Abstract
Bacterial cell wall is a polymer of considerable complexity that is in constant equilibrium between synthesis and recycling. AmpDh3 is a periplasmic zinc protease of Pseudomonas aeruginosa, which is intimately involved in cell-wall remodeling. In this report we document the hydrolytic reactions that this enzyme performs on the cell wall. The process removes the peptide stems from the peptidoglycan, the major constituent of the cell wall. We document that the majority of the reactions of this enzyme takes place on the polymeric insoluble portion of the cell wall, as opposed to the fraction that is released from it. We show that AmpDh3 is tetrameric both in crystals and in solution. Based on the X-ray structures of the enzyme in complex with two synthetic cell-wall-based ligands, we present for the first time a model for a multivalent anchoring of AmpDh3 onto the cell wall, which lends itself to its processive remodeling.
Bacterial growth is linked to integrity of the bacterial envelope, which in Gram-negative organisms is comprised of the outer membrane, the cell wall and the inner membrane. Cell wall, also known as the sacculus, is a unique bacterial structure made up of polymeric repeats of the disaccharide N-acetylglucosamine (NAG)-N-acetylmuramic acid (NAM) with stem peptides linked to the NAM unit. The full-length stem peptide is L-Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala (DAP for diaminopimelate; Figure 1) in many Gram-negative bacteria, but various lengths for the stem peptide are found in the sacculus. This building unit is crosslinked to its neighbors through the stem peptides in the course of its biosynthesis.1 Cell wall imparts critical structural integrity that is required for the survival of the bacterium, hence cell wall itself and its biosynthetic machinery are targets of antibiotics.2
Figure 1.
The reactions of AmpDh2 and AmpDh3. OM and IM stand for the outer and inner membranes.
Cell wall is a dynamic polymer in that it experiences synthesis, recycling and remodeling in the course of bacterial existence.3 The recycling events lead to degradation of the peptidoglycan to result in compounds that are collectively called muropeptides. A portion of these muropeptides is internalized for the purpose of recycling into peptidoglycan building units. Yet, some muropeptides are involved in signaling events that lead to manifestation of antibiotic resistance3 and/or virulence responses.4 In this vein, the system in Pseudomonas aeruginosa, a problematic human pathogen, is of particular interest. This organism has evolved two zinc proteases, annotated as AmpDh2 and AmpDh3, which turn over the cell wall and muropeptides (Figure 1).5–7 Expression of the genes for the two enzymes is constitutive.7 Indeed, when the genes for both proteins are knocked out, P. aeruginosa becomes less virulent.8
We have shown that both AmpDh2 and AmpDh3 are periplasmic enzymes, and that they have the ability to turn over minimalist synthetic surrogates of cell wall and certain muropeptides involved in the cell-wall recycling.5 More recently, we have documented that AmpDh2 is a dimeric protein that inserts itself into the inner leaflet of the outer membrane, where it interacts and hydrolyzes the amide bond between the stem peptide and the NAM moiety of the sacculus.6 We identified distinct turnover products for the reaction of AmpDh2 with the cell wall. The enzyme is capable of turning over both the crosslinked and non-crosslinked stem peptides.6 In the present report, we document that AmpDh3 is a tetrameric protein both in solution and in crystals, whose X-ray structure was also elucidated. We determine the nature of the reaction products of AmpDh3 with the cell wall and show that the enzyme interacts preferentially with the polymeric and insoluble component of the cell wall. The present study documents that AmpDh2 and AmpDh3 complement each other in remodeling of the cell wall.
The gene for AmpDh3 was cloned and the protein was purified to homogeneity, as reported earlier.5 We have previously shown that AmpDh3 turns over minimalist synthetic peptidoglycan-mimetic substrates and presented an argument that this enzyme exists in the periplasm.5 When we explored the possibility of the reaction of AmpDh3 with the sacculus, we observed very little products, but those that were detected had their peptide stems removed. The minute quantities were detected by LC/MS using the extracted-ion chromatograms (EICs) of target products at the limit of low picomole.9 We were keen on knowing whether the reaction products were indeed present predominantly on the insoluble polymeric sacculus itself, but also, to make a side-by-side comparison to the case of the reactions of the aforementioned paralogous protease AmpDh2. We set up two experiments. One was to use pre-degraded sacculus by the reaction of MltE of Escherichia coli, a lytic transglycosylase that performs predominantly endolytic reactions (cleavage in the middle of the peptidoglycan strand).9 The resultant mixture included peptidoglycan strands, both crosslinked and non-crosslinked, which would provide greater access to AmpDh3 than would the intact sacculus. Under this condition, we detected 15 fragments of the peptidoglycan by LC/MS total-ion chromatogram (TIC) that had anywhere from n = 0 (compound 1) to n = 14 (compound 15; Figure 2A). All these fragments lacked the peptide stem—crosslinked or non-crosslinked—due to the hydrolytic reaction of AmpDh3. The reactions were complete and the abundance of products was now significant. A control reaction of MltE alone with the sacculus indicated that the hydrolytic activity was that of AmpDh3 (Figure 2B). The chemical structures of the products were further analyzed by LC/MS/MS with comparison to the mass spectrometric outcome of the analysis for the authentic synthetic standard 16 (Figure S1, Supporting Information).
