Abstract
Our knowledge of gene function has increasingly lagged behind gene discovery, hindering our understanding of the genetic basis of microbial phenotypes. Recently, however, massively parallel sequencing has been combined with traditional transposon mutagenesis in techniques referred to as transposon sequencing (Tn-seq), high-throughput insertion tracking by deep sequencing (HITS), insertion sequencing (INSeq) and transposon-directed insertion site sequencing (TraDIS), making it possible to identify putative gene functions in a high-throughput manner. Here, we describe the similarities and differences of these related techniques and discuss their application to the probing of gene function and higher-order genome organization.
In the mid 1970s, researchers surmised that transposons could be used as tools “to manipulate the genes of bacteria, phage and plasmids in ways which are otherwise difficult or impossible” (REF. 1). Transposons are genetic elements that can move within or between genomes by either replicative or ‘cut-and-paste’ mechanisms mediated by an enzyme called transposase. This enzyme recognizes the inverted repeats at the ends of the transposon and also recognizes the target sequence, in which it makes a double-strand break and inserts the transposon2. Transposons were originally discovered as “controlling elements” in maize by Barbara McClintock and have since been found in every kingdom of life3. They have important roles, such as in the evolution of speciation and antibiotic resistance in microorganisms3,4. The most frequently used application of transposons has been insertional mutagenesis, in which a library of bacterial strains, each containing a single randomly located transposon, is constructed. With the addition of transposons to the molecular toolbox, it became much easier to generate mutants and to identify their phenotypes by transductional crosses and complementation assays5.
An enormous advance in the use of transposon mutagenesis was made after the first microbial genomes were sequenced6–8. For the first time, it became possible to directly link an observed phenotype to a genotype (for example, a gene disruption resulting from a transposon insertion) by sequencing of the genome. It is also now possible to sequence many transposon mutants simultaneously, thus allowing genome-wide analyses. At around the same time that this genome sequencing began, signature-tagged mutagenesis was invented9, and together these methods established transposons as the most frequently used tool for genome-wide genotype–phenotype studies10. Such studies led to the identification of thousands of virulence genes in different bacteria, including Salmonella enterica subsp. enterica serovar Typhimurium9, Staphylococcus aureus11, Vibrio cholerae12, Streptococcus agalactiae13, Listeria monocytogenes14 andStreptococcus pneumoniae15–17.
With the advent of massively parallel sequencing (MPS; also referred to as second-generation sequencing)18, microbial genomes continue to flood the public databases. Unfortunately there has been no corresponding explosion in our knowledge of gene function. Today, the situation is so challenging that if one were to sequence the genome of a new microorganism, the percentage of genes of unknown function would be similar to that obtained a decade ago (approximately 30–40%)19–21. To bridge this gap, we need high-throughput approaches that can reveal genotype–phenotype relationships and are applicable to a range of species. This void has now been filled by the development of a group of similar techniques, referred to here collectively as transposon sequencing, that combine transposon mutagenesis with MPS. The approach requires the construction of a transposon insertion library in which most or all non-essential genes contain insertions, followed by growth of the library in defined in vitro conditions or in vivo (infection of a host). The relative frequency of each mutant in the population is determined at the start and at the end of the experiment by means of MPS of the transposon junctions. From this data, the fitness contribution of each gene in each condition can be quantified.
Here, we describe the similarities and differences between the four main transposon sequencing approaches. We also discuss the emerging applications of this technology, such as the elucidation of higher-order genome organization, and the identification of small RNAs (sRNAs) and genes required for pathogenicity.
Transposon sequencing methods
With the recent advent of MPS technologies, it seemed only a matter of time before a genome-wide method that could accurately link genotypic changes to specific phenotypes became established. In 2009, four research groups independently published transposon sequencing methods for this purpose22–26 (FIG. 1). MPS generates short sequence reads of millions of DNA molecules simultaneously, allowing whole genomes to be sequenced in a single experiment27–29. In addition, it is an effective technology for digital-counting applications, including RNA-seq (RNA sequencing)30, sRNA-seq (sRNA sequencing) 31, ChIP–seq (chromatin immunoprecipitation followed by sequencing)32, promoter assays33, assessing histone occupancy34 and, most recently, RNAi-target profiling35. The basic principle of all four transposon sequencing methods involves purification of genomic DNA from a pooled population of mutants, cleavage of the DNA (using either specific enzymes or random shearing), attachment of one or more adaptors to the DNA fragments to facilitate PCR amplification of the fragments containing transposon sequences, and finally, MPS of the amplified fragments to determine the location of the transposon and the relative abundance of mutants containing a transposon at this site.
