Summary/Abstract
Uncoating is an essential step in the retrovirus life cycle about which little is known. Uncoating is defined as the specific dissociation of the capsid shell from the viral core in the host cell cytoplasm. In this chapter, biochemical assays for studying HIV-1 uncoating in vitro are described. These techniques have proven useful for characterizing HIV-1 mutants that exhibit defects in the uncoating step of infection.
Keywords: Uncoating, retrovirus, HIV-1, core, capsid, disassembly, reverse transcription, assay
1. Introduction
The term uncoating has been used to refer to the early postentry steps in virus infection immediately following fusion of the viral and cellular membranes and delivery of the viral core into the cytoplasm. For most enveloped viruses, particularly retroviruses, the details of uncoating are essentially unknown. Uncoating can be defined more specifically as the shedding of the viral capsid from the retroviral core (reviewed in [1]). For HIV-1, this appears to involve disassembly of the conical capsid and the release of the CA protein into a soluble form (Fig.1). The intracellular location and timing of HIV-1 uncoating during infection are not known.
Fig. 1.

Schematic of HIV-1 uncoating. During incubation at 37°C, purified HIV-1 cores spontaneously release the CA and RT proteins into a soluble form. The extent of uncoating is determined by p24 ELISA after separating free from core-associated CA by ultracentifugation.
Retroviruses assemble as immature particles that undergo maturation to become infectious. Maturation requires cleavage of the viral Gag and Pol polyproteins by the viral protease into individual proteins; these rearrange to form the mature viral core. For lentiviruses, such as HIV-1, the shell of the core is formed by a conical capsid composed of CA protein molecules arranged in lattice of hexagons. The conical shape of lentiviral cores suggests that some unique aspect of their biology, such as the ability to infect nondividing cells, may be dependent on a specific uncoating mechanism. Studies of HIV-1 mutants containing substitutions in CA have revealed that infection is critically dependent on the proper stability of the HIV-1 capsid [2]. Species-specific restriction factors potently inhibit HIV-1 infection by targeting the viral capsid. These studies indicate that uncoating is a key step in HIV-1 infection that may be attractive for targeted antiviral therapy.
Historically, retroviral uncoating has been difficult to study due to the lack of sensitive and specific assays for this process. The perception that a high percentage of HIV-1 particles are defective may also have intimidated researchers from developing assays for uncoating due to potential difficulties in interpreting experimental outcomes. Here we describe a quantitative method for assaying HIV-1 uncoating in vitro by using purified viral cores. The approach involves purifiying HIV-1 cores by sedimentation of intact virions through a detergent. Samples of the cores are then incubated at 37°C, and the extent of CA release is quantified by p24 ELISA after the cores have been pelleted in an ultracentrifuge. This approach has been employed successfully to analyze the effects of viral mutations on HIV-1 capsid stability [2-4] and may also prove useful for identifying cellular activities that influence HIV-1 uncoating [5]. The procedures are based on original methods for isolating intact cores from virions of retroviruses other than HIV-1 [6-13].
2. Materials
2.1. Cells and Media
Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum and penicillin/streptomycin.
293T cells (American Type Culture Collection).
2.2. 293T Transfection
2X Bes-buffered saline solution (2X BBS): 50 mM BES (pH 6.95), 280 mM NaCl, 1.5 mM Na2HPO4. Adjust pH to 6.95 at room temperature with NaOH. Filter sterilize and store in 10 ml aliquots at −20°C.
2.5 M CaCl2 solution: Dissolve 36.8 g of CaCl2·2H2O in purified water (MilliQ or equivalent) and adjust the volume to 100 ml. Sterilized the solution by vacuum filtration through a 0.22 micron pore-size filter, and store it in aliquots at −20°C.
Dulbecco’s phosphate-buffered saline, sterile (PBS).
HIV-1 proviral plasmid DNA (pNL4-3 or equivalent, purified by Qiagen maxi-prep or cesium chloride density gradient centrifugation).
2.3. Density Gradient Ultracentrifugation
STE buffer: 10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA, diluted from a 10X concentrated stock solution. Adjusted the pH to 7.4 with HCl after diluting to the final (1X) concentration.