Figure 2.
LC/MS total-ion chromatograms of the reaction of AmpDh3 with the MltE-predigested sacculus (A), of MlE alone (B), of mutanolysin alone (C), and of AmpDh3, followed by that of mutanolysin (D).
The question remains whether AmpDh3 could access its substrates on the intact sacculus. As such, we incubated AmpDh3 with the sacculus first, followed by centrifugation and reconstitution of the pelleted sacculus into a solution with the enzyme mutanolysin. Mutanolysin hydrolyzes the glycosydic bond between NAM and NAG to produce [NAG-NAM(peptide)]n (compounds 17–21 in Figure 2C). If there were a reaction by AmpDh3 on the insoluble polymeric sacculus, incubation with mutanolysin would produce a NAG-NAM sugar backbone ladder without the peptide stems. Figure 2D shows LC/MS TIC of the AmpDh3 reaction, followed by that of mutanolysin. The reducing-end sugar was reduced by sodium borohydride to eliminate complexity due to the anomeric mixture. The most abundant product was 1′. This is evidence of the ability of AmpDh3 to react with the intact sacculus. The additional products are oligomers of NAG-NAM (2′–6′, for n = 0 to 5). As minor products, the ones with 1,6-anhydromuramic acid at the end were also found (compounds 1–4, for n = 0 to 3).
For the side-by-side comparison, we performed the same experiments with homogeneously purified AmpDh2 and identified the products. The results for both enzymes are given in Table 1. There are several important observations. The first is on specific activities of these enzymes. Insofar as the amount of enzyme, the amount of sacculus and the length of time for incubation were held the same in all these reactions, the quantities of the products would correlate as a first approximation with the specific activities of AmpDh2 and AmpDh3 in their reactions with the sacculus. AmpDh3 is a more active enzyme than is AmpDh2 (by roughly three-fold). Second, both cross-linked and non-crosslinked peptidoglycans are substrates for both enzymes. Notwithstanding that AmpDh3 produces more products for complete hydrolysis (higher specific activity), AmpDh2 produced significantly more partial-reaction products. This includes partial reaction products for crosslinked peptides, when AmpDh2 cleaves one end, but not the other. Third, and strikingly, AmpDh3 exhibits most of its reactions in the insoluble polymeric sacculus fraction, whereas those of AmpDh2 are mostly found in the soluble fractions (Table 1). The profound significance of this last finding is addressed in the context of the X-ray structure for AmpDh3, discussed below.
Table 1.
Comparison of Reactions of AmpDh2 and AmpDh3 with sacculus.a
| Solubleb | Insolubleb | ||
|---|---|---|---|
| AmpDh2 | Products | 3,900,000 (53%) | 310,000 (86%) |
| Partial-reaction | 3,300,000 (45%) | 50,000 (14%) | |
| Substrates | 150,000 (2%) | Not detected | |
|
| |||
| Total | 7,400,000 (100%) | 360,000 (100%) | |
|
| |||
| AmpDh3 | Products | 300,000 (100%) | 19,000,000 (93%) |
| Partial-reaction | Not detected | 1,100,000 (5%) | |
| Substrates | Not detected | 390,000 (2%) | |
|
| |||
| Total | 300,000 (100%) | 20,000,000 (100%) | |
Amounts are expressed as the total EIC peak areas.
Souble and insoluble portions were separated by centrifugation of reaction mixtures with sacculus, before digestion by mutanolysin.
Sedimentation velocity (SV) data for AmpDh3 in solution were fit using numerical solutions of the Lamm equation.10 The SV data showed a single peak corresponding to a tetrameric species with an apparent mass, Mapp = 117,000 ± 7000 Da (based on the amino-acid sequence, the tetramer should be 115,000 Da) with the corresponding s20,w value at 6.12 ± 0.24 (Figures S2 and S3). Hence, AmpDh3 is a tetramer in solution. As described below, it is also so in crystals.
