Abstract
Background
Estimates of the heritability of plasma fibrinogen concentration, an established predictor of cardiovascular disease (CVD), range from 34 to 50%. Genetic variants so far identified by genome-wide association (GWA) studies only explain a small proportion (< 2%) of its variation.
Methods and Results
We conducted a meta-analysis of 28 GWA studies, including more than 90,000 subjects of European ancestry, the first GWA meta-analysis of fibrinogen levels in 7 African Americans studies totaling 8,289 samples, and a GWA study in Hispanic-Americans totaling 1,366 samples. Evaluation for association of SNPs with clinical outcomes included a total of 40,695 cases and 85,582 controls for coronary artery disease (CAD), 4,752 cases and 24,030 controls for stroke, and 3,208 cases and 46,167 controls for venous thromboembolism (VTE). Overall, we identified 24 genome-wide significant (P<5×10−8) independent signals in 23 loci, including 15 novel associations, together accounting for 3.7% of plasma fibrinogen variation. Gene-set enrichment analysis highlighted key roles in fibrinogen regulation for the three structural fibrinogen genes and pathways related to inflammation, adipocytokines and thyrotrophin-releasing hormone signaling. Whereas lead SNPs in a few loci were significantly associated with CAD, the combined effect of all 24 fibrinogen-associated lead SNPs was not significant for CAD, stroke or VTE.
Conclusion
We identify 23 robustly associated fibrinogen loci, 15 of which are new. Clinical outcome analysis of these loci does not support a causal relationship between circulating levels of fibrinogen and CAD, stroke or VTE.
Keywords: Fibrinogen, cardiovascular disease, genome-wide association study
Introduction
Fibrinogen plays a major role in wound healing and thrombosis. Circulating levels of fibrinogen are upregulated in inflammatory conditions, consequently serving as an important marker of inflammation. Fibrinogen is a well-established predictor of cardiovascular disease (CVD) outcomes, such as myocardial infarction,1, 2 stroke3 and venous thromboembolism (VTE) .4, 5
It is estimated that 34 (extended pedigrees study) to 44% (twins study) of the inter-individual variation in fibrinogen levels is heritable,6, 7 indicating a substantial influence of genetics. Two recent meta-analyses of genome-wide association (GWA) studies, conducted in cohorts of European ancestry, identified several genetic variants affecting fibrinogen levels.8, 9 These variants account only for a small proportion (< 2%) of plasma fibrinogen variation, suggesting that additional genetic variants with more modest effects may remain to be detected.
There is now increasing evidence that a substantial proportion of consequential genetic variation for many phenotypes is tagged by common SNPs10, although most of these SNPs cannot pass the restrictive genome-wide significance level of p<5×10−8 in a typical association study. To overcome this limitation, increased sample sizes are needed. We conducted a large meta-analysis of 28 GWA studies including more than 90,000 individuals of European ancestry, a 4-fold increase in sample size compared to prior meta-analyses.8, 9 We included data from an additional 8,423 samples from the first GWA studies of African Americans and 1,447 Hispanic individuals to also explore whether ethnic differences exist in the genetic regulation of plasma fibrinogen concentration. To further elucidate possible biological mechanisms underlying fibrinogen regulation, we examined genome-wide significant loci in relation to expression levels of nearby genes, and in gene pathway analyses. Finally, we examined whether fibrinogen related genes affect risk of coronary artery disease (CAD), stroke and VTE.11–16
Methods
Cohorts and Plasma Fibrinogen Measurements
Twenty-eight studies contributed to the discovery GWA study meta-analysis of European-ancestry individuals. Characteristics of all participating studies are provided in Supplementary Methods and Supplementary Table S1. In 7 cohorts, with 33,745 individuals, plasma fibrinogen concentration was measured by an immunonephelometric method.17 For the other 21 European-ancestry cohorts ( 57,578 individuals), plasma fibrinogen levels were determined by a functional method (based on the Clauss method).18 Seven African-American cohorts with GWA data, including a total of 8,423 individuals (5,937 with Clauss and 2,486 with immunonephelometric measures)and one cohort of 1,447 Hispanics with immunonephelometric fibrinogen measures was also analyzed (Supplementary Methods and Supplementary Table S2). Exclusion criteria applied in individual cohorts are provided in Supplementary Methods. All studies were approved by the relevant research ethics committees.
Genotyping, Quality Control of Genotype Data and Imputation
Commercial arrays were used for genome-wide genotyping in all cohorts, and quality control (QC) filtering of SNP genotype data was generally performed in individual cohorts by call rate, minor allele frequency (MAF) and deviation from Hardy-Weinberg equilibrium (HWE) (Supplementary Methods, Supplementary Tables S3 and S4). Approximately 2.5 million autosomal SNPs were imputed cohorts using the HapMap II Caucasian (CEU, Centre d’Etude du Polymorphisme Humain) sample as reference panel for the European-ancestry cohorts, a combined CEU+YRI reference panel for the African-American cohorts, and a combined CEU+YRI+CHB+JPT reference panel for the Hispanic sample. MACH or IMPUTE software19–21 were used in the imputation (Supplementary Tables S3 and S4).
Meta-Analysis of GWA Studies
Values of plasma fibrinogen concentration were natural logarithm-transformed prior to analysis. Association analyses were conducted in each cohort of measured and imputed autosomal SNP allele dosage with fibrinogen values, using a linear regression model assuming additive genetic effects adjusted for age and sex. Additional adjustments for principal components or multi-dimensional scaling, country, or center were made, when necessary, by individual cohorts to account for population stratification (see Supplementary Methods). Relatedness was accounted for in family studies by applying linear mixed-effect models. Genotype-phenotype association results from the 28 cohorts were then meta-analyzed by using an inverse-variance model with fixed effects in METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html).22 In order to identify additional independent association signals in the genome-wide significant loci, conditional GWA analysis was performed as described in Supplementary Methods. Overall, we selected for further analysis only SNPs from genome-wide significantly associated loci, including the lead SNP for each locus in the initial meta-analysis along with one additional lead SNP representing a new clear signal identified in the conditional analysis.
In order to identify genes that regulate fibrinogen levels in other ethnic groups, we conducted a separate GWA meta-analysis using 7 separate GWA scans in African Americans totaling 8,289 samples, and a single GWA analysis in a cohort of Hispanic-Americans totaling 1,366 samples
The threshold of genome-wide significance was set at P=5.0×10−8 for the primary analyses of GWA with plasma fibrinogen levels and their heterogeneity measures, as well as for the conditional meta-analysis. We used Bonferroni correction for the exploration of the 24 lead-SNPs in African-American and Hispanic samples, and for the lookups in clinical outcomes (P<0.002).
Genetic risk score
A genetic risk score (GRS) was computed using data from 88,251 European-ancestry individuals to model the increase in fibrinogen levels according to number of fibrinogen-raising alleles for each of the lead SNPs. Methods are further described in Supplementary Methods.
Multivariable adjusted model
We re-analyzed the association with plasma fibrinogen concentration of the lead SNPs, using a linear model with further adjustment for BMI and smoking, in addition to sex and age and the extra covariates used in each cohort in the discovery analyses. Association results from all cohorts were then meta-analyzed using inverse-variance weighted fixed-effects meta-analysis implemented in METAL.
Pathway Analyses
MAGENTA and GRAIL23, 24 were used to assess putative relationships between the lead SNPs and to infer genes and pathways underlying SNP associations with plasma fibrinogen levels. MAGENTA v. 2 analysis was performed as described,24 including gene sets from Gene Ontology (GO), KEGG, PANTHER, and Ingenuity downloaded in June 2011 (http://www.broadinstitute.org/mpg/magenta/). Gene set statistics were determined for an empirically derived 95th percentile threshold of gene-wide adjusted P values. Only gene sets meeting a false discovery rate (FDR) < 0.05 were considered for further inspection. Candidate SNPs were identified in the MAGENTA analysis as SNPs with nominal locus-wide corrected P-values (corrected P<0.05) mapping to genes in gene sets that met FDR<0.05. GRAIL analysis was performed as described (http://www.broadinstitute.org/mpg/grail/) using the pair-wise similarity metric compiled from the literature in December 2006 to limit bias, as recommended 25.
Association with Gene Expression in Human Liver
The lead SNPs and their perfect proxies (r2=1) were further analyzed with respect to association with expression levels of nearby genes (located within ±200 kilobases (kb) of the SNP). Global gene expression data from human liver were obtained from the Advanced Study of Aortic Pathology (ASAP).26 Details of the ASAP biobank and the methods for gene expression analysis and genotyping are provided in the Supplementary Methods. Further queries were made against significant results from four other liver eQTL analyses whose methods were previously published.27–30
Associations with Clinical Outcomes
We examined associations of the 24 lead SNPs with prevalent CAD, stroke and VTE. Genotype-CAD association results for the selected SNPs were obtained from the Coronary ARtery DIsease Genome-wide Replication And Meta-analysis (CARDIoGRAM) and Europe South Asia Coronary Artery Disease Genetics (C4D) consortia, including a total of 40,695 CAD cases and 85,582 controls. Lead SNP associations with stroke were explored in data generated from four large cohorts composing the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium, including 1544 incident strokes (1164 ischemic strokes) developed over an average follow-up of 11 years, and 18,058 controls, and in data generated from four cohorts comprising the Welcome Trust Case Control Consortium (WTCCC), including 3,548 cases with ischemic stroke and 5,972 controls. The SNP genotype-VTE association results were generated in 3,208 VTE cases and 46,167 controls from the French MARTHA and the CHARGE studies. Definitions of the disease phenotypes adopted in each individual study are detailed elsewhere.11–15, 31 Each of the 24 fibrinogen-associated SNPs was tested for association with each of the clinical outcomes by logistic regression, adjusting for age and sex. The log-odds-ratios and their standard errors for each SNP were standardized for direction and magnitude to correspond to the change in allele dosage that accounted for a 3.1% relative increase in circulating fibrinogen level (fibrinogen-effect associated with the FGB variant rs1800789). These harmonised effect estimates were then pooled by fixed-effects (inverse-variance weighted) meta-analysis (stroke and VTE) or by random-effects meta-analysis (for CAD, due to significant heterogeneity in both direction and magnitude of the harmonised log-odds-ratios).