STE buffer containing 30% (w/vol) sucrose: prepare by adding 30 g sucrose to 10 ml of 10X STE buffer. Add purified water to final volume (100 ml). Adjust pH to 7.4. Store at room temperature.
STE containing 70% (w/vol) sucrose: Add 70 g sucrose to 10 ml of 10X STE buffer. Add purified water to final volume (100 ml). Adjust pH to 7.4. Store at room temperature.
1% Triton X-100 in STE buffer. Add 1 ml of 10% Triton X-100 (in water) to 9 ml of 1X STE buffer. Store at 4°C.
2.4. p24 ELISA
PBS, non-sterile: Prepare a 10X concentrated stock by dissolving 80 g NaCl, 2 g KCl, 6.1 g Na2HPO4, 2 g KH2PO4 in 900 ml of deionized water. Adjust the volume to 1000 ml. Make 1:10 dilution in water prior to use.
ELISA sample diluent: mix 10 ml of newborn calf serum and 5 ml of 10% Triton X-100 with 85 ml of PBS. This solution should be prepared fresh. Alternatively, a larger quantity can be prepared and the solution filter sterilized and stored at 4°C.
Blocking buffer: 5% calf serum in PBS. Mix 5 ml of newborn calf serum with 95 ml sterile PBS (sufficient for blocking 4 plates). This solution should be prepared fresh.
ELISA wash solution: 0.2% Tween 20 in PBS. Add 2 ml Tween 20 to 1000 ml PBS.
Coating antibody: monoclonal antibody to p24 (183-H12-5C; NIH AIDS Research and Reference Reagent Program) produced in Fibercell hollow fiber bioreactor (Fibercell Systems, Inc.). Dilute to 4 ug/ml in PBS immediately prior to use (10 ml per plate required).
Primary antibody: Add 5 μl of a 1:10 dilution of HIV-Ig (hyperimmune human patient serum, NIH AIDS Research and Reference Reagent Program) to 10 ml of ELISA sample diluent.
Secondary antibody: add 2 μl ImmunoPure Goat Anti-Human IgG (H+L), peroxidase conjugated (0.4 mg/ml; Pierce) to 10 ml of ELISA sample diluent.
Recombinant p24 protein or p24 standard from a commercial ELISA kit.
HRP substrate (TMB Microwell Peroxidase Substrate System; KPL, Inc.)
4N H2SO4 solution in water. Slowly add 100 ml of 18N H2SO4 to 350 ml water with stirring. Allow to cool to room temperature.
Immulon 2HB 96 well plates (Thermo Electron Corp.).
2.5. Specialized Equipment Needed
1. SW28Ti and SW41Ti rotors and compatible ultracentrifuge (Beckman Instruments, Inc.)
2. TLA45 rotor and tabletop ultracentrifuge (Beckman Instuments, Inc.)
2. Highspeed microfuge tubes (Beckman Instruments, Inc.)
3. Auto Densi-Flow density gradient fractionator (Labconco Corp., Kansas City, MO) or other peristaltic pump
4. 20 ml linear gradient former (GM-20; CBS Scientific Co., Inc., Solana Beach, CA)
5. Refractometer (optional; Leica Abbe Mark II Plus or equivalent)
3. Methods
The procedure for isolating HIV-1 cores involves transfection of 293T cells to produce HIV-1 particles, concentration of the virus by ultracentrifugation, and ultracentrifugation of the concentrated virus particles through a layer of Triton X-100 into a linear density gradient. Exposure of the enveloped virus particles to detergent results in release of the viral cores which sediment to an equilibrium density of 1.24-1.27 g/ml. Because the density of intact HIV-1 particles is significantly less (~1.16 g/ml), the presence of CA protein in the denser fractions is a good indication that the viral membrane has been disrupted. A significant fraction of the viral CA protein is not associated with the viral core, and separation of the free and core-associated CA ultracentrifugation allows quantification of the percentage of CA protein associated with the cores by p24 ELISA. In our experience, the yield of p24 in the cores fractions is a highly sensitive measure of the stability of the viral capsid.