We resorted to solving the X-ray structure of AmpDh3 to address these observations. The apo enzyme was crystallized and we soaked two synthetic substrates, compounds 2211 and 23,12 into the crystals. As anticipated, the apo enzyme is tetrameric. Crystals of AmpDh3 turned over 22 to produce a complex of the enzyme and the saccharide and peptide products (AmpDh3:22 complex), whose structure was solved (Table S1). The complex of AmpDh3 with synthetic sample 23 (AmpDh3:23 complex) was also solved (Table S1), but under the conditions that the zinc ion was removed, hence giving the intact compound 23 bound to the active site.
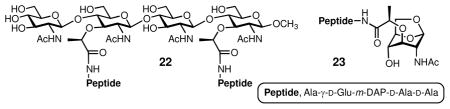
The tetrameric structure for AmpDh3 showed two monomers in the asymmetric unit (A and B) related by a pseudo-two-fold axis that orients the two active sites to opposite sides of the quaternary structure (Figure 3A). The two symmetry-related monomers A′ and B′ complete the tetrameric structure (Figure 3A). Both monomers in the asymmetric unit are nearly identical (backbone root-mean-square deviation value of 0.38 Å), with a typical metal-binding coordination sphere for zinc proteases. Each monomer presents an N-terminal coiled-coil loop (residues 1–19), a catalytic domain (residues 20–173) and a globular C-terminal domain (residues 174–255, Figure S4). The closest structural homolog of AmpDh3 is the zinc protease AmpDh2, also from P. aeruginosa (Figure S5). The significant difference that stands out between the two enzymes is the conformations of the N-terminal regions and the presence of an extra helix (α2) in the AmpDh3 structure (Figure S5). An L-shaped active site defines a ~22 Å-long peptide-stem binding segment and a ~26 Å-long extended binding site for the sugar backbone. The catalytic zinc ion is sequestered between the two binding sites (Figure S4). The pentapeptide product of hydrolysis of the synthetic substrate 22 by AmpDh3 is found in the complex in all four monomers, with the sole exception that the terminal D-Ala is not seen in the density, with the implication that it is mobile. The saccharide product (three of the four sugar rings were seen) was found bound, mapping out the saccharide-binding surfaces of the tetramer (Figures 3, S4, and S6). It is known that AmpDh3 can hydrolyze both the cell wall and the 1,6-anhydroMur-containing muropeptides, with strong preference for the former.5 The complexes AmpDh3:22 (Figures S4 and S6) and AmpDh3:23 (Figure S7) support these observations. The difference between the apo enzyme and the complexes is in the β5–β6 protrusion, where the sugar backbone is accommodated (Figure S8).
Figure 3.
(A) The tetrameric X-ray structure of AmpDh3 in complex with the reaction products for turnover of 22. Each subunit is colored differently to show the embrace by the four in the middle of the tetramer. The reaction products seen in the crystal structure are shown for monomers B and B′, whose active sites are facing the viewer (the peptide stems in green and the saccharides in magenta). The other two active sites are on the opposite side of the tetramer. The NMR-based structure of the crosslinked peptidoglycan (in black) is superimposed on the coordinates for the products from the X-ray structure. (B) A cartoon for the complex showing the perspective down the axis of the peptidoglycan saccharide backbone (shown in open circles; viewed from 12 o’clock down in panel A). The peptides are shown symbolically as black lines. The three-fold symmetry for the peptidoglycan is shown down the axis of the saccharide backbones. The processivity of the action of AmpDh3 tetramer is shown by the arrows, as the pivoting of the enzyme on the saccharide backbone takes it to the next sites of reaction on the opposite face of the tetramer.
Notwithstanding the monomer similarities of AmpDh2 and AmpDh3, they exhibit profound differences in the quaternary structures. The former is a dimeric protein that anchors to the membrane through its N-terminus, yet the latter is a soluble tetramer. A strong network of interactions in the AmpDh3 N-terminal coiled-coil loop (Figure S9) links the four monomers together. These distinct quaternary structures determine the unique activities of AmpDh2 and AmpDh3, notwithstanding the fact that they both hydrolyze the stem peptide from the peptidoglycan. The AmpDh3 structure exhibits an excellent accommodation of the solution structure of the peptidoglycan, which we solved recently by NMR.13 The three-dimensional structure of the peptidoglycan exhibits a right-handed helical arrangement for the sugar backbone, which sequesters the stem peptides at 120 degrees of each other, a three-fold symmetry. If we map this three-dimensional structure onto the X-ray structure of the tetramer of AmpDh3, two backbones of the saccharide chains coincide precisely with the X-ray positions for the bound sugars, simultaneously with overlapping of the crosslinked peptides with those seen for peptides in the X-ray structure (Figure 3A). This is an extended surface for binding of both the peptide and the saccharide segments. The two active sites on one side of the tetramer can perform catalysis on two peptidoglycan strands. The conformation for the solution NMR structure of the peptidoglycan13 does not allow occupation of all four active sites simultaneously. On completion of the reaction with the two bound peptidoglycans, the tetramer would release the product strands and by a rotation of 60 degrees the two active sites on the opposite side of the tetramer will be aligned to engage two neighboring strands for additional catalysis in a processive manner (Figure 3B). The three-dimensional arrangements are entirely consistent and coincident with the three-fold symmetry of the peptidoglycan backbone and the spatial disposition of the pendant stem peptides. Furthermore, this model explains the existence of the majority of the product sites for AmpDh3 in the insoluble polymeric sacculus fraction, to which the AmpDh3 tetramer latches on.