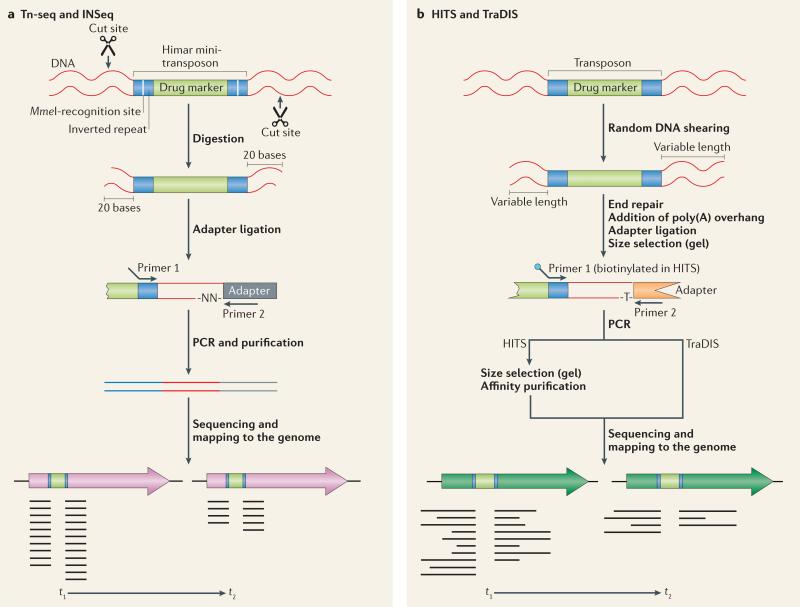
Figure 1. Four methods of massively parallel sequencing of transposon insertions.
Each of four methods of transposon sequencing is illustrated, starting from pooled genomic DNA from the transposon insertion library and ending with sequencing of the left and right transposon junctions. The number of sequences (reads) for each junction can differ between the start of the experiment (t1) and the end (t2), after a selection has been carried out on the library of transposon insertion mutants. In both examples shown, the transposon insertion mutation decreases fitness during growth under the conditions tested, as indicated by there being fewer reads at the end of the experiment than at the start. a | The Tn-seq (named for transposon sequencing) and insertion sequencing (INSeq) methods are highly similar, but INSeq includes a PAGE gel purification step following adaptor ligation and PCR, whereas Tn-Seq includes an agarose gel purification at this point. A recent study66 introduced additional steps to the original INSeq protocol: a linear-PCR step using a biotinylated primer and subsequent purification of the product with magnetic streptavidin beads were added following adapter ligation. These steps reduce both the amount of sample and the amount of enzymes needed. Although they make the protocol more laborious, the results suggest that these modifications increase the sensitivity of the technique. b | The high-throughput insertion tracking by deep sequencing (HITS) and transposon-directed insertion site sequencing (TraDIS) methods are more similar to each other than to Tn-seq and INSeq: after shearing of the DNA, the DNA ends are repaired, and a poly(A) tail is added. However, the methods diverge after the PCR step; in HITS, the PCR products undergo size selection (on a gel) and affinity purification before sequencing, whereas in TraDIS, the PCR products are sequenced directly.
The work described in each of the four original transposon sequencing papers was innovative and demonstrated the power and versatility of the technology by using different bacterial species to address different types of question. The high-throughput insertion tracking by deep sequencing (HITS) method was applied to a library of ~75,000 Haemophilus influenzae mutants22. In a mouse lung infection model, several virulence genes were identified, including those involved in lipopolysaccharide synthesis, metabolism and transport. Another group23 generated a library of 370,000 Salmonella enterica subsp. enterica serovar Typhi mutants with, on average, an insertion every 13 bp in the genome. Owing to this high coverage, the transposon-directed insertion site sequencing (TraDIS) method that was used was able to determine that ~8% of S. Typhi genes are essential for growth in a rich medium. In addition, this study identified genes with an advantageous or disadvantageous growth effect in rich media, as well as 169 genes that are involved in tolerance to bile, a substance that S. Typhi encounters when colonizing the human gall bladder. The insertion sequencing (INSeq) method24 was used to determine whether the human symbiont Bacteroides thetaiotaomicron harbours specific genes that are necessary for survival in the colon. The data showed that colonization is partly influenced by microbial community composition and competition for nutrients such as vitamin B12. The final method, termed Tn-seq (for transposon sequencing, following the terminology of RNA-seq, sRNA-seq and ChIP–seq), was applied to the pathogen S. pneumoniae to identify genes that are essential for growth in rich medium, to determine the fitness effect (advantageous or disadvantageous) of each non-essential gene and to identify 97 genetic interactions between five query genes and the rest of the genome25.
Choice of transposon
Tn-seq and INSeq are nearly identical methods that make use of the Himar I Mariner transposon (FIG. 1). The teams that developed these two methods realized that a single nucleotide change in the terminal inverted repeats of Mariner introduced MmeI recognition sites24,25. MmeI is a type IIS restriction endonuclease that makes a 2 bp staggered cut 20 bases downstream of its recognition site36. Thus, when DNA from a library of transposon insertion mutants is digested with MmeI, fragments comprising the left and right transposon ends plus 16 bp of flanking genomic DNA are produced (the MmeI recognition site is located 4 bp before the end of each terminal repeat). This 16 bp is enough to accurately pinpoint the location of the transposon insertion in the bacterial genome. The 2-base overhangs facilitate the ligation of an adaptor; using a primer specific for this adaptor and another one specific for the transposon, the sequence is amplified by PCR, followed by agarose gel or PAGE purification to isolate the 120 bp product. Finally MPS is used to determine the flanking 16 bp sequences.
