0.809 and P > 0.99, respectively). Median cyst size was similar in the GNAS mutant group (2.2 cm), compared with the wild type GNAS group (2.5 cm). When considering a dimensional cutoff of 3 cm, no statistically significant difference was found between GNAS mutant IPMNs versus GNAS wild type IPMNs (Table 1). Nine IPMNs with mutant GNAS (26 %) had an associated invasive cancer, while 16 (31 %) IPMNs with wild type GNAS harbored an associated invasive component (P = 0.809). Furthermore, IPMNs located in the head–neck–uncinate process had similar prevalence of GNAS mutations compared with those arising in the body or tail of the gland (Table 1).
We also examined the pattern of GNAS mutations in the different histologic subtypes of IPMNs. The pancreatobiliary subtype was observed in 7 % of IPMNs harboring a GNAS mutation compared with a prevalence of 11 % in the wild type group (P = 0.694) (Table 1). The gastric subtype was more frequently observed in IPMNs with wild type GNAS (87 %) compared to IPMNs harboring mutant GNAS (61 %); this difference was statistically significant (P = 0.02). Conversely, the intestinal subtype was frequently observed within the GNAS mutant group (27 %) but never observed in IPMNs with wild type GNAS alleles (P = 0.001). These statistically significant differences persisted in the pairwise comparison between histological subtypes, with 100 % of intestinal and only 51 % of gastric type IPMNs demonstrating GNAS mutations (P = 0.001). With the exception of borderline significance when the intestinal subtype was compared to the oncocytic (P = 0.011), the other pairwise comparisons did not reach statistical significance (Table 2). Finally, Kaplan–Meier survival analysis revealed no significant difference in survival between tumors harboring GNAS mutations and tumors wild type for GNAS (Fig. 2).
FIG. 2.
Kaplan–Meier survival analysis of patients with IPMN according to GNAS mutational status
DISCUSSION
The entire exomes of the four most common cystic neoplasms of the pancreas have been sequenced, and mutations specific for each cyst type have been discovered, suggesting that the clinical evaluation of pancreatic cysts could greatly benefit from new gene-based biomarkers.8,9,17 Among the genetic alterations identified in IPMNs, activating GNAS mutations harbor the most clinically relevant potential, as they are highly prevalent (approximately two thirds of cases) and are limited to a single hot spot codon, 201.
The GNAS gene, located on human chromosome 20q13.3, encodes the alpha subunit (Gsα) of a stimulatory G protein. Mutations in the codon 201 of GNAS result in the constitutive activation of Gsα and its effector adenylate cyclase, ultimately leading to autonomous synthesis of cyclic AMP (cAMP) and uncontrolled growth signaling.14 Somatically acquired GNAS-activating mutations have been identified in several tumor types other than IPMN, such as kidney, thyroid, pituitary, liver, colorectal, and more recently in villous adenomas of the colorectum.18–23
In light of the recognition of GNAS as a definite mountain in the mutational landscape of IPMNs, we endeavored to determine the implications of this mutation on clinical and pathological traits in a series of well-characterized surgically resected IPMNs. We utilized data from our previously published series that assessed GNAS mutation status in IPMN cyst fluid samples using a sensitive PCR/ligation assay (historical group), as well as a separate cohort of IPMNs, in which GNAS codon 201 mutations were analyzed by pyrosequencing (validation group) in order to increase the sample size.8 Overall, the combined mutational prevalence of codon 201 alterations in our series was 61 % (52 of 86 IPMNs). Despite this high prevalence of point mutations in GNAS, most clinicopathological factors examined, as well as survival analysis, did not correlate with either the presence or absence of a mutant allele. Notably, only the pattern of differentiation of the neoplastic epithelium was significantly associated with presence or absence of GNAS mutations. We confirmed that 100 % of the intestinal type IPMNs harbored a codon 201 mutation, while only 51 % of gastric IPMNs (P = 0.001) and 71 % of pancreatobiliary IPMNs (P = 0.123) had mutant GNAS. The association between GNAS mutations and an intestinal differentiation is not unique to IPMNs; a recent study has reported that 83 % of villous adenomas of the colorectum (which, by virtue of their lush fingerlike papillae, are the closest histological mimic of intestinal-type IPMNs) also demonstrate GNAS mutations.23 In that series, none of the tubular adenomas and only 3 % of tubulovillous adenomas/colorectal adenocarcinomas harbored GNAS mutations, emphasizing the notion that shared genetic traits might underlie the readily observed morphological similarities between colorectal villous adenomas and intestinal-type IPMNs.23
Some limitations of our study should be noted. First, the prevalence of GNAS mutations in our current series as well as in our prior publication is based on IPMNs that reached clinical criteria for surgical resection (Sendai criteria).8 Because the majority of IPMNs are not resected, it is possible that we are overestimating the frequency of altered GNAS in IPMNs. This limitation could be overcome by including cysts that are sampled by endoscopy but not resected. In addition, the lack of correlation with natural history might also reflect the fact that all of the samples were surgically resected. A more biologically heterogeneous group of resected and unresected IPMNs might elucidate a tangible clinical correlate. Finally, we only assessed codon 201 for mutations in GNAS, as it has been historically reported as the unique hot spot codon for mutations in IPMNs.8,9,17 Thus, although unlikely, IPMNs with a wild type codon 201 might harbor oncogenic mutations at other codons of GNAS, copy number alterations of chromosome 20q (the GNAS locus), or aberrant activation of downstream effectors of Gsa, such as protein kinase A, none of which were assessed in this study.24–26
In conclusion, the high prevalence of GNAS mutations in IPMNs, irrespective of grade of dysplasia, suggests that it is an early driver gene in IPMN pathogenesis. Furthermore, our data support the correlation of GNAS mutations and intestinal differentiation in IPMNs.