3.1. Isolation of HIV-1 cores
The first step in studying HIV-1 uncoating in vitro is to isolate HIV-1 cores by brief detergent treatment of virions followed by ultracentrifugation (see Note 1). This involves the following steps:
- Production of HIV-1 particles. This is typically performed by transient transfection of 293T cells with HIV-1 proviral DNA {e.g. pNL4-3 [14], available through the NIH AIDS Research and Reference Reagent Program}. Most molecular virology labs are familiar with this technique. Because the procedure varies considerably among laboratories, we describe our standard calcium phosphate-based approach here [15].
- 2.1. Culture 293T cells in DMEM containing 10% FBS and antibiotics (penicillin and streptomycin) in an incubator calibrated at 37°C and 5% CO2. Cells are detached from nearly confluent dishes with trypsin-EDTA. Two million cells are seeded in 9 ml medium in 100 mm plastic culture dishes and cultured for 1 day prior to transfection. The next day, cultures will exhibit approximately 25% confluence. Because 30 ml of virus is typically used to isolate cores, we typically transfect 6 dishes per virus to be analyzed.
- 2.2. For each dish of cells to be transfected, mix 20 μg of proviral plasmid DNA with sterile water to a total volume of 450 μl. Add 50 μl 2.5 CaCl2 solution and mix well. Add 0.5 ml 2X BBS solution. Mix well by pipetting up and down. Incubate transfection mixture for 10-20 minutes at room temperature.
- 2.3. Pipet transfection mixture directly onto one dish of cultured 293T cells. Add the mixture dropwise over most of the culture area. The color of the medium will change when the drops contact the media. After adding the entire 1 ml volume, rock the plate back and forth and side-to-side and place the dish overnight (~16 h) in an incubator calibrated to 35°C and 3% CO2.
- 2.4. Aspirate medium and rinse gently with 4 ml PBS.
- 2.5. Aspirate PBS and add 5 ml fresh medium.
- 2.6. Culture in an incubator calibrated at 37°C and 5% CO2 for 24-48 hr.
- 2.7. Withdraw supernatant by pipetting; transfer to conical centrifuge tube and centrifuge for 5 min at 1500 xg to pellet cells and debris. Clarify supernatant by passing through 0.45 μm pore-size syringe filter {for volumes greater than 30 ml, a vacuum filtration unit (Nalgene or equivalent) may be employed}.
- Concentration of HIV-1 particles by ultracentrifugation.
- 3.1. Place 30 ml virus suspension in a 38.5 ml polyallomar centrifuge tube (for SW28 rotor). Using a 5 ml pipet, carefully underlay the virus with 5 ml of a solution of 20% sucrose in PBS.
- 3.2. Centrifuge for 2.5 h at 28,000 rpm (141,000 xg at rmax) to pellet virus particles.
- 3.3. Aspirate supernatant; resuspend pellet in 0.5 ml STE buffer. Pipet gently to resuspend, taking care to avoid foaming as much as possible. Transfer to 1.5 screw cap eppendorf tube and place at 4°C for 1-3 hours to allow small clumps of virus to disperse.
- 3.4. Gently pipet the suspension up-and-down several more times. Centrifuge concentrated virus suspension for 1 minute at 8000 rpm in an Eppendorf centrifuge to remove residual clumps.
- “Spin-thru” detergent treatment of virions.
- 4.1. Prepare a ~12 ml linear gradient of 30-70% sucrose in STE buffer for the SW41 Ti rotor. We use a 20 ml gradient former and place 6 ml of 70% sucrose on the near side (closest to outlet port) and 6 ml of 30% sucrose solution on the far side. It is important to prime the channel between the two chambers prior to filling the second one to permit free flow from one chamber to the other. Using the Auto Densi-Flow gradient former, pump the gradient from the bottom to the top of the tube. It is recommended that the procedure be practiced a few times to ensure consistency. Balance tubes by adding 30% sucrose in STE buffer until the masses of the tubes are equivalent. Place gradients at 4°C until cooled (2-4 hrs).
- 4.2. Overlay the gradient with 0.25 ml of 1% Triton X-100 dissolved in 15% sucrose/STE buffer. This step must be performed carefully to maintain distinct layers (see Note 2.)