HITS22 and TraDIS23 are similar methods, and both use transposons that lack type IIS restriction sites within their inverted repeats. Therefore, to generate transposon end fragments, the DNA is sheared, after which several extra steps are necessary before an adaptor can be ligated (FIG. 1). In addition, affinity purification is carried out in the HITS method to remove extraneous DNA before sequencing. TraDIS uses a derivative of the Tn5 transposon, which is commercially available and, similarly to the Mariner transposons used in the other three methods, is active in different species.
HITS and TraDIS have the advantage of being applicable to any transposon or insertional element. On the other hand, Tn-seq and INSeq have the advantages of a shorter sample preparation protocol and the generation of a final product of precise length. By contrast, the DNA shearing used in HITS and TraDIS results in a range of PCR product sizes, potentially allowing for PCR bias (shorter DNA templates are preferentially amplified over longer templates).
Data analysis
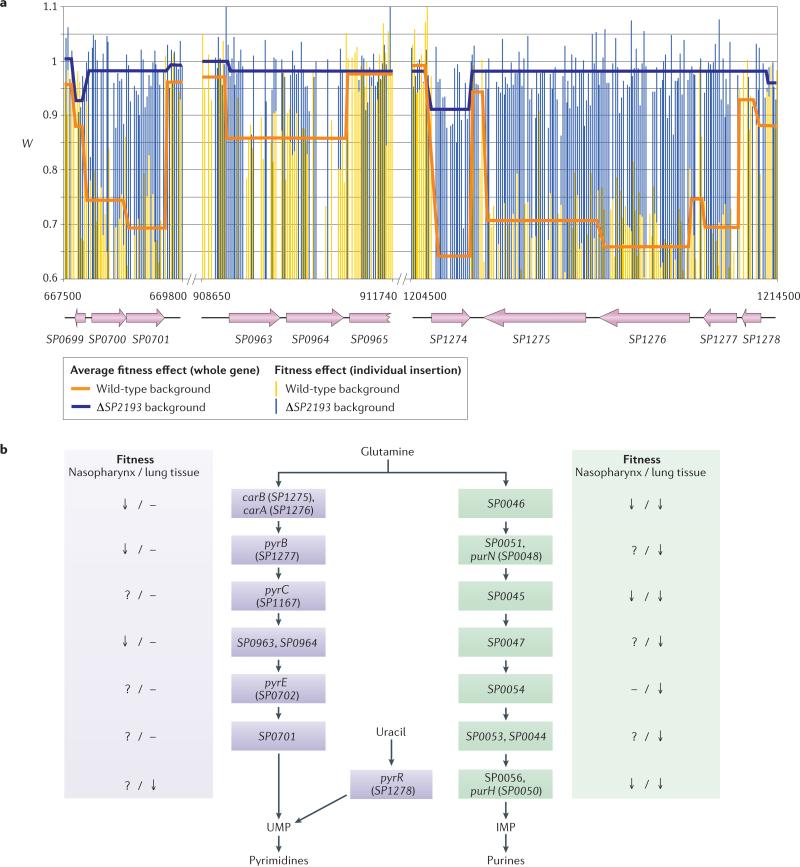
As well as the choice of transposon and the sample preparation method, data analysis is the other main difference between the four methods. All four are digital-counting methods, in which the number of sequence reads for a particular insertion corresponds to the frequency of that insertion mutant in the population. The change in insertion frequency after selection correlates with the fitness effect of the insertion mutation during the selection. Thus, if the frequency of a particular insertion does not change, this would indicate that the gene into which the transposon is inserted is neutral (that is, it does not contribute to fitness (W)) in the condition tested. If a particular insertion disappears from the population, this would indicate that the corresponding gene is essential under the experimental conditions. Conversely, if a mutant increases in frequency, disruption of the gene is beneficial. There is a large gradient of fitness effects between neutral (W = 1) and essential (W <<1), so a precise and quantitative estimate of the fitness cost is crucial to capture such differences in fitness. Because of the digital and therefore highly accurate nature of MPS, a sensitive measure of fitness that correlates with the growth rate was developed25. By accounting for growth of the population during the selection, one can determine the effect of each disrupted gene on the growth rate (BOX 1). An example is shown in FIG. 2a of Tn-seq data for individual transposon insertions at three loci that are involved in pyrimidine biosynthesis in S. pneumoniae. By calculating the average fitness effect from all insertions within a gene (BOX 1; FIG. 2a), mutants that differ by as little as 5% in their growth rates can be accurately distinguished from each other. This means that it is possible to differentiate between, for instance, a wild-type strain and mutant strain that double every 35 and 37 minutes, respectively25 (BOX 1). The other three methods estimate fitness from the change in the ratio of mutants over time. Although this provides a semiquantitative estimate of fitness, it is challenging to obtain robust statistical results and to compare fitness values across experiments and environments.