Results
Meta-analysis in European-Ancestry Samples
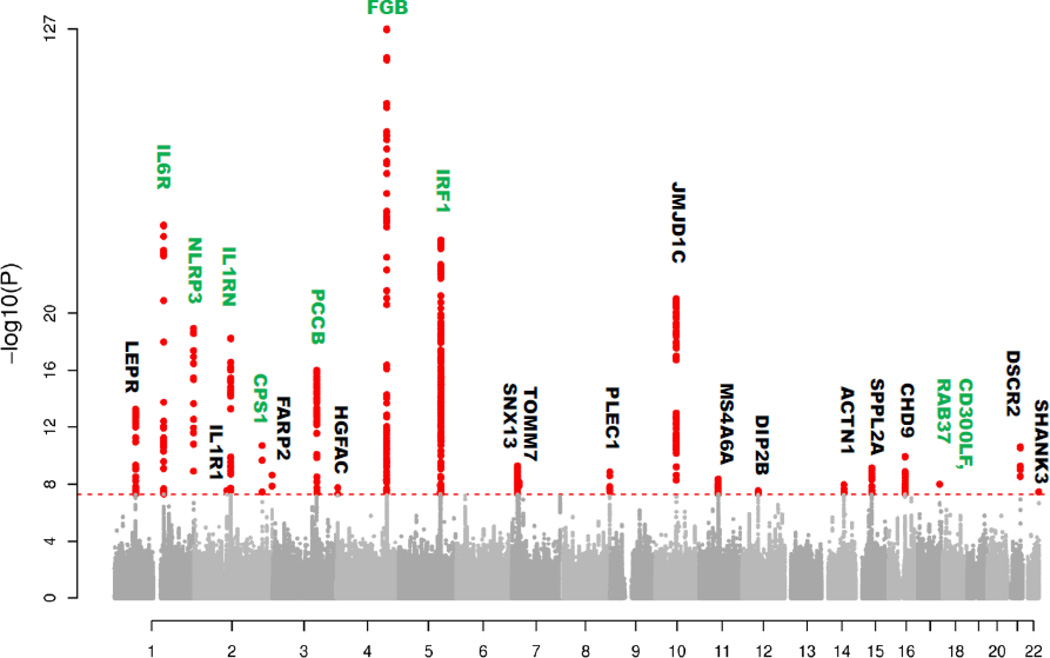
Meta-analysis was performed for 2,515,567 SNPs on individual GWA study results generated in 28 European-ancestry cohorts including a total of 91,323 individuals. A total of 985 SNPs, located in 23 chromosomal loci, passed the genome-wide significance threshold of P=5.0×10−8 (Figure 1). Among the 23 loci (designated according to nearest gene), 8 (IL6R, NLRP3, IL1RN, CPS1, PCCB, FGB, IRF1 and CD300LF) represent replications of previously identified fibrinogen-associated loci and 15 are novel associations (JMJD1C, LEPR, PSMG1, CHD9, SPPL2A, PLEC1, FARP2, MS4A6A, TOMM7/IL6, ACTN1, HGFAC, IL1R1, DIP2B and SHANK3/CPT1B). More information about these genes is provided in Supplementary Table S5. Further information about the lead SNPs and their association with fibrinogen levels is listed in Table 1.
Figure 1.
Manhattan plot of the association P-values for plasma fibrinogen concentration in the meta-analysis performed on European-ancestry samples. Analyzed SNPs are plotted on the X-axis ordered by chromosomal position. Y-axis plots the logarithm of the P-values. Gene loci labeled in green were previously known; gene loci labeled in black are novel discoveries in this meta-analysis. The dotted line indicates the threshold for genome-wide significance (P=5×10−8).
Table 1.
Details of the 24 lead SNPs and association β and P-values for the original meta-analysis performed with the European-ancestry cohorts (adjusted for age and sex) together with the corresponding values of the values obtained in the same cohorts with further adjustments for BMI and smoking.
| Original meta-analysis | Further adjustment for BMI and smoking |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Band | Position | Closest gene | In gene |
Distan ce (bp) |
A 1 |
A 2 |
Freq | Beta | SE | P | Het P | N | Beta | SE | P |
| rs1938492 | 1p31.3 | 65890417 | LEPR | 14653 | A | C | 0.62 | 0.008 | 0.001 | 5.28×10−14 | 0.438 | 89330 | 0.008 | 0.001 | 1.12×10−15 | |
| rs4129267 | 1q21.3 | 152692888 | IL6R | intron | T | C | 0.39 | −0.011 | 0.001 | 5.97×10−27 | 0.724 | 91419 | −0.011 | 0.001 | 4.57×10−30 | |
| rs10157379 | 1q44 | 245672222 | NLRP3 | intron | T | C | 0.62 | 0.010 | 0.001 | 1.15×10−19 | 0.416 | 86730 | 0.010 | 0.001 | 3.12×10−22 | |
| rs12712127 | 2q11.2 | 102093093 | IL1R1/IL1R2 | 43740 | A | G | 0.41 | 0.006 | 0.001 | 2.72×10−08 | 0.097 | 91406 | 0.006 | 0.001 | 3.66×10−10 | |
| rs6734238 | 2q13 | 113557501 | IL1F10/IL1RN | 7603 | A | G | 0.58 | −0.009 | 0.001 | 5.77×10−19 | 0.487 | 91426 | −0.010 | 0.001 | 6.66×10−22 | |
| rs715 | 2q34 | 211251300 | CPS1 | exon | T | C | 0.68 | 0.009 | 0.001 | 1.98×10−11 | 0.153 | 74715 | 0.011 | 0.001 | 3.95×10−19 | |
| rs1476698 | 2q37.3 | 241945122 | FARP2 | intron | A | G | 0.65 | 0.007 | 0.001 | 2.24×10−09 | 0.420 | 91419 | 0.007 | 0.001 | 1.44×10−10 | |
| rs1154988 | 3q22.3 | 137407881 | MSL2/PCCB | 10503 | A | T | 0.78 | −0.010 | 0.001 | 9.64×10−17 | 0.154 | 91416 | −0.012 | 0.001 | 2.98×10−24 | |
| rs16844401 | 4p16.2 | 3419450 | HGFAC/LRPAP1 | exon | A | G | 0.08 | 0.015 | 0.003 | 1.74×10−08 | 0.077 | 74680 | 0.014 | 0.002 | 7.07×10−09 | |
| rs1800789 | 4q32.1 | 155702193 | FGB | 1388 | A | G | 0.21 | 0.031 | 0.001 | 1.68×10−127 | 0.001 | 91301 | 0.031 | 0.001 | 1.94×10−140 | |
| rs11242111 | 5q31.1 | 131783957 | C5orf56/IRF1 | intron | A | G | 0.05 | 0.023 | 0.002 | 1.60×10−21 | 0.353 | 91423 | 0.024 | 0.002 | 1.14×10−23 | |
| rs2106854 | 5q31.1 | 131797073 | C5orf56/IRF1 | intron | T | C | 0.21 | −0.019 | 0.001 | 1.72×10−48 | 0.082 | 91406 | −0.019 | 0.001 | 1.93×10−54 | |
| rs10226084 | 7p21.1 | 17964137 | SNX13/PRPS1L1 | 17481 | T | C | 0.52 | −0.007 | 0.001 | 5.05×10−10 | 0.441 | 91403 | −0.007 | 0.001 | 6.68×10−11 | |
| rs2286503 | 7p15.3 | 22823131 | TOMM7 | intron | T | C | 0.36 | −0.006 | 0.001 | 6.88×10−09 | 0.845 | 91413 | −0.005 | 0.001 | 2.26×10−07 | |
| rs7464572 | 8q24.3 | 145093155 | PLEC1 | intron | C | G | 0.60 | −0.007 | 0.001 | 1.33×10−09 | 0.123 | 82730 | −0.006 | 0.001 | 7.41×10−09 | |
| rs7896783 | 10q21.3 | 64832159 | JMJD1C | intron | A | G | 0.48 | −0.010 | 0.001 | 8.90×10−22 | 0.754 | 91412 | −0.009 | 0.001 | 4.43×10−20 | |
| rs1019670 | 11q12.1 | 59697175 | MS4A6A | EXON | A | T | 0.36 | −0.007 | 0.001 | 4.37×10−09 | 0.696 | 9018 | −0.006 | 0.001 | 8.09×10−08 | |
| rs7968440 | 12q13.13 | 49421008 | DIP2B | intron | A | G | 0.64 | 0.006 | 0.001 | 2.74×10−08 | 0.360 | 91405 | 0.006 | 0.001 | 1.37×10−09 | |
| rs434943 | 14q24.1 | 68383812 | ACTN1 | 26780 | A | G | 0.31 | 0.007 | 0.001 | 1.08×10−08 | 0.014 | 86189 | 0.008 | 0.001 | 1.73×10−10 | |
| rs12915708 | 15q21.2 | 48835894 | SPPL2A | intron | C | G | 0.30 | −0.007 | 0.001 | 6.87×10−10 | 0.625 | 91434 | −0.007 | 0.001 | 3.45×10−11 | |
| rs7204230 | 16q12.2 | 51749832 | CHD9 | intron | T | C | 0.70 | 0.008 | 0.001 | 1.18×10−10 | 0.493 | 82835 | 0.008 | 0.001 | 6.40×10−12 | |
| rs10512597 | 17q25.1 | 70211428 | CD300LF | intron | T | C | 0.18 | −0.008 | 0.001 | 9.92×10−09 | 0.108 | 86737 | −0.009 | 0.001 | 4.23×10−11 | |
| rs4817986 | 21q22.2 | 39387382 | PSMG1 | 81871 | T | G | 0.28 | −0.008 | 0.001 | 2.46×10−11 | 0.539 | 85293 | −0.009 | 0.001 | 3.39×10−14 | |
| rs6010044 | 22q13.33 | 49448804 | SHANK3/ARSA | 11131 | A | C | 0.80 | −0.008 | 0.001 | 3.41×10−08 | 0.582 | 89138 | −0.008 | 0.001 | 7.07×10−09 | |
The closest gene is indicated in bold. Beta values and frequencies refer to allele 1 (A1).
To search for further independent association signals within the 23 loci, we repeated the individual GWA analyses, conditioning on the 23 lead SNPs. This analysis revealed two genome-wide significant SNPs located, respectively, in the FGA gene (rs2070016, P=3.9×10−8) and on chromosome 5 (rs11242111, P=1.60X10−21) (Supplementary Figure S1). Accordingly, rs11242111 was added to the list of independent lead SNPs selected for further analyses (Table 1). The rs2070016, in FGA, showed evidence of correlation with the lead SNP rs1800789 in FGB (r2 =0.364 according to 1000 Genomes Map Pilot 1); hence, we did not select this SNP for further analyses. After adjusting for number of tests, none of the 24 lead SNPs showed significant heterogeneity across European-ancestry cohorts. Regional association plots for the 24 loci are shown in Supplementary Figure S2.
Further adjustment for body mass index (BMI) and smoking, which together explained 5.3% of the variation in plasma fibrinogen level amongst 81,511 individuals from the European-ancestry meta-analysis, resulted in stronger associations for most of the lead SNPs but no new discoveries (Table 1).
Meta-analysis and Validation of European-ancestry Loci in African-American and Hispanic Samples
The Manhattan and QQ plots (λ=1.012) reporting the results for the African-American samples are shown in the Supplementary Figure S3. Only the FGA/FGB/FGG locus on chromosome 4 reached genome-wide significance in the African American meta-analysis, with the most strongly associated SNP being rs4463047, P=4.63×10−10, at 12,790 bp from rs1800789 (P=4.02×10−7). No single SNP attained genome-wide significance in the Hispanic samples (Supplementary Figure S3).
We tested the association of the 24 European-ancestry lead SNPs in the African American meta-analysis (Supplementary Table S6). After correcting for 24 statistical tests (P-value threshold < 0.002) only the two lead-SNPs, rs1800798 (FGB) and rs6734238 (ILRN) passed the significant threshold. However, 5 other lead-SNPs, located in the IRF1, IL6R, CHD9, JMJD1C and MS4A6A loci, were associated at P<0.05, with consistent directions of effect in both populations (Supplementary Table S7). Furthermore, at 20 of the 24 lead SNPs the direction of the beta estimate was the same in the European and African-American samples (P=0.00077, sign test). In the Hispanic samples, 3 European-ancestry lead SNPs, in FGB (rs1800798), IL6R (rs6734238) and CHD9 (rs7204230), passed the significance threshold (24 SNPs / P < 0.002) for association, and 3 additional lead SNPs were associated at a nominally significant threshold of P<0.05, with consistent directions of effect in both populations (Supplementary Table S6). In addition, the direction of the beta estimate at 20 of the 24 lead SNPs was the same in the European and Hispanic samples (P=0.00077, sign test).