- 4.3. Overlay with 0.25 ml of a solution of 7.5% sucrose/STE buffer. This will serve as a “barrier” layer to minimize mixing of the virus suspension and the detergent-containing layer until centrifugation.
- 4.4. Gently overlay with up to 0.5 ml of virus suspension. This step is critical; too much disturbance will result in mixing of the layers and a low yield of cores. A useful method is to widen the bore of a 1 ml pipet tip by trimming with scissors. This will reduce the velocity of the solution as it is pipeted, thus minimizing the mixing of the barrier and detergent layers. See Fig. 2 for a diagram depicting the gradient prior to ultracentrifugation.
- 4.5. Place the tubes in precooled SW41Ti buckets and ultracentrifuge at 35,000 rpm (210,000 xg at rmax) for 16-20 hr at 4°C.
- Recovery of cores from the dense fractions of the gradient.
- 5.1. Following ultracentrifugation, the gradients are fractionated from the top of the gradient with the Auto-Densi-Flow.
- 5.2. Fractions of 1 ml are taken from the top of the gradient, and the tubes are placed in an ice bucket immediately upon collection. The purified viral cores are typically present in the bottom half of the gradient whereas soluble CA protein is present at the top fractions.
- 5.3. Withdraw 50 μl of each fraction and set aside for p24 ELISA (see Method 3.3) and/or reverse transcriptase activity assay.
- 5.4. After collecting fractions, add 0.5 ml of cold STE buffer to each fraction and mix several times by inversion. This procedure reduces the viscosity of the solution, minimizing sampling errors.
- Localization of HIV-1 cores and storage of samples. The remainder of the cores fraction can be used immediately for uncoating assays or flash frozen in liquid nitrogen for future use.
- 6.1. The fractions containing intact cores must be identified prior to further analysis. This can be performed either by assaying the fractions for p24 by ELISA or by measuring reverse transcriptase activity. Both methods should reveal a peak near fraction 10, but it is best to determine this empirically until reproducibility has been established. Once an investigator has become proficient with the method, a rapid approach to predict the fractions containing the cores is to determine the density of each fraction by refractometry or gravimetric analysis. Lentiviral cores typically have a density of 1.24-1.27 g/ml (see Fig. 3).
- 6.2. Quantification of CA in the gradient fractions. The recovery of the CA protein in the cores has been linked to core stability. By assaying the percentage of CA in the gradient that is present in fractions of HIV-1 cores, one may glean information about the particular virus (see Note 3). For this purpose, assay a sample of each fraction for p24 by ELISA [16]. This procedure is described under Method 3.3, and an example is shown in Fig. 3.
- 6.3. If uncoating assays are not to be performed the same day, the fractions containing the HIV-1 cores can be pooled and aliquots flash-frozen in liquid nitrogen and stored at −80°C. The yield of cores is typically 1-2 micrograms of p24 per ml depending on the quantity of virus loaded on the gradient. The uncoating assay requires approximately 50 ng of p24. The purified cores may be frozen in 0.2-0.3 ml aliquots each of which is sufficient for performing several uncoating reactions.
- Analysis of pelleted cores by immunoblotting using HIV-1-specific antibodies.
- 7.1. To confirm that HIV-1 cores have been isolated, proteins present in the dense fractions are characterized by immunoblotting.
- 7.2. Following dilution of the fractions by addition of STE buffer (see 5.3), the tubes are subjected to ultracentrifugation for 30 min at 100,000 xg (45,000 rpm in a Beckman TLA-45 rotor).
- 7.3. Supernatants are removed by aspiration, and the pellets are solubilized in SDS-PAGE loading buffer and are subjected to SDS-PAGE and immunoblotting with monoclonal antibodies specific for HIV-1 CA, gp120, and gp41 {183-H12-5C, 902, and Chessie 8, respectively (NIH AIDS Research and Reference Reagent Program)}. HIV-1 cores are substantially free of gp41 and gp120 relative to intact virions.
- 7.4. The fractions identified as HIV-1 cores should also contain viral RNA. RNA can be extracted from the fractions and the HIV-1 RNA quantified by RT-PCR.