Figure 2. Pathway analysis in vitro and in different host niches.
a | Fitness (W) of individual Streptococcus pneumoniae transposon insertion mutants with insertions in three loci involved in pyrimidine biosynthesis. The genes and genome coordinates of the insertions are indicated. In vitro fitness was calculated during growth in a semi-defined minimal glucose medium42. The data show that SP2193, which encodes a response regulator (a type of transcription factor) and positive regulator of pyrimidine biosynthesis genes, is a suppressor of the fitness defects that result from insertion mutations in each gene involved in the pyrimidine biosynthesis pathway, as determined by genetic interaction analysis. b | Three metabolic pathways in S. pneumoniae lead to the production of inosine monophosphate (IMP) and uridine monophosphate (UMP), which are essential precursors for purine and pyrimidine synthesis, respectively. The effect of disrupting each gene in these three pathways, in terms of the change in bacterial fitness, is indicated for infections of the nasopharynx and of the lung. A question mark indicates that there is no data available. The pathway leading to the synthesis of IMP and purines is required for colonization of the nasopharynx and lung. For the synthesis of UMP and pyrimidines, the pathway that uses glutamine as a precursor is required only for colonization of the nasopharynx, whereas the alternative pathway, which uses uracil as a precursor, can be used for UMP synthesis in the lung.
Applications of transposon sequencing
The development of transposon sequencing and its compatibility with non-model organisms mean that large-scale, quantitative phenotypic profiling is now possible for any microbial species to which transposon mutagenesis can be applied. In addition, with the amount of MPS data that can be obtained per experiment rapidly increasing (it currently stands at ~108 reads on the Illumina HiSeq 2000) and the ability to multiplex samples by incorporating a barcode within the adaptor25, the cost of doing large-scale screens has decreased dramatically. Below, we highlight various biological questions that can be addressed using transposon sequencing.
Identification of new gene functions
Several groups have recently begun to sample the vast ‘genotype versus environment’ space with transposon sequencing. By screening in several in vitro conditions, phenotypes are revealed that help in identifying new gene functions. For example, transposon sequencing has been used in Pseudomonas aeruginosa to identify antibiotic resistance genes37. In this study, Tn-seq was modified slightly: after genomic shearing and adaptor ligation, the Tn5 transposon was enzymatically cut and, together with the genomic DNA flanking the transposon, circularized in a ligation step. Non-circularized products were then removed by exonucleases, and the sequencing target was amplified by PCR. Tn-seq has also been used in Mycobacterium tuberculosis38 to identify essential genes and pathways that are involved in the utilization of cholesterol (a crucial carbon source for M. tuberculosis during infection). Furthermore, a number of conditionally essential genes (genes that are essential for growth under specific conditions) have been identified in S. Typhimurium using Tn-seq39. Transposon sequencing was also used to identify an alternative citrate synthase and a novel dynamic branching pathway of the anaerobic tri carboxylic acid cycle in Shewanella oneidensis40, and to identify essential genes in the periodontal pathogen Porphyromonas gingivalis41. Finally, by screening 17 different in vitro growth conditions for S. pneumoniae, more than 1,800 genotype–phenotype interactions were discovered42. From this data set, genes involved in differential carbon source utilization and in resistance against specific stresses were identified. The data from this study were used to carry out an overlap analysis, which involves combining condition-specific in vitro data with in vivo virulence data to reveal previously unknown selective pressures that S. pneumoniae encounters in specific niches in the host42. At the gene level, the overlap analysis also served to provide leads on gene function for many hypothetical genes.
Identification of virulence genes
The original HITS22 and INSeq24 studies examined the fitness contribution of genes using infection and colonization models, respectively, and the application of these techniques for such studies has since grown. For example, a re-analysis of an Escherichia coli transposon library in cattle revealed that transposon sequencing is superior to the original signature- tagged mutagenesis analysis in terms of sensitivity, allowing for the identification of hundreds of additional virulence genes43,44. Another study45 identified 17 genes that are essential for the growth of Yersinia pseudotuberculosis inside a mouse host. The authors of this study went on to show that MrtAB, a previously uncharacterized efflux pump, is essential for Y. pseudotuberculosis colonization of the mesenteric lymph nodes.
Such in vivo screens are extremely valuable for discovering previously unrecognized virulence mechanisms and for probing pathogen metabolism. However, a limitation to the use of transposon sequencing methods in vivo is that a substantial number of bacteria are often killed or removed stochastically during the establishment of infection. Such population bottlenecks are problematic because it is difficult to determine whether transposon mutants disappear by chance or because they are less fit. In such cases, it is necessary to adjust the number of different mutants present in the inoculum (that is, the complexity of the inoculated population) so that it is less than the bottleneck. For example, in an infection model that allows 104 bacteria to colonize the host regardless of inoculum size (thus, with a bottleneck equal to 104), an inoculum containing less than 104 unique transposon mutants should be used (from experience, we recommend a range of 10–100 cells per insertion mutant). By separately analysing a set of neutral insertions that should not disappear from the library (because the transposon has a neutral impact on fitness; for example, insertions in degenerate genes), it is possible to estimate such bottlenecks on a per animal basis and correct for them in the fitness calculations. This strategy, in combination with a method to calculate the bacterial generation time in vivo, facilitates a sensitive genome-wide analysis of fitness from in vivo experiments42 (BOX 1).