AmpDh2, as indicated earlier, is sequestered on the inner leaflet of the outer membrane as a dimer. This positioning regulates its activity. It operates on the peripheries of the peptidoglycan, whereby its products are largely found in the soluble product fraction (released from the polymeric entity; Table 1). Its specific activity is lower and it would appear to exhibit selectivity in its reaction, as it shows the presence of a significant partially turned over peptidoglycan (cuts one side of the cross-linked stem peptide). The contrary is true for AmpDh3. This enzyme is soluble, it has higher specific activity and it does not leave behind partially hydrolyzed peptidoglycan. We do not know presently how its activity is regulated. However, likely as a function of the tetrameric structure, AmpDh3 anchors to the peptidoglycan itself in a multivalent manner (Figure 3A). This multivalency of interaction, in conjunction with two active sites on either side of the tetrameric structure, allows for the processive sequence of reactions that limits the activity of AmpDh3 primarily to the larger insoluble polymeric sacculus (Table 1). What is clear from our experiments is that AmpDh2 and AmpDh3 complement each other in the turnover and/or maturation of the cell wall.
Supplementary Material
Acknowledgments
This work was supported by a grant from the US National Institutes of Health (GM61629) and by grants BFU2011-25326 (the Spanish Ministry of Economy and Competitiveness) and S2010/BMD-2457 (the Government of Community of Madrid). The Mass Spectrometry & Proteomics Facility of the University of Notre Dame is supported by grant from the US National Science Foundation (CHE0741793).
Footnotes
The authors declare no competing financial interest.
The crystallographic coordinates are deposited in the Protein Data Bank (PDB codes 4bxj, 4bxd and 4bxe for AmpDh3, AmpDh3:22 complex and AmpDh3:23 complex, respectively). Experimental details for structural determination, sedimentation analyses, and MS/MS analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Litzinger S, Mayer C. The Murein Sacculus. In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds. Springer; Berlin Heidelberg: 2010. pp. 3–52. [Google Scholar]; (b) Suvorov M, Fisher JF, Mobashery S. Bacterial Cell Wall: Morphology and Biochemistry. In: Goldman E, Green LH, editors. Practical Handbook of Microbiology. 2. CRC Press; 2009. [Google Scholar]
- 2.(a) von Nussbaum F, Brands M, Hinzen B, Weigand S, Häbich D. Angew Chem, Int Ed. 2006;45:5072–5129. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]; (b) Green DW. Expert Opin Ther Targets. 2002;6:1–19. doi: 10.1517/14728222.6.1.1. [DOI] [PubMed] [Google Scholar]
- 3.(a) Johnson JW, Fisher JF, Mobashery S. Ann NY Acad Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Park JT, Uehara T. Microbiol Mol Biol Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boudreau MA, Fisher JF, Mobashery S. Biochemistry. 2012;51:2974–2990. doi: 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Lee M, Hesek D, Lastochkin E, Boggess B, Mobashery S. J Am Chem Soc. 2013;135:4950–4953. doi: 10.1021/ja400970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez-Caballero S, Lee M, Artola-Recolons C, Carrasco-López C, Hesek D, Spink E, Lastochkin E, Zhang W, Hellman LM, Boggess B, Mobashery S, Hermoso JA. J Am Chem Soc. 2013;135:10318–10321. doi: 10.1021/ja405464b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juan C, Moya B, Perez JL, Oliver A. Antimicrob Agents Chemother. 2006;50:1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moya B, Juan C, Alberti S, Perez JL, Oliver A. Anti-microb Agents Chemother. 2008;52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, Mobashery S. J Am Chem Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M, Hesek D, Shah IM, Oliver AG, Dworkin J, Mobashery S. ChemBioChem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. J Am Chem Soc. 2009;131:8742–8743. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Proc Natl Acad Sci USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