Fig. 2.
Construction of density gradients for isolation of HIV-1 cores. A 30-70% sucrose density gradient is prepared and successively overlaid with a layer of detergent, a barrier layer to prevent premature mixing of virus and detergent, and the concentrated virus suspension. Upon ultracentrifugation, virions pass through the detergent layer, releasing HIV-1 cores that then sediment to their equilibrium density.
Fig. 3.
Distribution of CA in the density gradient after ultracentifugation. Fractions from a gradient were collected from top to bottom and analyzed for p24 by ELISA and density by refractometry. In this experiment, a quantity of HIV-1 corresponding to approximately 250 μg of p24 was applied to the gradient.
3.2. Kinetic assay of HIV-1 uncoating in vitro.
Once HIV-1 cores have been purified and characterized, the stability of the capsid may be determined by studying the rate of CA dissociation upon warming to 37°C. The extent of CA release is quantified by p24 ELISA after separating the cores from the soluble p24 by centrifugation. (see Note 4.)
Pre-dilute cores into cold STE buffer to reduce viscosity, thus avoiding sampling errors.
Further dilute samples by adding 50 μl into 0.5-1.0 ml cold STE buffer. Mix gently but thoroughly by inversion several times. Do not vortex. Flicking of the inverted tubes may be necessary to achieve thorough mixing. Place tubes in a 37°C water bath for various time periods (typically 20-30 min). Be sure to immerse the tubes to their internal liquid level to ensure thorough warming. Invert the tubes periodically throughout the incubation period (every 5-10 minutes). As a time zero control, dilute cores into cold buffer and keep on ice for the same time period (typically 20-30 min).
Following the incubations, rapidly chill the samples by placing in an ice-water bath for 10 min.
Pellet the cores by ultracentrifugation for 20 minutes at 125,000 xg (RCFmax for Beckman TLA-55 rotor at 45,000 rpm). The rotor should be precooled to 4°C and the tubes wiped thoroughly before placing into the rotor. In our experience, it is also helpful to wipe down the rotor before placing in the centrifuge. This removes the condensation on the outside of the rotor, reducing the time necessary for the centrifuge chamber to attain a sufficient vacuum.
Remove the supernatant and transfer to a clean microfuge tube. Resuspend the pellet in 200 μl of ELISA sample diluent.
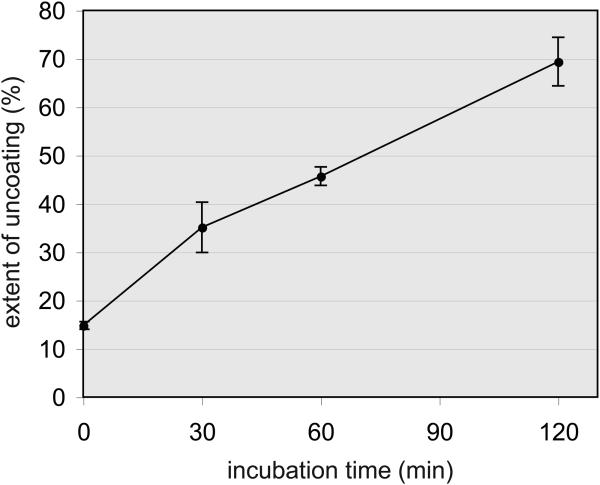
Quantify p24 in both pellet and supernatant fractions by ELISA. Calculate the percentage of the total p24 present in the supernatant (see Note 5.) The extent of uncoating should increase with increasing incubation time (see Fig. 4).
Fig. 4.
Uncoating of HIV-1 cores occurs spontaneously during incubation at 37°C. Samples of purified HIV-1 cores were diluted in buffer and incubated for the indicated times. Reactions were chilled and the cores were pelleted by ultracentrifugation. p24 concentrations in the pellets and supernatants were determined by ELISA, and the extent of uncoating was calculated. Error bars correspond to standard deviations from the mean values from triplicate reactions.