Using transposon sequencing in several in vivo environments has the potential to reveal the requirement for specific genes in specific niches. For example, by applying Tn-seq to S. pneumoniae, it was found that the core biosynthetic genes for pyrimidines and purines have differential roles in a mouse lung infection model and in a nasopharynx model of asymptomatic colonization42 (FIG. 2b). The genes in the inosine monophosphate (IMP) pathway, which are required for the synthesis of purines from glutamine, are important for survival in both niches, whereas the genes involved in synthesis of uridine monophosphate (UMP) and pyrimi-dines from glutamine are important only for survival in the nasopharynx. Further analysis showed that pyrR (SP1278), which is required for the alternative pathway of UMP synthesis from uracil, is important in the lung. This example illustrates two key points. First, in vivo niches present different challenges that the bacterium must cope with, and thus impose different selective pressures; in the nasopharynx, UMP is preferably synthesized from glutamine, whereas in the lung, it is synthesized through uracil by means of PyrR. Second, the essentiality of metabolic pathways differs according to the type of in vivo niche, and this in turn necessitates different treatment strategies. In the example described, although disabling the IMP pathway would reduce bacterial survival in both niches, inhibition of the core UMP pathway (synthesis from glutamine) would reduce the ability of the bacterium to colonize the nasopharynx, but would not compromise the ability of S. pneumoniae to cause pneumonia.
Genetic interactions uncover higher-order genome organization
The identification of a genetic mutation that confers a phenotype is only the first step towards understanding the function of the gene and its broader genetic context. Genes do not function in isolation and are typically integrated in highly interconnected networks. Such genetic networks can be deciphered by screening for genetic interactions between genes, wherein the combination of two mutations in a double mutant results in a strain with a fitness value that deviates from the expected fitness value given by the product of fitness values for each single mutant (that is, Wij ≠ Wi × Wj). Such dependencies between genes have been extensively explored in Saccharomyces cerevisiae and have revealed the functional relevance, organization and transcriptional regulation of individual genes and pathways involved in diverse cellular processes46–55. Genetic interaction networks can also be explored using transposon mutagenesis by creating a transposon library in a strain that lacks a gene of interest (the so-called query gene). Previously, genetic interactions were successfully identified in M. tuberculosis56 and E. coli57 using DNA microarray-based tracking of transposon insertions. Recently, Tn-seq was used to elucidate genetic interactions between five query genes and the rest of the genome in S. pneumoniae, revealing both aggravating and alleviating genetic interactions25. One of the query genes, the catabolite control protein A gene (ccpA), was shown to interact with 64 other genes, suggesting that ccpA is a master regulator of complex carbohydrate metabolism in this pathogen.
Data from genetic interaction mapping can readily be combined with other types of data such as protein–protein interaction data or gene expression data to reveal the higher-order genome organization. For example, a genetic interaction screen was recently combined with gene expression data to determine the regulatory relationship between the response regulator SP2193 (a type of transcription factor) and the core pyrimidine biosynthesis genes42. Although mutations in any of the genes leading to UMP synthesis in the pyrimidine branch caused a severe fitness defect in glucose medium, these defects were fully alleviated by mutations in SP2193 (FIG. 2a). Further analysis confirmed that SP2193 is a positive regulator of the genes in this pathway, leading to the hypothesis that mutations in any one of the genes in this pathway result in a loss of energy or in toxicity owing to the accumulation of toxic intermediates42.
Examining the role of non-coding DNA regions
Transposon sequencing is not restricted to screening for the fitness effects of gene knockouts. In one study58, this technology was used to construct a dense library of Caulobacter crescentus insertion mutants (with 8 bp resolution). In addition to the identification of essential genes, 402 regulatory sequences and 130 non-coding elements from intergenic regions were also shown to be important for growth. Intergenic regions required for optimal growth were identified in M. tuberculosis59 using a similar approach. By combining RNA-seq with Tn-seq, 56 new putative sRNAs in non-coding regions of S. pneumoniae60 were identified. RNA-seq was carried out to ascertain the complete transcriptional activity of the genome and to predict the presence of sRNAs in intergenic regions. Subsequently, Tn-seq was used in mouse models of colonization (infection of the nasopharynx), pneumonia (infection of the lung) and bacteraemia (intravenous infection) to reveal the contribution of the predicted sRNAs to fitness in vivo.
Transposon sequencing in mammalian cells
A method based on the same principle as transposon sequencing has been developed for loss-of-function screens in mammalian cells. Because mammals have two copies of each gene, inactivation of one gene copy rarely leads to a severe phenotype; thus, compared with haploid organisms, cell lines derived from mammals are relatively robust in their tolerance to gene loss. In principle, transposon mutagenesis is therefore not an effective way to screen for genotype–phenotype relationships in diploid cells. However, using a derivative of the human KBM7 chronic myeloid leukaemia cell line, which is haploid for all chromosomes except chromosome 8, the phenotypic effects of insertion mutations are testable61. This so-called haploid genetic screen has been used to identify genes that are important for infection by influenza viruses, for the biosynthesis of diphth amide (which is required for the cytotoxic effects of diphtheria toxin and exotoxin A) and for the action of cytolethal distending toxin (including the gene encoding a cell surface protein that interacts with the toxin)61. This approach has also been used to screen for and identify other host factors, including the membrane receptor that mediates uptake of Clostridium difficile toxin62, and several genes that are important for attachment of Chlamydia trachomatis to its target cell63.