GRS and Proportion of Variance Explained
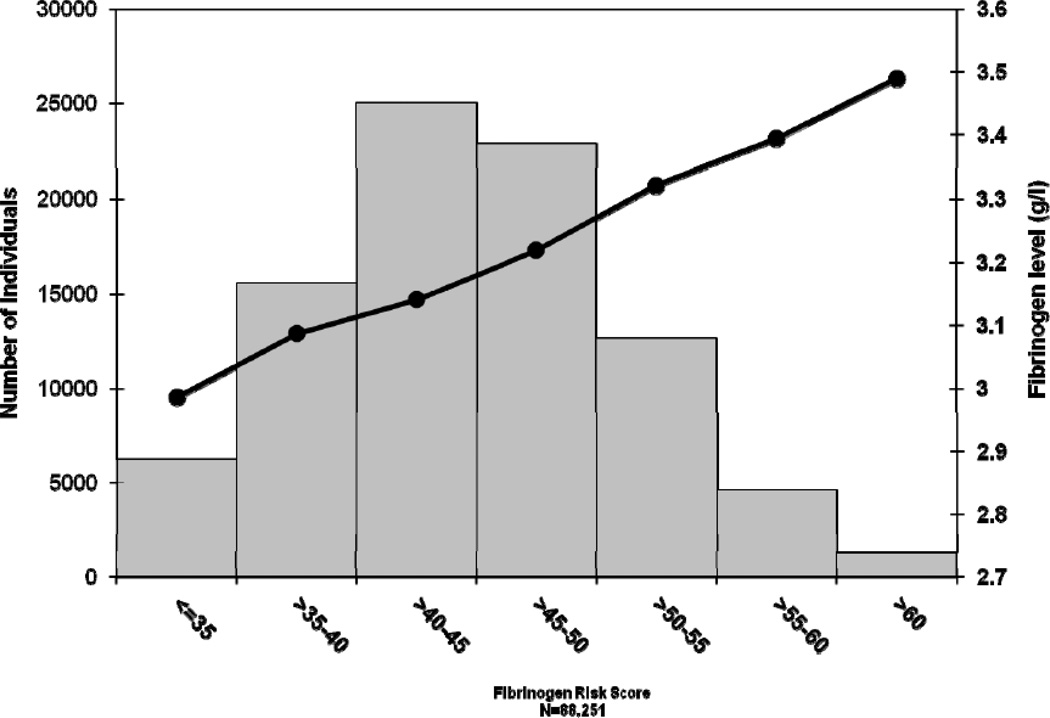
Figure 2 presents the average fibrinogen values (in g/l) across categories of the GRS. The mean percentage of residual variance (after adjustment for age and sex) explained by 24 lead-SNPs was 3.7% in all European-ancestry cohorts (range 1.4–7.6% in individual cohorts). The heritability of plasma fibrinogen concentration estimated from the family cohorts within this study (NTR, CROATIA-Vis, CROATIA-Korcula, ORCADES, FHS and SardiNIA) ranged from 15% to 51% (mean(SD)=31(15)%) (Supplementary Results). The proportion of variance in fibrinogen levels explained by common SNPs (MAF>0.01) was calculated in one of our participant cohorts (WGHS, n=21,336) using the method proposed by Yang and Visscher10. Results showed that 16% (SE=0.017) of the variance in fibrinogen levels was explained by common SNPs.
Figure 2.
Mean values for plasma fibrinogen concentration in g/l (right Y-axis) plotted by categories of fibrinogen-associated single nucleotide polymorphism (SNP) score (X-axis), represented by the black dots. Number of individuals in each category is represented by the grey bars (left Y-axis).
Finally, the GRS was strongly associated with levels of fibrinogen in the combined African-American cohorts (P= 1.5×10−8) and the Hispanic cohort (P=3.8×10−15).
Pathway and Expression QTL Analyses
We performed additional in silico pathway analyses using GRAIL and MAGENTA (Supplementary Table S8). The GRAIL results identified 6 SNPs (rs6734238, rs12712127, rs8192284, rs10157379, rs1938492 and rs6831256) that were located within or near genes (IL1RN, IL1R1, IL6R, NLRP3, LEPR and LRPAP1) with significantly related function among all of the genes in the vicinity of the 24 lead SNPs, suggesting that these genes should be prioritized as the most plausible functional candidate genes within the associated loci. Gene-set enrichment analysis using MAGENTA (based on the whole genome-wide genetic dataset) identified several gene sets and pathways that were enriched in the analysis (Supplementary Table S9). Apart from the three structural genes, the most represented pathways were related to inflammation (acute-phase response, interleukin signaling), adipocytokine signaling and thyrotrophin-releasing hormone signaling. According to these results, several genes (LEPR, IL6R, IL1R, IL1F10/IL1F5/IL1F8/IL1RN, FGA/FGB, ACTN1 and CPT1B) were prioritized as plausible candidate genes within our 23 genomic regions. A comprehensive SNP list, which includes both the 24 lead SNPs and the SNPs selected by either GRAIL or MAGENTA on the whole genome-wide genetic dataset, is reported in Supplementary Table S9.
We then interrogated the 24 lead SNPs and their perfect proxies with respect to their associations with expression levels of nearby genes (located within ±200 kb of the lead SNP) in 5 human liver databases. Expression levels of LEPR, PCCB, MSL2L1, NGFRAP1, FGB and TOMM7 were significantly associated with allelic differences in one of the 24 lead SNPs (results are shown in Supplementary Table S8). Finally, to assess the functional role of SNPs in Fibrinogen genes we also studied the eQTL associations of all SNPs within 100Kb of the fibrinogen genes cluster. The highest association with expression of fibrinogen transcripts within the fibrinogen cluster was found for SNP rs4220 (P=1.38×10−20), causing a missense mutation in the FGB gene. All positive associations with fibrinogen transcripts are shown in Supplementary Table S10.
Associations with Clinical Outcomes
After correction for multiple testing (P<0.002 threshold), rs4129267 located in the IL6R locus, rs6734238 in the IL1F10/IL1RN locus and rs1154988 in the PCCB locus were found to be significantly associated with CAD; however, the directions of the effects on CAD and fibrinogen levels were consistent only for rs4129267 in the IL6R locus . The pooled association for the 24 lead SNPs with CAD was not significant (OR(CI95%)= 1.00 (0.97,1.03)). None of the fibrinogen-associated lead SNPs was significantly associated with stroke or VTE after correction for multiple testing. The pooled results were suggestive for stroke (stroke OR(CI95%)= 1.03 (1.00,1.07); but not for VTE OR(CI95%)= 0.96 (0.92,1.01)) (Table 2). Additional results from the WTCCC stroke consortium, generated according to clinical subphenotypes, are shown in Supplementary Table S11. No significant associations with stroke subphenotypes were found after correction for multiple hypothesis testing.
Table 2.
Association results for the 24 lead SNPs with coronary artery disease (CAD), stroke and venous thromboembolism (VTE).
| CAD* | Stroke** | VTE*** | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Band | Allele1 | Allele2 | Freq1 | Closest gene | OR | SE | P | OR | SE | P | OR | SE | P |
| rs1938492 | 1p31.3 | A | C | 0.597 | LEPR | 0.98 | 0.011 | 0.038 | 0.98 | 0.025 | 0.405 | 1.00 | 0.032 | 0.892 |
| rs4129267 | 1q21.3 | T | C | 0.378 | IL6R | 0.96 | 0.011 | 1.73×10−05 | 0.97 | 0.024 | 0.212 | 1.01 | 0.032 | 0.838 |
| rs10157379 | 1q44 | T | C | 0.603 | NLRP3 | 1.00 | 0.011 | 0.883 | 1.02 | 0.025 | 0.329 | 1.04 | 0.032 | 0.204 |
| rs12712127 | 2q11.2 | A | G | 0.451 | IL1R1/IL1R2 | 1.00 | 0.011 | 0.985 | 0.98 | 0.025 | 0.423 | 1.00 | 0.032 | 0.909 |
| rs6734238 | 2q13 | A | G | 0.589 | IL1F10/IL1RN | 1.04 | 0.011 | 9.44×10−05 | 1.00 | 0.025 | 0.974 | 1.01 | 0.032 | 0.702 |
| rs715 | 2q34 | T | C | 0.685 | CPS1 | 1.03 | 0.013 | 0.011 | 1.01 | 0.029 | 0.822 | 0.91 | 0.054 | 0.081 |
| rs1476698 | 2q37.3 | A | G | 0.615 | FARP2 | 1.00 | 0.011 | 0.873 | 1.02 | 0.026 | 0.388 | 1.06 | 0.033 | 0.089 |
| rs1154988 | 3q22.3 | A | T | 0.778 | MSL2/PCCB | 1.04 | 0.013 | 0.002 | 0.95 | 0.029 | 0.100 | 0.95 | 0.037 | 0.186 |
| rs16844401 | 4p16.2 | A | G | 0.089 | HGFAC/ LRPAP1 | 1.03 | 0.024 | 0.263 | 1.01 | 0.052 | 0.848 | 0.92 | 0.082 | 0.285 |
| rs1800789 | 4q32.1 | A | G | 0.2 | FGB | 1.00 | 0.014 | 0.939 | 0.99 | 0.031 | 0.828 | 0.89 | 0.04 | 0.004 |
| rs11242111 | 5q31.1 | A | G | 0.101 | C5orf56/IRF1 | 0.95 | 0.024 | 0.02 | 1.09 | 0.057 | 0.145 | 0.97 | 0.079 | 0.72 |
| rs2106854 | 5q31.1 | T | C | 0.267 | C5orf56/IRF1 | 0.98 | 0.012 | 0.068 | 0.99 | 0.030 | 0.671 | 1.05 | 0.039 | 0.191 |
| rs2286503 | 7p15.3 | T | C | 0.397 | TOMM7 | 0.97 | 0.011 | 0.005 | 0.97 | 0.025 | 0.173 | 0.99 | 0.033 | 0.641 |
| rs10226084 | 7p21.1 | T | C | 0.543 | SNX13/PRPS1L1 | 1.01 | 0.011 | 0.497 | 1.02 | 0.024 | 0.379 | 0.98 | 0.032 | 0.614 |
| rs7464572 | 8q24.3 | C | G | 0.624 | PLEC1 | 1.02 | 0.011 | 0.03 | 0.98 | 0.028 | 0.526 | 0.99 | 0.041 | 0.724 |
| rs7896783 | 10q21.3 | A | G | 0.508 | JMJD1C | 1.02 | 0.01 | 0.14 | 0.98 | 0.024 | 0.449 | 0.98 | 0.032 | 0.512 |
| rs1019670 | 11q12.1 | A | T | 0.381 | MS4A6A | 1.01 | 0.012 | 0.311 | 0.96 | 0.028 | 0.173 | 1.02 | 0.036 | 0.597 |
| rs7968440 | 12q13.13 | A | G | 0.69 | DIP2B | 1.00 | 0.012 | 0.825 | 1.00 | 0.025 | 0.989 | 1.01 | 0.033 | 0.819 |
| rs434943 | 14q24.1 | A | G | 0.305 | ACTN1 | 1.01 | 0.013 | 0.314 | 1.03 | 0.027 | 0.256 | 0.97 | 0.035 | 0.366 |
| rs12915708 | 15q21.2 | C | G | 0.3 | SPPL2A | 0.98 | 0.012 | 0.063 | 1.00 | 0.027 | 0.889 | 1.00 | 0.034 | 0.915 |
| rs7204230 | 16q12.2 | T | C | 0.682 | CHD9 | 0.99 | 0.012 | 0.419 | 1.01 | 0.029 | 0.721 | 0.96 | 0.044 | 0.401 |
| rs10512597 | 17q25.1 | T | C | 0.202 | CD300LF | 1.02 | 0.014 | 0.218 | 1.00 | 0.032 | 0.909 | 0.99 | 0.041 | 0.781 |
| rs4817986 | 21q22.2 | T | G | 0.268 | PSMG1 | 1.02 | 0.013 | 0.182 | 1.00 | 0.027 | 0.928 | 1.03 | 0.035 | 0.486 |
| rs6010044 | 22q13.33 | A | C | 0.777 | SHANK3/ARSA | 0.97 | 0.014 | 0.012 | 0.97 | 0.030 | 0.364 | 0.96 | 0.042 | 0.368 |
Abbreviations: Freq1= frequency of allele1; OR= Odds ratio; SE= Standard error;
Joint meta-analysis of results from the Coronary ARtery DIsease Genome-wide Replication And Meta-analysis (CARDIoGRAM) and Europe South Asia Coronary Artery Disease Genetics (C4D) consortia.