3.3. Assay for CA protein by p24 ELISA
Many commercial p24 assay kits are available and can be used for studies described herein. For in-depth studies, use of the homemade p24 sandwich ELISA described here will result in significant cost savings. The procedure has been adapted from a previous report [16] and can be performed using capture and detector antibodies available from the NIH AIDS Research and Reference Reagent Program.
Coat 96-well Immulon 2HB plates with monoclonal antibody to p24 (183-H12-5C) diluted to 4 μg/ml in PBS (100 μl per well). Plates are sealed with adhesive film and incubated at 37°C overnight to allow efficient binding.
Rinse wells twice with PBS and add 0.25 ml of blocking solution. Seal plate and incubate for 1h at 37°C to block.
Rinse wells 3 times with ELISA wash buffer. This can be performed using an automated plate washer; we use a plastic wash bottle and slap the plate on a stack of paper towels after each wash to remove most of the liquid.
Add standards and samples (100 μl per well) diluted in ELISA sample diluent. The assay has a dynamic range of 0.06-1 ng/ml of p24. We typically employ a series of standards containing 0, 0.06, 0.12, 0.25, 0.5, and 1.0 ng/ml p24. Because transfected 293T cells typically yield between 200 and 2000 ng/ml, dilutions ranging from 1:100 to 1:10,000 are normally sufficient. For concentrated samples of cores, dilutions of up to 1:100,000 may be necessary.
Seal plate and incubate at 37°C for 2 hr.
Wash plate 3 times with ELISA wash buffer.
Add 100 μl primary antibody solution.
Seal plate and incubate 1 hr at 37°C.
Wash plate 3 times with ELISA wash buffer.
Add 100 μl secondary antibody.
Seal plate and incubate 1 hr at 37°C.
Wash plate 3 times with ELISA wash buffer. After the final wash, slap the plate several times on paper towels to remove residual wash solution.
Add 100 μl substrate solution (made by mixing equal volumes of TMB Peroxidase Substrate and Peroxidase Substrate Solution B just prior to use).
Incubate at room temperature until blue color is medium strong in the well corresponding to the 1 ng/ml standard (usually 10-20 min). Do not overincubate—this will result in extreme nonlinearity in the assay.
Terminate the color development reaction by adding 100 μl of 4N H2SO4 solution. The color will change from blue to yellow.
Read absorbance of each well at 450 nm with 650 nm reference in an ELISA microplate reader (e.g. Molecular Devices Emax using Softmax Pro software). Plot absorbance vs. p24 concentration for the standards to obtain the standard curve. Calculate unknown concentrations by interpolation. (Note 6.)
Acknowledgments
The author thanks the former members of his laboratory Alex Kotov and Brett Forshey who contributed to the development of the HIV-1 uncoating assay, along with the present members who assisted in compiling the methods. The NIH AIDS Research and Reference Program is gratefully acknowledged for distributing reagents. Work in the author’s laboratory was supported by NIH grants AI040364 and AI050423.
Footnotes
Although our laboratory prefers to use the equilibrium ultracentrifugation to purify HIV-1 cores, other investigators have employed more rapid methods involving direct pelleting of cores [5, 17-19].
Because HIV-1 uncoating is accelerated at elevated temperature, it is important to precool all solutions and the centrifuge buckets used for isolation of the cores. We store the ultracentrifuge buckets in the refrigerator, but usually leave the rotor itself at room temperature. Likewise, it is important to fractionate the gradients while they are still cold and to place the fractions in an ice-water bath immediately upon collection. We also cool the TLA45 rotor prior to use.
The yield of cores is subject to experimental variation. While we have not identified all of the relevant variables, it appears that the care taken in application of the layers to the gradients, and in keeping the samples cold, are significant factors. It is often difficult for novices to avoid mixing the virus and detergent layers during application of the virus suspension onto the gradient. Members of the laboratory who have practiced the technique were able to master it, but some individuals obtained higher yields of cores than others. With repetition, an individual can obtain reproducible yields of cores. We have not observed a significant dependence of the recovery of HIV-1 cores on the quantity of virus nor of the rate of uncoating in vitro on the amount of cores in the reaction, but we cannot exclude possible effects of these variables.