ACKNOWLEDGMENT
The authors thank Michael Rolfe and the Lustgarten Foundations for their support.
Footnotes
CONFLICT OF INTEREST The authors declare no conflict of interest.
REFERENCES
- 1.Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–87. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 2.Longnecker DS, Adsay NV, Fernandez-del Castillo C, et al. Histopathological diagnosis of pancreatic intraepithelial neoplasia and intraductal papillary–mucinous neoplasms: interobserver agreement. Pancreas. 2005;31:344–9. doi: 10.1097/01.mpa.0000186245.35716.18. [DOI] [PubMed] [Google Scholar]
- 3.Adsay NV, Merati K, Basturk O, et al. Pathologically and biologically distinct types of epithelium in intraductal papillary mucinous neoplasms: delineation of an “intestinal” pathway of carcinogenesis in the pancreas. Am J Surg Pathol. 2004;28:839–48. doi: 10.1097/00000478-200407000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Adsay NV. Cystic neoplasia of the pancreas: pathology and biology. J Gastrointest Surg. 2008;12:401–4. doi: 10.1007/s11605-007-0348-z. [DOI] [PubMed] [Google Scholar]
- 5.Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12:81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 6.Correa-Gallego C, Ferrone CR, Thayer SP, et al. Incidental pancreatic cysts: do we really know what we are watching? Pancreatology. 2010;10:144–50. doi: 10.1159/000243733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salvia R, Malleo G, Marchegiani G, et al. Pancreatic resections for cystic neoplasms: from the surgeon's presumption to the pathologist's reality. Surgery. 2012;152:S135–42. doi: 10.1016/j.surg.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Jiao Y, Dal Molin M, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA. 2011;108:21188–93. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthaei H, Norris AL, Tsiatis AC, et al. Clinicopathological characteristics and molecular analyses of multifocal intraductal papillary mucinous neoplasms of the pancreas. Ann Surg. 2012;255:326–33. doi: 10.1097/SLA.0b013e3182378a18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hruban RH, Klimstra DS, Pitman MB. Tumors of the pancreas. American Registry of Pathology; Washington, DC: 2006. [Google Scholar]
- 12.Furukawa T, Kloppel G, Volkan Adsay N, et al. Classification of types of intraductal papillary–mucinous neoplasm of the pancreas: a consensus study. Virchows Arch. 2005;447:794–9. doi: 10.1007/s00428-005-0039-7. [DOI] [PubMed] [Google Scholar]
- 13.R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 14.Weinstein LS, Liu J, Sakamoto A, Xie T, Chen M. GNAS: normal and abnormal functions (minireview). Endocrinology. 2004;145:5459–64. doi: 10.1210/en.2004-0865. [DOI] [PubMed] [Google Scholar]
- 15.Kanda M, Knight S, Topazian M, et al. Mutant GNAS detected in duodenal collections of secretin-stimulated pancreatic juice indicates the presence or emergence of pancreatic cysts. Gut. 2013;62(7):1024–33. doi: 10.1136/gutjnl-2012-302823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G, Olson MT, O'Neill A, et al. A virtual pyrogram generator to resolve complex pyrosequencing results. J Mol Diagn. 2012;14:149–59. doi: 10.1016/j.jmoldx.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa T, Kuboki Y, Tanji E, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1:161. doi: 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalfa N, Lumbroso S, Boulle N, et al. Activating mutations of Gsa in kidney cancer. J Urol. 2006;176:891–5. doi: 10.1016/j.juro.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Collins MT, Sarlis NJ, Merino MJ, et al. Thyroid carcinoma in the McCune-Albright syndrome: contributory role of activating Gs alpha mutations. J Clin Endocrinol Metab. 2003;88:4413–7. doi: 10.1210/jc.2002-021642. [DOI] [PubMed] [Google Scholar]
- 20.Freda P, Chung W, Matsuoka N, et al. Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumors: correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary. 2007;10:275–82. doi: 10.1007/s11102-007-0058-2. [DOI] [PubMed] [Google Scholar]
- 21.Nault JC, Fabre M, Couchy G, et al. GNAS-activating mutations define a rare subgroup of inflammatory liver tumors characterized by STAT3 activation. J Hepatol. 2012;56:184–91. doi: 10.1016/j.jhep.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 23.Yamada M, Sekine S, Ogawa R, et al. Frequent activating GNAS mutations in villous adenoma of the colorectum. J Pathol. 2012;228:113–8. doi: 10.1002/path.4012. [DOI] [PubMed] [Google Scholar]
- 24.Freda PU, Chung WK, Matsuoka N, et al. Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumors: correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary. 2007;10:275–82. doi: 10.1007/s11102-007-0058-2. [DOI] [PubMed] [Google Scholar]
- 25.Nishihara E, Amino N, Maekawa K, et al. Prevalence of TSH receptor and Gsalpha mutations in 45 autonomously functioning thyroid nodules in Japan. Endocr J. 2009;56:791–8. doi: 10.1507/endocrj.k09e-073. [DOI] [PubMed] [Google Scholar]
- 26.Tominaga E, Tsuda H, Arao T, et al. Amplification of GNAS may be an independent, qualitative, and reproducible biomarker to predict progression-free survival in epithelial ovarian cancer. Gynecol Oncol. 2010;118:160–6. doi: 10.1016/j.ygyno.2010.03.010. [DOI] [PubMed] [Google Scholar]