Joint meta-analysis of results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium and the Wellcome Trust Case-Control Consortium (WTCCC).
Meta-analysis result from the French MARseille THrombosis Association (MARTHA) Consortium and the CHARGE Consortium Studies on Venous Thrombosis.
Discussion
The present study represents the largest effort to identify novel gene loci regulating plasma fibrinogen levels. Overall, we identified 24 independent genome-wide significant SNPs in 23 loci, including 15 loci with newly discovered fibrinogen associations. Using our genetic findings, we found no evidence for a causal role of fibrinogen in CAD, stroke, and VTE.
The proportion of variance in plasma fibrinogen level accounted for by all 24 fibrinogen-associated lead SNPs increased to 3.7% (a detailed description of the novel nearby candidate genes is presented in Supplementary Table S5). These results support the notion that regulation of plasma fibrinogen levels is driven by multiple genes, each having a modest effect on the phenotype. It is likely that even more loci with smaller effects remain to be discovered.
Relevance of the Fibrinogen-Related Loci in Non-European Ancestry Individuals
We performed the first meta-analysis of GWA studies on African-American samples and we provide evidence for significant association of a weighted SNP score based on the 24 lead SNPs from the European-ancestry meta-analysis with levels of fibrinogen in both African-Americans (P=1.5×10−8) and Hispanics (P=3.8×10−15). Thus, despite differences in allele frequencies and/or differences in the relative impact of covariates associated with fibrinogen among populations, loci identified in European-ancestry samples collectively contribute to the regulation of plasma fibrinogen in African-American and Hispanic populations. 20 of 24 lead SNPs showed the same direction of effect when comparing the European sample with either the African-American or the Hispanic samples. The substantially smaller size of the African-American and Hispanic cohorts compared to the total sample with European ancestry restricted available power and may have limited the significance of the candidate SNP associations in these populations (see power calculations in Supplementary Methods).
Pathways Involved in Regulation of Plasma Fibrinogen Level
It is interesting to note that several of the genome-wide significant loci identified in the present study harbor inflammatory genes, a remarkable set of which relate to the IL1 pathway, indicating the importance of this pathway in the regulation of fibrinogen. Most of these inflammatory genes have been previously reported in relation to other inflammation-related phenotypes and diseases. For example, IL6R, NLRP3, IL1RN/ILF10, and IRF1 were recently identified in a GWA study meta-analysis of C-reactive protein (CRP) conducted on European samples.32 Both fibrinogen and CRP are acute-phase proteins whose levels are largely influenced by inflammatory triggers. It is thus not surprising that they are both partly regulated by a common group of genes that are implicated in the immune response. These results are also consistent with our in silico gene-set enrichment analyses, which showed that inflammation-related pathways, including acute-phase response and interleukin signaling, were most enriched for fibrinogen-associated genes. In this regard, interesting new plausible candidate genes could be discerned within the newly identified loci, including IL6, located in the TOMM7-IL6 locus on chromosome 7, and IL1R1, located in the cytokine receptor gene cluster on chromosome 2.
Our gene-set enrichment analysis also highlighted genes regulating fat metabolism as important in the control of plasma fibrinogen concentration, as indicated by the strong representation of adipocytokine signaling genes. This is consistent with our observation that smoking and BMI contributed about 5.3% of the plasma fibrinogen variation and with data from The Fibrinogen Studies Collaboration, reporting that 7% of the variation in plasma fibrinogen concentration was accounted for by smoking, BMI and high-density lipoprotein (HDL) cholesterol33
Relations to Cardiovascular Disease
Although plasma fibrinogen concentration has been identified as a predictor of incident CAD events,1, 34 it has been argued that increased plasma fibrinogen levels in population subgroups at increased CAD risk could be due to other mechanisms, including existing atherosclerosis, which might induce a pro-inflammatory state with a subsequent increase in acute-phase reactants such as fibrinogen or CRP. Given the associations of fibrinogen levels with other established CAD risk factors (such as smoking and BMI), it remains uncertain whether these other factors may confound the association of fibrinogen with disease risk. Prior studies that assessed the causality of the association between plasma fibrinogen concentration and risk of CAD by Mendelian randomization (MR), using 2 common SNPs located in the promoter region of the FGB gene, found no significant association of this locus with CAD, concluding that the relationship was non-causal.35, 36 One limitation of these studies is that this single locus might have biologically unusual effects on measured fibrinogen levels. 35, 36 Our analysis of 23 other fibrinogen-associated SNPs offers a broader perspective, and thus a more robust and generalisable evaluation of the causal relationship between fibrinogen and cardiovascular events. A further strength of our study is that we present estimates of the effects on risk of clinical outcomes individually for each SNP as well as globally for all SNPs combined.
Our results do not support a causal relationship between plasma fibrinogen level and CAD. In fact, consistent with the negative results from previous MR, the lead SNP located in the FGB gene showed no association with CAD. Whereas SNPs rs4129267, rs6734238, and rs1154988, located in the IL6R, IL1F10/IL1RN and PCCB loci, respectively, were significantly associated with CAD in CARDIoGRAM and C4D, the direction of effect was consistent only for SNP located in the IL6R locus (i.e., the allele that lowered the plasma fibrinogen concentration also lowered CAD risk). Furthermore, the global effect of all 24 fibrinogen-associated SNPs was not associated with CAD risk (OR(CI95%)= 1.00 (0.97,1.03).
Overall, our results suggest that systemic inflammation both causes raised fibrinogen level and (by a different mechanism) is associated with increased risk of CAD. The lack of overlap between the top CAD-associated SNPs from the literature and the fibrinogen-associated SNPs identified in our study further argues against a reverse causality hypothesis, where inflammation caused by the atherosclerosis process would raise the fibrinogen level.
Although not as consistent as for CAD or MI, some studies have also suggested that an elevated fibrinogen concentration is a risk factor for stroke.3, 37–39 In the present study, none of the fibrinogen-associated SNPs were significantly associated with stroke. Our findings suggest that similar to what we observed for CAD, a raised fibrinogen concentration is not causally related to stroke, although a positive trend was observed that warrants further investigation. Similarly, our results show that none of the fibrinogen-associated SNPs was significantly associated with VTE after correction for multiple testing, although rs1800789G in the fibrinogen gene cluster, which is associated with higher fibrinogen level in our discovery study, showed a clear trend (P=0.004). However, given the small sample size of the VTE cases examined, the power for detection of VTE association in our data is substantially lower than for stroke and CAD (Supplementary Methods).
Conclusions
The present meta-analysis of fibrinogen GWA studies, based on a 4-fold greater sample size than previous meta-analyses (≈91,500 individuals), identified 24 independent signals in 23 loci (of which 15 are new) and increased the proportion of variance of plasma fibrinogen level accounted for by all lead SNPs in genome-wide significant loci from <2% to 3.7%. For some of these loci, our pathway and eQTL analyses provided supporting evidence regarding the most plausible candidate genes. Finally, our study does not support causal involvement of fibrinogen in CVD, particularly in clinically apparent CAD. Functional studies are needed to confirm and characterize candidate genes suggested by the in silico analyses presented here.
Future studies aimed at explaining the substantial missing heritability of plasma fibrinogen concentration should focus on exploring gene-gene and gene-environment interactions as well as on applying resequencing technologies to elucidate the role of rare variants.
Supplementary Material
Clinical Perspective.
Plasma fibrinogen concentration is a predictor of cardiovascular disease independent of other traditional risk factors, and variation in fibrinogen concentration has a substantial heritable component. We conducted a meta-analysis of 28 genome-wide association studies, including more than 90,000 subjects of European ancestry and substantial numbers of African Americans and Hispanic-Americans. We identified 24 genome-wide significant (P<5×10−8) independent single nucleotide polymorphisms (SNPs) in 23 genetic loci, including 15 novel associations, together accounting for 3.7% of plasma fibrinogen variation. Gene-set enrichment analysis highlighted potential key roles in fibrinogen regulation for the known structural fibrinogen genes as well as inflammation and other candidate pathways. However, in an evaluation for associations of the top fibrinogen SNPs with coronary artery disease, stroke and venous thromboembolism in very large case-control genomewide studies, there was no evidence for association with any of these clinical outcomes of either the single SNP most closely related to fibrinogen level (in the fibrinogen gene) or the combined effect of all 24 fibrinogen-associated SNPs (across 23 distinct loci). Our findings in a very large total study population provide comprehensive data for new and known genetic variants underlying fibrinogen concentration in human populations including multiple ethnic groups. Our findings highlight potential pathways for future study of the role of fibrinogen in the pathophysiology of atherosclerosis and cardiovascular disease. Clinical outcome analysis does not support a strong causal relationship between circulating levels of fibrinogen and coronary artery disease, stroke or venous thromboembolism.