When assaying uncoating, reactions are typically performed in duplicate. If the values are not within 15% of one another, the experiment should be performed in triplicate until the consistence is improved.
Although the extent of uncoating is reflected in the calculated value (percentage of total CA in the supernatant), it is important to note both the supernatant and pellet values. In some cases, an apparent increase in uncoating may result from a loss of CA from the pellet if the samples are handled improperly. This outcome could also reflect degradation of CA in the reactions.
In our experience, the p24 standard values fit best to a quadratic formula, which is an option in the Softmax Pro software. It is important not to let the color reaction proceed too long or the reaction will become saturated. For most uncoating studies, the sensitivity and dynamic range of the ELISA is ideal. However, the sensitivity of the assay can be enhanced by substituting additional standards (16 and 32 pg/ml) for the high standards (0.5 and 1.0 ng/ml) and extending the color reaction time.
References
- 1.Aiken C. Viral and cellular factors that regulate HIV-1 uncoating. Curr Opin HIV AIDS. 2006;1:194–199. doi: 10.1097/01.COH.0000221591.11294.c1. [DOI] [PubMed] [Google Scholar]
- 2.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wacharapornin P, Lauhakirti D, Auewarakul P. The effect of capsid mutations on HIV-1 uncoating. Virology. 2007;358:48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 4.Forshey BM, Aiken C. Disassembly of human immunodeficiency virus type 1 cores in vitro reveals association of nef with the subviral ribonucleoprotein complex. J Virol. 2003;77:4409–4414. doi: 10.1128/JVI.77.7.4409-4414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auewarakul P, Wacharapornin P, Srichatrapimuk S, Chutipongtanate S, Puthavathana P. Uncoating of HIV-1 requires cellular activation. Virology. 2005;337:93–101. doi: 10.1016/j.virol.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Menendez-Arias L, Risco C, Pinto da Silva P, Oroszlan S. Purification of immature cores of mouse mammary tumor virus and immunolocalization of protein domains. J Virol. 1992;66:5615–5620. doi: 10.1128/jvi.66.9.5615-5620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart L, Schatz G, Vogt VM. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts MM, Oroszlan S. The preparation and biochemical characterization of intact capsids of equine infectious anemia virus. Biochem. Biophys. Res. Commun. 160:486–494. doi: 10.1016/0006-291x(89)92459-5. [DOI] [PubMed] [Google Scholar]
- 9.Chrystie IL, Almeida JD. The recovery of antigenically reactive HIV-2 cores. J. Med. Virol. 1989;27:188–195. doi: 10.1002/jmv.1890270303. [DOI] [PubMed] [Google Scholar]
- 10.Benzair AB, Rhodes-Feuillette A, Lasneret J, Emanoil-Ravier R, Peries J. Purification and characterization of simian foamy virus type I structural core polypeptides. Arch Virol. 1986;87:87–96. doi: 10.1007/BF01310545. [DOI] [PubMed] [Google Scholar]
- 11.Yoshinaka Y, Luftig RB. Murine leukemia virus morphogenesis: cleavage of P70 in vitro can be accompanied by a shift from a concentrically coiled internal strand (“immature”) to a collapsed (“mature”) form of the virus core. Proc Natl Acad Sci USA. 1977;74:3446–3450. doi: 10.1073/pnas.74.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furmanski P, Loeckner CP, Longley C, Larson LJ, Rich MA. Identification and isolation of the major core protein from the oncornavirus-like particle in human milk. Cancer Res. 1976;36:4001–4007. [PubMed] [Google Scholar]
- 13.Bader JP, Brown NR, Bader AV. Characteristics of cores of avian leukosarcoma viruses. Virology. 1970;41:718–728. doi: 10.1016/0042-6822(70)90436-8. [DOI] [PubMed] [Google Scholar]
- 14.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Okayama H. High-efficiency tranformation of mammalian cells by plasmid DNA. Mol. Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 17.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Krausslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. Embo J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Accola MA, Ohagen A, Gottlinger HG. Isolation of human immunodeficiency virus type 1 cores: retention of vpr in the absence of p6(gag) J Virol. 2000;74:6198–6202. doi: 10.1128/jvi.74.13.6198-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]