Conclusions and perspectives
Owing to the high-throughput nature of transposon sequencing and the sensitivity of this technique to even small fitness differences, this approach has emerged as the method of choice for examining genotype–phenotype interactions on a large scale. When used in a variety of environmental conditions, the technology has the potential to reveal putative functions for most non-essential genetic elements within an organism. It has been suggested that transposon sequencing methods lack the usefulness of ordered gene knockout arrays, the most important argument being that it is nearly impossible to fish out a particular gene knockout from a transposon insertion library. However, it has been shown that a transposon insertion library is conveniently turned into an ordered gene knockout array using robotic arraying of strains followed by MPS24.
Despite the applicability of transposon sequencing, the technique has limitations. The gene function leads must be followed up with other, often lower-throughput approaches (for example, microscopy and biochemical studies) to determine the details of how the gene functions. In addition, although we have reached high sensitivity and can obtain a quantitative measure of fitness, there is room for improvement. Fitness differences of less than 5% are not detectable with the current methods, but they are important from the standpoint of population biology and evolution.
Transposon sequencing has the potential to be used for other purposes. For example, probing for differences in genotype–phenotype interactions between strains or closely related species has the potential to reveal divergent evolutionary trajectories for shared genes or pathways, such as functional pathways that have undergone reconfiguration either in terms of their regulation or to accomplish new tasks. Although transposon sequencing is ideally suited to bacterial strains for which the complete genome sequence is available, it should work nearly as well in strains with unfinished (draft) genomes.
Although it has been applied almost exclusively to bacterial species, transposon sequencing could be expanded to study other microorganisms. For example, it could be used to probe gene function and gene networks in viruses. All that is needed is a means of introducing transposon insertions into the viral genome. This could be accomplished by building an inducible transposition system within the host, such that insertions are introduced into the viral genome during viral replication. Alternatively, for viruses that are able to establish infection after transformation, transposon insertions could be introduced into a cloned copy of the complete or partial viral genome, such as on a bacterial artificial chromosome. It might also be possible to extend transposon sequencing to diploid mammalian cell lines using either transposons, or retroviral or lentiviral vectors, and to rely on haploinsufficiency to produce phenotypes. Haploinsufficiency generally results in mild phenotypes, but the sensitivity of transposon sequencing might allow the detection of such phenotypes.
One of the most difficult environments to probe is the infected mammalian host, wherein host–pathogen interactions are complex and dynamic. Although transposon sequencing has been used to identify genotype–phenotype interactions in the pathogen, little has been learned about the host environment per se, such as the host parameters that influence pathogen behaviour. However, it should be possible to use transposon sequencing to probe the host environment through the ‘eyes’ of the pathogen. For example, it would be possible to compare gene fitness data for a pathogen following infection of a wild-type host and an immune-deficient host. Because the pathogen would be the constant and the host the variable in this scenario, differences in pathogen gene fitness would help to identify host genes that are involved in pathogenesis. Analyses of this type could easily be expanded to other types of change in the host, such as diet, environment and chemical or drug treatment. In conclusion, the application of transposon sequencing has proved to be highly valuable for the probing of gene function, and further technological advances are likely to see it continue to be applied as a useful, high-throughput screening method for some time to come.
Box 1 | Measuring fitness quantitatively.
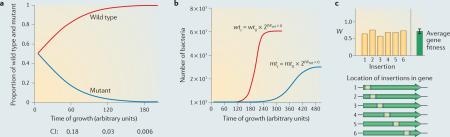
One approach that is commonly used to estimate the fitness cost of a mutation relies on calculating a competitive index (CI), which involves growing the mutant in competition with the wild-type strain. The CI is calculated by dividing the proportion of mutant cells at the end of the competition by the proportion at the start. In the depicted hypothetical example (see the figure, part a), the CI becomes smaller over time. The CI is thus a relative measure of growth, and its value depends on when the measurement is taken. In a quantitative sense, the CI is therefore not particularly informative; it can provide only a semiquantitative and time-dependent estimate of the fitness cost associated with a mutation. Growth curves for the same wild-type and mutant strains grown individually are shown (see the figure, part b), along with the exponential growth equations that define their growth (in which wt and mt are the number of wild-type and mutant cells, respectively; W is the growth rate; and t is time). The only difference between the equations is W; here, Wwt = 1 and Wmt = 0.5. In contrast to the CI, the growth rate is independent of when the measurement is taken and is therefore more meaningful; the mutant shown in this particular example grows half as fast as the wild type (Wwt / Wmt = 2). The most important assumption that is made here is that the measurements are taken during exponential growth; however, this is an assumption that holds for any calculation, including the CI (the carrying capacity, which refers to the maximum number of individuals that can be supported by the environment, is omitted from the equations, for simplicity).