Acknowledgments
Funding Sources: PROCARDIS was supported by the European Community Sixth Framework Program (LSHM-CT- 2007-037273), AstraZeneca, the British Heart Foundation, the Wellcome Trust (Contract No. 075491/Z/04), the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Swedish Heart-Lung Foundation, the Torsten and Ragnar Söderberg Foundation, the Strategic Cardiovascular and Diabetes Programs of Karolinska Institutet and Stockholm County Council, the Foundation for Strategic Research and the Stockholm County Council. Jemma C Hopewell and Robert Clarke acknowledge support from the BHF Centre of Research Excellence, Oxford. Bengt Sennblad acknowledge funding from the Magnus Bergvall foundation. Maria Sabater-Lleal is a recipient of a Marie Curie Intra European Fellowship within the 7th Framework Programme of the European Union (PIEF-GA-2009-252361) to investigate on the genetic regulation of plasma fibrinogen. FHS was partially supported by the National Heart, Lung, and Blood Institute’s (NHLBI's) Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. Partial investigator support was provided by the National Institute of Diabetes and Digestive and Kidney Diseases K24 DK080140 (JB Meigs), the National Institute on Aging and National Institute for Neurological Disorders and Stroke R01 AG033193, NS017950 (S Seshadri). The WGHS is supported by HL043851 and HL080467 from the National Heart, Lung, and Blood Institute and CA047988 from the National Cancer Institute, the Donald W. Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen.The SardiNIA (‘‘Progenia’’) team was supported by Contract NO1-AG-1–2109 from the NIA. We thank the many individuals who generously participated in this study, the Mayors and citizens of the Sardinian towns involved, the head of the Public Health Unit ASL4, and the province of Ogliastra for their volunteerism and cooperation. In addition, we are grateful to the Mayor and the administration in Lanusei for providing and furnishing the clinic site. We are grateful to the physicians Angelo Scuteri, Marco Orrù, Maria Grazia Pilia, Liana Ferreli, Francesco Loi, nurses Paola Loi, Monica Lai and Anna Cau who carried out participant physical exams; the recruitment personnel Susanna Murino; Mariano Dei, Sandra Lai, Antonella Mulas, Luca Usala, Andrea Maschio, Fabio Busonero for genotyping; Maria Grazia Piras and Monica Lobina for fibrinogen phenotyping. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research (NWO); the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. Jacqueline Witteman is supported by NWO grant (vici, 918-76-619). Abbas Dehghan is supported by NWO grant (veni, 916.12.154) and the EUR Fellowship. Dr. Ikram was supported by the Netherlands Heart Foundation (2009B102). SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg - West Pomerania. Genome- wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg West Pomerania. Computing resources have been made available by the Leibniz Supercomputing Centre of the Bavarian Academy of Sciences and Humanities (HLRB project h1231). The University of Greifswald is a member of the 'Center of Knowledge Interchange' program of the Siemens AG and the Caché Campus program of the InterSystems GmbH. This work is also part of the research project Greifswald Approach to Individualized Medicine (GANI_MED). The GANI_MED consortium is funded by the Federal Ministry of Education and Research and the Ministry of Cultural Affairs of the Federal State of Mecklenburg – West Pomerania (03IS2061A). The Coronary Artery Risk Development in Young Adults (CARDIA) study is funded by contracts N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, N01-HC-05187, N01-HC-45205, and N01-HC-45204 from the National Heart, Lung, and Blood Institute to the CARDIA investigators. Genotyping of the CARDIA participants was supported by grants U01-HG-004729, U01-HG-004446, and U01-HG-004424 from the National Human Genome Research Institute. Statistical analyses were supported by grants U01-HG-004729 and R01-HL-084099 to MF. PROSPER received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement n° HEALTH-F2-2009-223004. For a part of the genotyping we received funding from the Netherlands Consortium of Healthy Aging (NGI: 05060810). Measurement of serum fibrinogen was supported by a grant from the Scottish Executive Chief Scientist Office, Health Services Research Committee grant number CZG/4/306. This work was performed as part of an ongoing collaboration of the PROSPER study group in the universities of Leiden, Glasgow and Cork. Prof. Dr. J.W. Jukema is an Established Clinical Investigator of the Netherlands Heart Foundation (2001 D 032). This CHS research was supported by National Heart, Lung, and Blood Institute (NHLBI) contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center of Advancing Translational Technologies CTSI grant UL1TR000124 and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center and the Cedars-Sinai Board of Governors' Chair in Medical Genetics (JIR); support for genotyping in CHS African Americans was also provided by NHLBI R01-HL085251. Bruce M. Psaty is a member of the DSMB for a clinical trial of a device funded by the manufacturer (Zoll LifeCor), and a member of the Steering Committee for the Yale Open Data Access Project funded by Medtronic. We thank the LBC1936 and LBC1921 participants and research team members. We thank the nurses and staff at the Wellcome Trust Clinical Research Facility, where subjects were tested and the genotyping was performed. The whole genome association study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC; Ref. BB/F019394/1). The LBC1936 research was supported by a programme grant from Research Into Ageing and continues with programme grants from Help the Aged/Research Into Ageing (Disconnected Mind). The LBC1921 data collection was funded by the BBSRC. The study was conducted within the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology (http://www.ccace.ed.ac.uk/), supported by the BBSRC, Engineering and Physical Sciences Research Council (EPSRC), Economic and Social Research Council (ESRC), and Medical Research Council (MRC), as part of the cross-council Lifelong Health and Wellbeing Initiative. Lorna M. Lopez is the beneficiary of a post-doctoral grant from the AXA Research Fund. The MARTHA project was supported by a grant from the Program Hospitalier de la Recherche Clinique. Tiphaine Oudot-Mellakh was supported by a grant from the Fondation pour la Recherche Médicale. Statistical analyses conducted in MARTHA benefit from the C2BIG computing centre funded by the Fondation pour la Recherche Médicale and La Région Ile de France. The CROATIA-Split study was funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No. LSHG-CT-2006-018947) and Republic of Croatia Ministry of Science, Education and Sports research grants to I.R. (108-1080315-0302). We would like to acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to The University of Split and Zagreb Medical Schools and the Croatian Institute for Public Health. The SNP genotyping for the CROATIA-Split cohort was performed by AROS Applied Biotechnology, Aarhus, Denmark. The CROATIA-Korcula study was funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No. LSHG-CT-2006-018947) and Republic of Croatia Ministry of Science, Education and Sports research grants to I.R. (108-1080315-0302). We would like to acknowledge the invaluable contributions of the recruitment team in Korcula, the administrative teams in Croatia and Edinburgh and the people of Korcula. The SNP genotyping for the CROATIA-Korcula cohort was performed in Helmholtz Zentrum München, Neuherberg, Germany. The CROATIA-Vis study was funded by grants from the Medical Research Council (UK) and Republic of Croatia Ministry of Science, Education and Sports research grants to I.R. (108-1080315-0302). We would like to acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to The University of Split and Zagreb Medical Schools, the Institute for Anthropological Research in Zagreb and Croatian Institute for Public Health. The SNP genotyping for the CROATIA-Vis cohort was performed in the core genotyping laboratory of the Wellcome Trust Clinical Research Facility at the Western General Hospital, Edinburgh, Scotland. ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, the Medical Research Council Human Genetics Unit and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney. B58C acknowledges use of phenotype and genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. (http://www.b58cgene.sgul.ac.uk/). Genotyping for the B58C-WTCCC subset was funded by the Wellcome Trust grant 076113/B/04/Z. The B58C-T1DGC genotyping utilized resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. B58C-T1DGC GWAS data were deposited by the Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research (CIMR), University of Cambridge, which is funded by Juvenile Diabetes Research Foundation International, the Wellcome Trust and the National Institute for Health Research Cambridge Biomedical Research Centre; the CIMR is in receipt of a Wellcome Trust Strategic Award (079895). The B58C-GABRIEL genotyping was supported by a contract from the European Commission Framework Programme 6 (018996) and grants from the French Ministry of Research. The MONICA/KORA Augsburg studies (KORS) were financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany, and supported by grants from the German Federal Ministry of Education and Research (BMBF). Part of this work was financed by the German National Genome Research Network (NGFNPlus, project number 01GS0834) and through additional funds from the University of Ulm. Furthermore, the research was supported within the Munich Center of Health Sciences (MC Health) as part of LMU innovative. The InCHIANTI study baseline (1998–2000) was supported as a "targeted project" (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336); This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The Twins UK study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and (FP7/2007-2013), ENGAGE project grant agreement HEALTH-F4-2007-201413 and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also receives support from the Dept of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London. TDS is an NIHR senior Investigator. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant. (G20234) .The authors acknowledge the funding and support of the National Eye Institute via an NIH/CIDR genotyping project (PI: Terri Young). Genotyping of TwinsUK samples: We thank the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation, Quality Control and Genotyping led by Leena Peltonen and Panos Deloukas; Le Centre National de Génotypage, France, led by Mark Lathrop, for genotyping; Duke University, North Carolina, USA, led by David Goldstein, for genotyping; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie. Genotyping was also performed by CIDR as part of an NEI/NIH project grant. NS is supported by the Wellcome Trust (Core Grant Number 091746/Z/10/Z). SYS is supported by a Post-Doctoral Research Fellowship from the Oak Foundation. Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland (129255 and 126775), the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, European Science Foundation (EUROSTRESS), Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation, Juho Vainio Foundation, and Wellcome Trust (grant number WT089062). We thank all study participants as well as everybody involved in the Helsinki Birth Cohort Study. Netherland Twins study: Funding was obtained from the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904-61-090, 985-10-002,904-61-193,480-04-004, 400-05-717, Addiction-31160008, Middelgroot-911-09-032, Spinozapremie 56-464-14192), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL, 184.021.007), the VU University’s Institute for Health and Care Research (EMGO+ ) and Neuroscience Campus Amsterdam (NCA), the European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community's Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413); the European Science Council (ERC Advanced, 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH, R01D0042157-01A). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, the (NIMH, MH081802) and by the Grand Opportunity grants 1RC2MH089951-01 and 1RC2 MH089995-01 from the NIMH. ARIC is is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support is provided by grants and contracts N01 HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and RR-024156. Funding for CARe genotyping was provided by NHLBI Contract N01-HC-65226. The authors thank the participants of the MESA study, the Coordinating Center, MESA investigators, and study staff for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. GeneSTAR was supported by grants from the National Institutes of Health/National Heart, Lung, and Blood Institute (U01 HL72518, HL097698, HL59684, HL58625-01A1, HL071025-01A1), by grants from the National Institutes of Health/National Institute of Nursing Research (NR0224103, NR008153-01), and by a grant from the National Institutes of Health/National Center for Research Resources (M01-RR000052) to the Johns Hopkins General Clinical Research Center. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A listing of WHI investigators can be found at http://www.whiscience.org/publications/WHI_investigators_shortlist_2010-2015.pdf. A.P.R was supported by R01 HL71862, “Thrombosis Genetics, MI, and Stroke in Older Adults.” CSF was funded by NIH grant HL 463680 from the National Heart, Lung, and Blood Institute (NHLBI). CARe Acknowledgement. The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. The following parent studies contributed study data, ancillary study data, and DNA samples through the Broad Institute (N01-HC-65226) to create this genotype/phenotype data base for wide dissemination to the biomedical research community. Wellcome Trust Case Control consortium 2 (WTCCC2). The principal funding for this study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z). We also thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, M.L. Perez, C.R. Stribling, S. Taylor and A. Wilk of the Wellcome Trust Sanger Institute's Sample and Genotyping Facilities for technical assistance. We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02, and of the UK National Blood Service controls funded by the Wellcome Trust. A membership list of WTCCC2 can be found in Supplementary Material. The C4D Consortium comprises CHD cases and controls of European origin from PROCARDIS and the Heart Protection Study and of South Asian origin from the LOLIPOP and PROMIS studies. Data analyzed with respect to risk of CHD all relate to the European origin participants from PROCARDIS and HPS. We would like to acknowledge the UK Twins Study and WTCCC2-National Blood Service Collection for providing population controls. Jemma C Hopewell and Robert Clarke acknowledge support from the BHF Centre of Research Excellence, Oxford. CARDIOGRAM : The ADVANCE study was supported by a grant from the Reynold's Foundation and NHLBI grant HL087647.Genetic analyses of CADomics were supported by a research grant from Boehringer Ingelheim. Recruitment and analysis of the CADomics cohort was supported by grants from Boehringer Ingelheim and PHILIPS medical Systems, by the Government of Rheinland-Pfalz in the context of the “Stiftung Rheinland-Pfalz für Innovation”, the research program “Wissen schafft Zukunft” and by the Johannes-Gutenberg University of Mainz in the context of the “Schwerpunkt Vaskuläre Prävention” and the “MAIFOR grant 2001”, by grants from the Fondation de France, the French Ministry of Research, and the Institut National de la Santé et de la Recherche Médicale. The deCODE CAD/MI Study was sponsored by NIH grant, National Heart, Lung and Blood Institute R01HL089650-02. The German MI Family Studies (GerMIFS I-III (KORA)) were supported by the Deutsche Forschungsgemeinschaft and the German Federal Ministry of Education and Research (BMBF) in the context of the German National Genome Research Network (NGFN-2 and NGFN-plus), the EU funded integrated project Cardiogenics (LSHM-CT-2006-037593) , and the bi-national BMBF/ANR funded project CARDomics (01KU0908A). LURIC has received funding from the EU framework 6 funded Integrated Project “Bloodomics” (LSHM-CT-2004-503485), the EU framework 7 funded Integrated Project AtheroRemo (HEALTH-F2-2008-201668) and from Sanofi/Aventis, Roche, Dade Behring/Siemens, and AstraZeneca. The MIGen study was funded by the US National Institutes of Health (NIH) and National Heart, Lung, and Blood Institute’s STAMPEED genomics research program through R01 HL087676. Ron Do from the MIGen study is supported by a Canada Graduate Doctoral Scholarship from the Canadian Institutes of Health Research. Recruitment of PennCATH was supported by the Cardiovascular Institute of the University of Pennsylvania. Recruitment of the MedStar sample was supported in part by the MedStar Research Institute and the Washington Hospital Center and a research grant from GlaxoSmithKline. Genotyping of PennCATH and Medstar was performed at the Center for Applied Genomics at the Children’s Hospital of Philadelphia and supported by GlaxoSmithKline through an Alternate Drug Discovery Initiative research alliance award (M. P. R. and D. J. R.) with the University of Pennsylvania School of Medicine. The Ottawa Heart Genomic Study was supported by CIHR #MOP--82810 (R. R.), CFI #11966 (R. R.), HSFO #NA6001 (R. McP.), CIHR #MOP172605 (R. McP.), CIHR #MOP77682 (A. F. R. S.). The WTCCC Study was funded by the Wellcome Trust. Recruitment of cases for the WTCCC Study was carried out by the British Heart Foundation (BHF) Family Heart Study Research Group and supported by the BHF and the UK Medical Research Council. N. J. S. and S. G. B. hold chairs funded by the British Heart Foundation. The Age, Gene/Environment Susceptibility Reykjavik Study has been funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament).The Cleveland Clinic GeneBank study was supported by NIH grants P01 HL098055, P01HL076491-06, R01DK080732, P01HL087018, and 1RO1HL103931-01. The collection of clinical and sociodemographic data in the Dortmund Health Study was supported by the German Migraine- & Headache Society (DMKG) and by unrestricted grants of equal share from Astra Zeneca, Berlin Chemie, Boots Healthcare, Glaxo-Smith-Kline, McNeil Pharma (former Woelm Pharma), MSD Sharp & Dohme and Pfizer to the University of Muenster. Blood collection was done through funds from the Institute of Epidemiology and Social Medicine, University of Muenster. The EPIC-Norfolk study is supported by the Medical Research Council UK and Cancer Research UK. The EpiDREAM study is supported by the Canadian Institutes fo Health Research, Heart and Stroke Foundation of Ontario, Sanofi-Aventis, GlaxoSmithKline and King Pharmaceuticals. Funding for Andrew Lotery from the LEEDS study was provided by tha T.F.C. Frost charity and the Macular Disease Society. The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO) (175.010.2005.011, 911.03.012), the Netherlands Genomics Initiative (NGI)/ NWO project nr. 050-060-810 and Research Institute for Diseases in the Elderly (RIDE). Abbas Dehghan is supported by a grant from NWO (Vici, 918-76-619). The SAS study was funded by the British Heart Foundation. The Swedish Research Council, the Swedish Heart & Lung Foundation and the Stockholm County Council (ALF) supported the SHEEP study. SMILE was funded by the Netherlands Heart foundation (NHS 92345). Dr Rosendaal is a recipient of the Spinoza Award of the Netherlands Organisation for Scientific Research (NWO) which was used for part of this work. The Verona Heart Study was funded by grants from the Italian Ministry of University and Research, the Veneto Region, and the Cariverona Foundation, Verona. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022. The authors thank the staff and participants of the ARIC study for their important contributions. The KORA (Kooperative Gesundheitsforschung in der Region Augsburg) research platform was initiated and financed by the Helmholtz Zentrum München - National Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria. Part of this work was financed by the German National Genome Research Network (NGFN-2 and NGFNPlus) and within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ.
Appendix
The following is a list of the institutional affiliations for the authors of this article:
PROCARDIS controls: Cardiovascular Genetics and Genomics Group, Atherosclerosis Research Unit, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Solna, Stockholm, Sweden (M.S-L, A.S, B.S, R.S, A.H).
PROCARDIS cases: Cardiovascular Genetics and Genomics Group, Atherosclerosis Research Unit, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Solna,Stockholm, Sweden (A.M), Department of Cardiovascular Medicine, University of Oxford, John Radcliffe Hospital, Headington, Oxford, United Kingdom, Department of Cardiovascular Medicine, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom (A.G, H.W), Clinical Trial Service Unit, University of Oxford, United Kingdom (R.C), Department of Cardiovascular Research, Istituto di Ricerche Farmacologiche Mario Negri, Milan, Italy (M.G.F), Leibniz-Institut für Arterioskleroseforschung an der Universität Münster, Münster, Germany (U.S).
FHS: National Heart, Lung and Blood Institute’s Framingham Heart Study, Framingham , MA USA National Heart, Lung and Blood Institute Division of Intramural Research, Bethesda MD USA (J.H, A.D.J, C.J.O, S.S), Royal North Shore Hospital, University of Sydney, Australia (G.H.T), Department of Biostatistics, Boston University, Boston, MA, USA (M-H.C). Department of Neurology, Boston University School of Medicine, Boston, MA, USA (S.S).
WGHS: Division of Preventive Medicine, Brigham and Women’s Hospital (D.I.C, L.M.R, P.M.R) and Harvard Medical School (DIC, PMR); 900 Commonwealth Avenue, East, Boston, MA 02215
SardiNIA: Istituto di Ricerca Genetica e Biomedica, Consiglio Nazionale delle Ricerche, Cagliari, Italy (S.N, M.S, S.S, F.C), Intramural Research Program, National Institute on Aging, 5600 Nathan Shock Drive, Baltimore, MD, USA (K.T, D.S).
The Rotterdam Study Department of Epidemiology, Erasmus Medical Center, Rotterdam, the Netherlands (A.D, J.CM.W, A.H, O.H.F, M.A.I). Department of Internal Medicine, Erasmus Medical Center, Rotterdam, the Netherlands (A.G.U, F.R); Department of Radiology and Neurology, Erasmus MC University Medical Center, Rotterdam, the Netherlands (M.A.I); Member of the Netherlands Consortium on Healthy Aging (NCHA), Leiden, The Netherlands (A.D, J.CM.W, A.H, A.G.U, F.R, O.H.F).
SHIP: Ernst-Moritz-Arndt University Greifswald, Interfaculty Institute for Genetics and Functional Genomics, Department for Functional Genomics, Friedrich-Ludwig-Jahn-Straße 15A, D-17487 Greifswald (A.T, G.H), University Medicine Greifswald, Institute of Clinical Chemistry and Laboratory Medicine, Ferdinand Sauerbruchstrasse, D-17487 Greifswald (A.G, H.W), Ernst-Moritz-Arndt-University of Greifswald, Institute for Community Medicine, Section Study of Health in Pomerania (SHIP), Walther-Rathenau-Straße 48, D-17475 Greifswald (H.V), University Medicine Greifswald, Policlinics for Restorative Dentistry, Periodontology and Endodontology, Department of Periodontology Rotgerberstraße 8, D-17475 Greifswald (T.K).
CARDIA: Department of Epidemiology, University of Washington, Seattle, WA (A.R.), Brown Foundation Institute of Molecular Medicine, Division of Epidemiology, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX, USA.(M.F). Division of Hematology/Oncology, Northwestern University Feinberg School of Medicine, Chicago, Ill, USA (D.G), Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN (M.G).
PROSPER/PHASE: Department of Cardiology and Department of Gerontology and Geriatrics, Leiden University Medical Center, Leiden, The Netherlands (S.T), ; Institute of Cardiovascular and Medical Sciences, School of Medicine, University of Glasgow, UK (D.J.S), BHF Glasgow Cardiovascular Research Centre, Faculty of Medicine, Glasgow, UK (N.S), Department of Pharmacology and Therapeutics, University College Cork, Ireland (B.M.B), Department of Cardiology, Leiden University Medical Center, Leiden, The Netherlands; Durrer Center for Cardiogenetic Research, Amsterdam, The Netherlands; Interuniversity Cardiology Institute of the Netherlands, Utrecht, The Netherlands (J.W.J).
CHS: Cardiovascular Health Research Unit, Departments of Medicine, Epidemiology, and Health Services, University of Washington, Seattle WA USA (J.C.B, B.M.P); Group Health Research Institute, Group Health Cooperative, Seattle WA, USA (B.M.P), Department of Biostatistics, University of Washington, Seattle WA USA (B.McK, T.L), Medical Genetics Institute, Cedars-Sinai Medical Center, Los Angeles CA USA (K.D.T, J.I.R), Department of Epidemiology, University of Washington, Seattle WA USA; Seattle Epidemiologic Research and Information Center, Office of Research and Development, Seattle WA USA; Group Health Research Institute, Group Health Cooperative, Seattle WA, USA (N.L.S).
LBC1936 & LBC1921: Centre for Cognitive Ageing and Cognitive Epidemiology, The University of Edinburgh, 7 George Square, Edinburgh, EH8 9JZ, UK. (L.M.L, G.D, S.E.H, D.C.L, J.M.S, I.J.D), Department of Psychology, The University of Edinburgh, 7 George Square, Edinburgh, EH8 9JZ, UK. (L.M.L, G.D, I.J,D), Medical Genetics Section, The University of Edinburgh Molecular Medicine Centre, Institute of Genetics and Molecular Medicine, Western General Hospital, Edinburgh, EH4 2XU, UK (S.E.H), Geriatric Medicine unit, University of Edinburgh, Royal Victoria Building, Western General Hospital, Crewe Road South, Edinburgh, UK. EH4 2XU (J.M.S).
MARTHA: Aix Marseille Université, Inserm, NORT, UMR_S 1062, 13005, Marseille, France (P-E.M); INSERM, UMR_S 937, F-75013, Paris, France (D-A.T., T. O-M); ICAN Institute for Cardiometabolism and Nutrition, Université Pierre et Marie Curie, F-75013, Paris, France (D-A.T.).