In order to extract this same value (that is, W) from a competition experiment, the following equation can be used64:
Nmt(t1) and Nmt(t2) are the proportions of the mutant in the population at the start (t1) and the end (t2) of the competition, respectively, and d represents the growth of the entire population over the course of the competition (number of all bacteria at t2 / number of all bacteria at t1). Applying this equation to the competition experiment shown in the figure, part a, provides growth rate estimates that are consistent with those obtained from the growth curves in the figure, part b25. Extrapolating to a transposon sequencing experiment in which tens of thousands of mutants are competing against each other at the same time works in the same way. The frequency of each mutant in the population is determined by massively parallel sequencing at t1 and after selection, at t2. In addition, d is determined by simply dividing the number of bacteria in the entire population at t2 by the number of bacteria at t1.
To be able to apply the same analysis to infection experiments in which growth of the bacterial population is not easily determined, a plasmid segregation system was used to estimate net growth42. By transforming Streptococcus pneumoniae with a temperature-sensitive plasmid that does not replicate above 30 °C and thus can be lost by segregation during growth at physiological temperatures, the replication rate of the bacterium can be estimated from the proportion of bacteria that maintain the plasmid. The rate of plasmid loss from the bacterial population in vivo is affected by both growth of the bacterial population and clearance of bacteria from the host (clearance can occur through the host immune system and theoretically should be unaffected by the presence or absence of the plasmid), and thus bacterial population65 and the population doubling time. d can then be determined from the following equations: infection time / doubling time = number of generations (n), and d = 2n). As an initial control, it was confirmed that in vitro estimates of the population growth rate could be accurately determined42. However, the complexity of in vivo environments might introduce unforeseen variation in the rate of plasmid loss, and this could make fitness estimates less accurate. In vivo validation of single knockout mutants indicated that fitness estimates were reasonably accurate, confirming that the approach was successful, at least under the tested conditions. This method was used to determine average gene fitness in a Tn-seq experiment (a type of transposon sequencing; see the figure, part c). To calculate fitness for a single gene, fitness must first be calculated for each transposon insertion, followed by averaging over all the insertions found within that gene to obtain a single fitness value (including a standard deviation). More than 150 genotype–phenotype relationships and genetic interactions obtained from in vitro and in vivo Tn-seq experiments have been validated using standard one-on-one competitions and growth curves25,42. Thus, this approach generates a highly meaningful quantitative fitness value that is reproducible across experiments and is a close approximation of the growth rate.
Acknowledgements
T.v.O. was supported by a postdoctoral fellowship from the Netherlands Organization for Scientific Research (Rubicon-NWO) and the Charles H. Hood Foundation. A.C. is an investigator of the Howard Hughes Medical Institute.
Glossary
- Bacterial artificial chromosome
A bacterial plasmid that contains a large eukaryotic DNA insertion (typically >150 kb) and can be used for cloning, genetic manipulation and transformation.
- ChIP–seq
(Chromatin immunoprecipitation followed by sequencing). A method that uses crosslinking of a protein to DNA followed by immunoprecipitation of the complex and subsequent sequencing of the bound DNA to reveal the binding site of the protein.
- Complementation assays
Assays in which a wild-type copy of a gene is reintroduced into a cell or organism that lacks the gene. This can confirm that the phenotype is caused by disruption or deletion of the gene in question, and that this phenotype can be reversed.
- Degenerate genes
Genes that once had a function, but through the accumulation of mutations, became inactive.
- Digital-counting applications
Methods that count the total number of reads obtained for a particular sequence after massively parallel sequencing (in contrast to hybridization-based methods of quantification).
- DNA microarray
A glass slide (or other surface) on which oligonucleotides or PCR products of defined sequence are spotted. These microarrays are used to quantify the nucleic acids within a sample by hybridization.
- Haploinsufficiency
The inability of a single functional copy of a gene to produce a wild-type phenotype in a diploid organism. This occurs when the second copy of the gene is inactivated by mutation.
- Overlap analysis
An analysis that indicates a putative function for a hypothetical virulence gene using fitness data for the gene obtained during growth in defined in vitro conditions.
- Promoter assays
Assays that measure the transcriptional activity of a gene promoter by converting the RNA transcripts to cDNAs and then using massively parallel sequencing to determine the number of cDNA molecules present.
- Signature-tagged mutagenesis
A technique in which a transposon is tagged with a specific DNA sequence (a bar-code) that is used to determine the presence of the transposon in a DNA pool (as the amplified and labelled tag hybridizes to a probe on a membrane).
- sRNA-seq
(Small-RNA sequencing). The discovery of non-coding sRNAs through direct sequencing of their cDNAs by massively parallel sequencing.
- Transductional crosses
Experiments in which DNA is transferred from one bacterium to another by means of a bacteriophage.
- Type IIS restriction endonuclease
An enzyme that cleaves DNA at a defined distance from an asymmetrical recognition site.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Contributor Information
Tim van Opijnen, Biology Department, Boston College, 140 Commonwealth Avenue, 420 Higgins Hall, Chestnut Hill, Massachusetts 02467, USA. tim.vanopijnen@bc.edu.