CROATIA-Split: Faculty of Medicine, University of Split, Soltanska 2, 21000 Split, Croatia (T.Z), Division of Hematology, Department of Medicine, Clinical Hospital Center Zagreb, Zagreb, Croatia, and Faculty of Medicine Osijek, J.J. Strossmayer University of Osijek, Osijek, Croatia (D.P), Centre for Population Health Sciences, University of Edinburgh, Teviot Place, Edinburgh, EH8 9AG, Scotland.(I.R).
CROATIA_Korcula: MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, Scotland (J.E.H, A.F.W, C.H).
CROATIA_Vis: MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, Edinburgh, EH4 2XU, Scotland (P.N), Department of Public Health, University of Split Medical School; Split, Croatia (I.K, O.P).
ORCADES: Centre for Population Health Sciences, University of Edinburgh, Teviot Place, Edinburgh, EH8 9AG, Scotland (H.C, S.H.W, J.F.W).
B58C: Division of Population Health Sciences and Education, St George’s, University of London, UK (D.P.S, A.R.R). Institute of Cardiovascular and Medical Sciences, University of Glasgow, UK (A.R, G.D.L), School of Social and Community Medicine, University of Bristol, UK (W.L.McA).
KORA: Institute of Epidemiology II, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (J.B), Institute of Epidemiology II, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Munich Heart Alliance, Munich, Germany (A.P), Institute of Genetic Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (C.G), Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Hannover Unified Biobank, Hannover Medical School, Hannover, Germany (T.I), Department of Internal Medicine II - Cardiology, University of Ulm Medical Center, Ulm, Germany (W.K).
InCHIANTI: Clinical Research Branch, National Institute on Aging, Baltimore MD 21250 (T.T, L.F), Unit, Azienda Sanitaria Firenze (ASF), Florence, Italy (S.B).
Twins UK: Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK (S-Y.S, N.S), Department of Twin Research and Genetic Epidemiology, King’s College London, London, UK (F.M.K.W, T.D.S), Division of Cardiovascular & Diabetes Research, Leeds University, Leeds, UK (P.J.G), MRC Centre for CAiTE, School of Social and Community Medicine, University of Bristol, Bristol, BS8 2BN, UK (S-Y.S).
HBCS: Institute of Behavioural Sciences, University of Helsinki, Helsinki, Finland (J.L, K.R), Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Finland (E.W), Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, UK. Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Finland. Department of Medical Genetics, University of Helsinki and University Central Hospital, Helsinki, Finland (A.P), National Institute for Health and Welfare, Finland. Department of General Practice and Primary health Care, University of Helsinki, Finland. Helsinki University Central Hospital, Unit of General Practice, Helsinki, Finland. Folkhalsan Research Centre, Helsinki, Finland. Vasa Central Hospital, Vasa, Finland (J.G.E).
The Netherlands Twin Registry (NTR): Department of Biological Psychology, VU University & EMGO+ institute, VU medical centre, Amsterdam, the Netherlands (JJ.H, J. van D., G.W., DI.B., EJC de G).
ARIC: Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN, USA (W.T, A.R.F); Division of Biostatistics, University of Minnesota, Minneapolis, MN, USA (S.B, J.S), Human Genetics Center and Institute of Molecular Medicine, University of Texas Health Science Center, Houston, TX, USA (E.B). Departments of Medicine (Geriatrics) and Neurology, University of Mississippi Medical Center, Jackson, MS, USA (T.H.M).
MESA: Medical Genetics Institute, Cedars-Sinai Medical Center, 8700 Beverly Blvd, Los Angeles, CA (X.G; J.Y; T.H); Center for Clinical and Translational Science, University of Vermont, VT (R.P.T). Department of Pathology, University of Vermont College of Medicine, Burlington, VT (N.S.J).
GeneSTAR: The Johns Hopkins University, School of Medicine, Division of General Internal Medicine, Baltimore, MD (L.R.Y, D.M.B, L.C.B.), The Johns Hopkins University, School of Medicine, Division of Cardiology, Baltimore, MD (L.C.B.).
WHI: Department of Epidemiology, University of Washington, Seattle, WA 98195, USA and Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA (A.R), Center for Primary Care and Prevention, Alpert Medical School, Brown University, Providence, RI, USA (C.B.E); Department of Epidemiology and Program on Genomics and Nutrition, School of Public Health, and Center for Metabolic Diseases Prevention, University of California Los Angeles, Los Angeles, CA (S.L); Departments of Epidemiology and Internal Medicine, University of Iowa College of Public Health, Iowa City, IA (R.W); John A. Burns School of Medicine, University of Hawaii and Pacific Health Research Institute, Honolulu, HI (J.D.C).
CSF: Department of Medicine, Harvard Medical School, Brigham and Women’s Hospital and Beth Israel Deaconess Medical Center, Boston, MA (S.R), Department of Medicine, Case Medical Center, Cleveland, OH (R.M).
ASAP: Atherosclerosis Research Unit, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Solna, Stockholm, Sweden (L.F, P.E), Cardiothoracic Surgery Unit, Department of Molecular Medicine and Surgery, Karolinska Institutet, Stockholm, Sweden (A.F-C).
C4D: Clinical Trial Service Unit, University of Oxford, United Kingdom (J.C.H), Department of Epidemiology & Biostatistics, Imperial College London, St Mary’s Campus, Norfolk Place, London, UK (J.C.C), Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK (J.D). Center for Non-Communicable Diseases, Karachi, Pakistan (D.S, J.D), Department of Biostatistics and Epidemiology and Department of Medicine, University of Pennsylvania, PA, USA (D.S); National Heart and Lung Institute, Imperial College London, London, UK (J.S.K).
CARDIOGRAM: Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Leicester,UK, Leicester National Institute for Health Research Biomedical Research Unit in Cardiovascular Disease, Glenfield Hospital, Leicester, UK (C.P.N, N.J.S), Universität zu Lübeck, Medizinische Klinik II, Lübeck, Germany, Deutsches Zentrum für Herz-Kreislauf-Forschung (DZHK), Universität zu Lübeck, Lübeck, Germany (J.E, H.S), Cardiovascular Institute, University of Pennsylvania Medical Center, Philadelphia, PA, USA, Division of Prevention and Population Sciences, National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD, USA (M.R), Cardiovascular Research Center and Center for Human Genetic Research, MA, USA; General Hospital and Harvard Medical School, Boston, MA, USA. Julius Center, Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA (S.K).
VTE consortium: LITE: Division of Epidemiology, University of Minnesota, Minneapolis MN 55455, USA (W.T), Womens Genome Health Study, Division of Preventive Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA, 02215 (D.C), Rotterdam Study, Department of Epidemiology, Erasmus Medical Center, 3000 CA Rotterdam, The Netherlands (M.T), Heart and Vascular Health Study, Departments of Epidemiology, University of Washington, Seattle WA USA 98101; Group Health Research Institute, Group Health Cooperative, Seattle, WA USA 98101; Seattle Epidemiologic Research and Information Center, Veterans Affairs Office of Research & Development, Seattle, WA USA 98108 (N.L.S).
WTCCC2: Stroke and Dementia Research Centre, St Georges, University of London, Cranmer Terrace, Tooting, London SW17 0RE (S.B, H.S.M); Institute for Stroke and Dementia Research, Klinikum der Universität München, Ludwig-Maximilians-Universität, Munich, Germany; Munich Cluster for Systems Neurology (SyNergy), Munich, Germany (M.D); Stroke Prevention Research Unit, Nuffield Department of Clinical Neuroscience, University of Oxford, Oxford, UK (P.M.R), Division of Clinical Neurosciences, University of Edinburgh, Edinburgh, UK (C.L.M.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures: None of the authors for this manuscript has disclosed a conflict of interest directly related to the manuscript. Martin Dichgans has declared receiving honoraria payments from Bayer Vital, Boehringer Ingelheim Pharma, Biologische Heitmittel heel, Bristol-Myers Squibb Lundbeck, Sanofi-Aventis Deustchland, Shire Deustchland, and the Deutsches Zentrum for Neurodegenerative Erkrankungen, and consults for Bayer Vital, Boehringer Ingelheim Pharma, Biologische Heitmittel heel, Bristol-Myers and Trommsdorff advisory boards. Eco de Geus has declared receiving honoraria payments for grant reviews and associate editorial functions. Bruce M. Psaty is a member of the DSMB for a clinical trial of a device funded by the manufacturer (Zoll LifeCor), and a member of the Steering Committee for the Yale Open Data Access Project funded by Medtronic. Numerous authors have noted their research is supported by government or non-profit agencies or foundations. Paul M. Ridker is a recipient of a research grant from Amgen. Henry Völzke is a recipient of a research grant from Siemens AG.
References
- 1.Danesh J, Lewington S, Thompson SG, Lowe GD, Collins R, Kostis JB, Wilson AC, Folsom AR, Wu K, Benderly M, Goldbourt U, Willeit J, Kiechl S, Yarnell JW, Sweetnam PM, Elwood PC, Cushman M, Psaty BM, Tracy RP, Tybjaerg-Hansen A, Haverkate F, de Maat MP, Fowkes FG, Lee AJ, Smith FB, Salomaa V, Harald K, Rasi R, Vahtera E, Jousilahti P, Pekkanen J, D'Agostino R, Kannel WB, Wilson PW, Tofler G, Arocha-Pinango CL, Rodriguez-Larralde A, Nagy E, Mijares M, Espinosa R, Rodriquez-Roa E, Ryder E, Diez-Ewald MP, Campos G, Fernandez V, Torres E, Marchioli R, Valagussa F, Rosengren A, Wilhelmsen L, Lappas G, Eriksson H, Cremer P, Nagel D, Curb JD, Rodriguez B, Yano K, Salonen JT, Nyyssonen K, Tuomainen TP, Hedblad B, Lind P, Loewel H, Koenig W, Meade TW, Cooper JA, De Stavola B, Knottenbelt C, Miller GJ, Bauer KA, Rosenberg RD, Sato S, Kitamura A, Naito Y, Palosuo T, Ducimetiere P, Amouyel P, Arveiler D, Evans AE, Ferrieres J, Juhan-Vague I, Bingham A, Schulte H, Assmann G, Cantin B, Lamarche B, Despres JP, Dagenais GR, Tunstall-Pedoe H, Woodward M, Ben-Shlomo Y, Davey Smith G, Palmieri V, Yeh JL, Rudnicka A, Ridker P, Rodeghiero F, Tosetto A, Shepherd J, Ford I, Robertson M, Brunner E, Shipley M, Feskens EJ, Kromhout D, Dickinson A, Ireland B, Juzwishin K, Kaptoge S, Memon A, Sarwar N, Walker M, Wheeler J, White I, Wood A. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: an individual participant meta-analysis. Jama. 2005;294:1799–1809. doi: 10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- 2.Ernst E, Resch KL. Fibrinogen as a cardiovascular risk factor: a meta-analysis and review of the literature. Ann Intern Med. 1993;118:956–963. doi: 10.7326/0003-4819-118-12-199306150-00008. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelmsen L, Svardsudd K, Korsan-Bengtsen K, Larsson B, Welin L, Tibblin G. Fibrinogen as a risk factor for stroke and myocardial infarction. N Engl J Med. 1984;311:501–505. doi: 10.1056/NEJM198408233110804. [DOI] [PubMed] [Google Scholar]
- 4.Koster T, Rosendaal FR, Reitsma PH, van der Velden PA, Briet E, Vandenbroucke JP. Factor VII and fibrinogen levels as risk factors for venous thrombosis. A case-control study of plasma levels and DNA polymorphisms--the Leiden Thrombophilia Study (LETS) Thromb Haemost. 1994;71:719–722. [PubMed] [Google Scholar]
- 5.Uitte de Willige S, de Visser MC, Houwing-Duistermaat JJ, Rosendaal FR, Vos HL, Bertina RM. Genetic variation in the fibrinogen gamma gene increases the risk for deep venous thrombosis by reducing plasma fibrinogen gamma' levels. Blood. 2005;106:4176–4183. doi: 10.1182/blood-2005-05-2180. [DOI] [PubMed] [Google Scholar]
- 6.de Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357:101–105. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 7.Souto JC, Almasy L, Borrell M, Gari M, Martinez E, Mateo J, Stone WH, Blangero J, Fontcuberta J. Genetic determinants of hemostasis phenotypes in Spanish families. Circulation. 2000;101:1546–1551. doi: 10.1161/01.cir.101.13.1546. [DOI] [PubMed] [Google Scholar]
- 8.Dehghan A, Yang Q, Peters A, Basu S, Bis JC, Rudnicka AR, Kavousi M, Chen MH, Baumert J, Lowe GD, McKnight B, Tang W, de Maat M, Larson MG, Eyhermendy S, McArdle WL, Lumley T, Pankow JS, Hofman A, Massaro JM, Rivadeneira F, Kolz M, Taylor KD, van Duijn CM, Kathiresan S, Illig T, Aulchenko YS, Volcik KA, Johnson AD, Uitterlinden AG, Tofler GH, Gieger C, Psaty BM, Couper DJ, Boerwinkle E, Koenig W, O'Donnell CJ, Witteman JC, Strachan DP, Smith NL, Folsom AR. Association of novel genetic Loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ Cardiovasc Genet. 2009;2:125–133. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danik JS, Pare G, Chasman DI, Zee RY, Kwiatkowski DJ, Parker A, Miletich JP, Ridker PM. Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome-wide association study of fibrinogen in 17 686 women: the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:134–141. doi: 10.1161/CIRCGENETICS.108.825273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preuss M, Konig IR, Thompson JR, Erdmann J, Absher D, Assimes TL, Blankenberg S, Boerwinkle E, Chen L, Cupples LA, Hall AS, Halperin E, Hengstenberg C, Holm H, Laaksonen R, Li M, Marz W, McPherson R, Musunuru K, Nelson CP, Burnett MS, Epstein SE, O'Donnell CJ, Quertermous T, Rader DJ, Roberts R, Schillert A, Stefansson K, Stewart AF, Thorleifsson G, Voight BF, Wells GA, Ziegler A, Kathiresan S, Reilly MP, Samani NJ, Schunkert H. Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: A Genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3:475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, Jackson CA, Traylor M, Strange A, Su Z, Band G, Syme PD, Malik R, Pera J, Norrving B, Lemmens R, Freeman C, Schanz R, James T, Poole D, Murphy L, Segal H, Cortellini L, Cheng YC, Woo D, Nalls MA, Muller-Myhsok B, Meisinger C, Seedorf U, Ross-Adams H, Boonen S, Wloch-Kopec D, Valant V, Slark J, Furie K, Delavaran H, Langford C, Deloukas P, Edkins S, Hunt S, Gray E, Dronov S, Peltonen L, Gretarsdottir S, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boncoraglio GB, Parati EA, Attia J, Holliday E, Levi C, Franzosi MG, Goel A, Helgadottir A, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Duncanson A, Jankowski J, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Worrall BB, Kittner SJ, Mitchell BD, Kissela B, Meschia JF, Thijs V, Lindgren A, Macleod MJ, Slowik A, Walters M, Rosand J, Sharma P, Farrall M, Sudlow CL, Rothwell PM, Dichgans M, Donnelly P, Markus HS. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germain M, Saut N, Greliche N, Dina C, Lambert JC, Perret C, Cohen W, Oudot-Mellakh T, Antoni G, Alessi MC, Zelenika D, Cambien F, Tiret L, Bertrand M, Dupuy AM, Letenneur L, Lathrop M, Emmerich J, Amouyel P, Tregouet DA, Morange PE. Genetics of venous thrombosis: insights from a new genome wide association study. PLoS One. 2011;6:e25581. doi: 10.1371/journal.pone.0025581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tregouet DA, Heath S, Saut N, Biron-Andreani C, Schved JF, Pernod G, Galan P, Drouet L, Zelenika D, Juhan-Vague I, Alessi MC, Tiret L, Lathrop M, Emmerich J, Morange PE. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood. 2009;113:5298–5303. doi: 10.1182/blood-2008-11-190389. [DOI] [PubMed] [Google Scholar]
- 15.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, Bos MJ, Beiser A, Cushman M, Launer LJ, Shahar E, Struchalin M, Du Y, Glazer NL, Rosamond WD, Rivadeneira F, Kelly-Hayes M, Lopez OL, Coresh J, Hofman A, DeCarli C, Heckbert SR, Koudstaal PJ, Yang Q, Smith NL, Kase CS, Rice K, Haritunians T, Roks G, de Kort PL, Taylor KD, de Lau LM, Oostra BA, Uitterlinden AG, Rotter JI, Boerwinkle E, Psaty BM, Mosley TH, van Duijn CM, Breteler MM, Longstreth WT, Jr, Wolf PA. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cremer P, Nagel D, Labrot B, Mann H, Muche R, Elster H, Seidel D. Lipoprotein Lp(a) as predictor of myocardial infarction in comparison to fibrinogen, LDL cholesterol and other risk factors: results from the prospective Gottingen Risk Incidence and Prevalence Study (GRIPS) Eur J Clin Invest. 1994;24:444–453. doi: 10.1111/j.1365-2362.1994.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 18.Clauss A. Rapid physiological coagulation method in determination of fibrinogen. Acta Haematol. 1957;17:237–246. doi: 10.1159/000205234. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 21.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Burtt NP, Chang M, Coblyn J, Costenbader KH, Criswell LA, Crusius JB, Cui J, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Kurreeman FA, Lee AT, Liu X, Li Y, Martin P, Morgan AW, Padyukov L, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth P, Altshuler D, Karlson EW, Toes RE, de Vries N, Begovich AB, Siminovitch KA, Worthington J, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folkersen L, van't Hooft F, Chernogubova E, Agardh HE, Hansson GK, Hedin U, Liska J, Syvanen AC, Paulsson-Berne G, Franco-Cereceda A, Hamsten A, Gabrielsen A, Eriksson P. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ Cardiovasc Genet. 2010;3:365–373. doi: 10.1161/CIRCGENETICS.110.948935. [DOI] [PubMed] [Google Scholar]
- 27.Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, Beaulaurier J, Zhang B, Castro V, Zhu J, Sieberts SK, Wang S, Molony C, Heymsfield SB, Kemp DM, Reitman ML, Lum PY, Schadt EE, Kaplan LM. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, Aldred SF, Trinklein ND, Schuetz E, Nickerson DA, Thummel KE, Rieder MJ, Rettie AE, Ratain MJ, Cox NJ, Brown CD. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroder A, Klein K, Winter S, Schwab M, Bonin M, Zell A, Zanger UM. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 2013;13:12–20. doi: 10.1038/tpj.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peden J, Hopewell JC, Saleheen D. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 32.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, Pellikka N, Wallaschofski H, Kettunen J, Henneman P, Baumert J, Strachan DP, Fuchsberger C, Vitart V, Wilson JF, Pare G, Naitza S, Rudock ME, Surakka I, de Geus EJ, Alizadeh BZ, Guralnik J, Shuldiner A, Tanaka T, Zee RY, Schnabel RB, Nambi V, Kavousi M, Ripatti S, Nauck M, Smith NL, Smith AV, Sundvall J, Scheet P, Liu Y, Ruokonen A, Rose LM, Larson MG, Hoogeveen RC, Freimer NB, Teumer A, Tracy RP, Launer LJ, Buring JE, Yamamoto JF, Folsom AR, Sijbrands EJ, Pankow J, Elliott P, Keaney JF, Sun W, Sarin AP, Fontes JD, Badola S, Astor BC, Hofman A, Pouta A, Werdan K, Greiser KH, Kuss O, Meyer zu Schwabedissen HE, Thiery J, Jamshidi Y, Nolte IM, Soranzo N, Spector TD, Volzke H, Parker AN, Aspelund T, Bates D, Young L, Tsui K, Siscovick DS, Guo X, Rotter JI, Uda M, Schlessinger D, Rudan I, Hicks AA, Penninx BW, Thorand B, Gieger C, Coresh J, Willemsen G, Harris TB, Uitterlinden AG, Jarvelin MR, Rice K, Radke D, Salomaa V, Willems van Dijk K, Boerwinkle E, Vasan RS, Ferrucci L, Gibson QD, Bandinelli S, Snieder H, Boomsma DI, Xiao X, Campbell H, Hayward C, Pramstaller PP, van Duijn CM, Peltonen L, Psaty BM, Gudnason V, Ridker PM, Homuth G, Koenig W, Ballantyne CM, Witteman JC, Benjamin EJ, Perola M, Chasman DI. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Associations of plasma fibrinogen levels with established cardiovascular disease risk factors, inflammatory markers, and other characteristics: individual participant meta-analysis of 154,211 adults in 31 prospective studies: the fibrinogen studies collaboration. Am J Epidemiol. 2007;166:867–879. doi: 10.1093/aje/kwm191. [DOI] [PubMed] [Google Scholar]
- 34.Palmieri V, Celentano A, Roman MJ, de Simone G, Best L, Lewis MR, Robbins DC, Fabsitz RR, Howard BV, Devereux RB. Relation of fibrinogen to cardiovascular events is independent of preclinical cardiovascular disease: the Strong Heart Study. Am Heart J. 2003;145:467–474. doi: 10.1067/mhj.2003.144. [DOI] [PubMed] [Google Scholar]
- 35.Keavney B, Danesh J, Parish S, Palmer A, Clark S, Youngman L, Delepine M, Lathrop M, Peto R, Collins R. Fibrinogen and coronary heart disease: test of causality by 'Mendelian randomization'. Int J Epidemiol. 2006;35:935–943. doi: 10.1093/ije/dyl114. [DOI] [PubMed] [Google Scholar]
- 36.Smith GD, Harbord R, Milton J, Ebrahim S, Sterne JA. Does elevated plasma fibrinogen increase the risk of coronary heart disease? Evidence from a meta-analysis of genetic association studies. Arterioscler Thromb Vasc Biol. 2005;25:2228–2233. doi: 10.1161/01.ATV.0000183937.65887.9c. [DOI] [PubMed] [Google Scholar]
- 37.Kannel WB, Wolf PA, Castelli WP, D'Agostino RB. Fibrinogen and risk of cardiovascular disease. The Framingham Study. JAMA. 1987;258:1183–1186. [PubMed] [Google Scholar]
- 38.Lee AJ, Lowe GD, Woodward M, Tunstall-Pedoe H. Fibrinogen in relation to personal history of prevalent hypertension, diabetes, stroke, intermittent claudication, coronary heart disease, and family history: the Scottish Heart Health Study. Br Heart J. 1993;69:338–342. doi: 10.1136/hrt.69.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qizilbash N, Jones L, Warlow C, Mann J. Fibrinogen and lipid concentrations as risk factors for transient ischaemic attacks and minor ischaemic strokes. BMJ. 1991;303:605–609. doi: 10.1136/bmj.303.6803.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.