Andrew Camilli, Howard Hughes Medical Institute and the Department of Molecular Biology and Microbiology, Tufts University School of Medicine, 136 Harrison Avenue, Boston, Massachusetts 02111, USA. andrew.camilli@tufts.edu.
References
- 1.Kleckner N, Roth J, Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J. Mol. Biol. 1977;116:125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- 2.Craig NL. Target site selection in transposition. Annu. Rev. Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 3.Kidwell MG, Lisch RD. Transposable elements, parasitic DNA and genome evolution. Evolution. 2001;55:1–25. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 4.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Kleckner N, Chan RK, Tye B-K, Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J. Mol. Biol. 1975;97:561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- 6.Smith V, Botstein D, Brown PO. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl Acad. Sci. USA. 1995;92:6479–6483. doi: 10.1073/pnas.92.14.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith V, Chou KN, Lashkari D, Botstein D, Brown PO. Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 8.Akerley BJ, et al. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl Acad. Sci. USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hensel M, et al. Simultneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 10.Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nature Rev. Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- 11.Mei JM, Nourbakhsh F, Ford CW, Holden DW. Identification of Staphylococcus aureusvirulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 12.Chiang SL, Mekalanos JJ. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 13.Jones AL, Knoll KM, Rubens CE. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 2000;37:1444–1455. doi: 10.1046/j.1365-2958.2000.02099.x. [DOI] [PubMed] [Google Scholar]
- 14.Autret N, Dubail I, Trieu-Cuot P, Berche P, Charbit A. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 2001;69:2054–2065. doi: 10.1128/IAI.69.4.2054-2065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau GW, et al. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 2001;40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 16.Polissi A, et al. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hava D, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 18.Loman NJ, et al. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nature Rev. Microbiol. 2012;10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 19.Bork P. Powers and pitfalls in sequence analysis: the 70% hurdle. Genome Res. 2000;10:398–400. doi: 10.1101/gr.10.4.398. [DOI] [PubMed] [Google Scholar]
- 20.Galperin MY, Koonin EV. From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 2010;28:398–406. doi: 10.1016/j.tibtech.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasif S, Steffen M. Biochemical networks: the evolution of gene annotation. Nature Chem. Biol. 2010;6:4–5. doi: 10.1038/nchembio.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl Acad. Sci. USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langridge GC, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Opijnen T, Camilli A. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr. Protoc. Microbiol. 2010;19:1E.3.1–1E.3.16. doi: 10.1002/9780471729259.mc01e03s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nature Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JM, et al. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 33.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nature Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nature Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 35.Alsford S, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan RD, Dwinell EA, Bhatia TK, Lang EM, Luyten YA. The MmeI family: type II restriction– modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res. 2009;37:5208–5221. doi: 10.1093/nar/gkp534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio. 2011;2:e00315–10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khatiwara A, et al. Genome scanning for conditionally essential genes in Salmonella enterica serotype Typhimurium. Appl. Environ. Microbiol. 2012;78:3098–3107. doi: 10.1128/AEM.06865-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brutinel ED, Gralnick JA. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol. Microbiol. 2012;86:273–283. doi: 10.1111/j.1365-2958.2012.08196.x. [DOI] [PubMed] [Google Scholar]
- 41.Klein BA, et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Opijnen T, Camilli A. A fine scale phenotype–genotype virulence map of a bacterial pathogen. Genome Res. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eckert SE, et al. Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J. Bacteriol. 2011;193:1771–1776. doi: 10.1128/JB.01292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS. Identification of Escherichia coli O157: H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology. 2004;150:3631–3645. doi: 10.1099/mic.0.27448-0. [DOI] [PubMed] [Google Scholar]
- 45.Crimmins GT, et al. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS Pathog. 2012;8:e1002828. doi: 10.1371/journal.ppat.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ooi S, Shoemaker D, Boeke J. DNA helicase gene interaction network defined ufsing synthetic lethality analyzed by microarray. Nature Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 47.Pan X, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 48.Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nature Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 49.Fiedler D, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins S, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 51.Tong A. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 52.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 53.St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nature Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dixon S, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu. Rev. Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 55.Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joshi SM, et al. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl Acad. Sci. USA. 2006;103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Girgis H, Liu Y, Ryu W, Tavazoie SA. Comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:e154. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christen B, et al. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:1–7. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang YJ, et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann B, et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012;8:e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 62.Papatheodorou P, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc. Natl Acad. Sci. USA. 2011;108:16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosmarin DM, et al. Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc. Natl Acad. Sci. USA. 2012;109:10059–10064. doi: 10.1073/pnas.1120244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Opijnen T, Boerlijst MC, Berkhout B. Effects of random mutations in the human immunodeficiency virus type 1 transcriptional promoter on viral fitness in different host cell environments. J. Virol. 2006;80:6678–6685. doi: 10.1128/JVI.02547-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamin WH, Hall P, Roberts SJ, Briles DE. The primary effect of the Ity locus is on the rate of growth of Salmonella typhimurium that are relatively protected from killing. J. Immunology. 1990;144:3143–3151. [PubMed] [Google Scholar]
- 66.Goodman A, Wu M, Gordon J. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nature Protoc. 2011;6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]