Abstract
Background
Modern genomic and proteomic studies reveal that many diseases are heterogeneous, comprising multiple different subtypes. The common notion that one biomarker can be predictive for all patients may need to be replaced by an understanding that each subtype has its own set of unique biomarkers, affecting how discovery studies are designed and analyzed.
Methods
We used Monte Carlo simulation to measure and compare the performance of eight selection methods with homogeneous and heterogeneous diseases using both single-stage and two-stage designs. We also applied the selection methods in an actual proteomic biomarker screening study of heterogeneous breast cancer cases.
Results
Different selection methods were optimal and more than 2-fold larger sample sizes were needed for heterogeneous diseases compared with homogeneous diseases. We also found that for larger studies, two-stage designs can achieve nearly the same statistical power as single-stage designs at significantly reduced cost.
Conclusions
We found that disease heterogeneity profoundly affected biomarker performance. We report sample size requirements and provide guidance on the design and analysis of biomarker discovery studies for both homogeneous and heterogeneous diseases.
Impact
We have shown that studies to identify biomarkers for the early detection of heterogeneous disease require different statistical selection methods and larger sample sizes than if the disease were homogeneous. These findings provide a methodological platform for biomarker discovery of heterogeneous diseases.
Keywords: biomarker discovery, heterogeneous disease, screening study design, power analysis, microarray analysis
INTRODUCTION
One major and underappreciated challenge in pre-diagnostic biomarker discovery is disease heterogeneity. Modern molecular analysis methods are increasingly revealing that what were once considered monotypic diseases instead comprise multiple molecular diseases that share a common clinical presentation(1-4). Breast cancer, for example, has many known molecular subtypes including the major subtypes luminal A and B, basal-like, and Erb-B2 over-expressed(5). These subtypes affect prognosis, response to treatment and recurrence (5-10). We hypothesize that disease heterogeneity may largely explain the low sensitivity of identified autoantibody biomarkers for breast cancer(11, 12). Heterogeneous diseases may not have a single biomarker that is predictive for all patients; each subtype may have its own set of unique biomarkers. Therefore, a biomarker that is excellent for detecting a particular subtype (e.g., 98% sensitivity) might only have 20% sensitivity for the disease overall because its sensitivity is capped by the prevalence of that subtype among those with the disease(13). Disease heterogeneity introduces new challenges for the discovery of biomarkers, such as the need for greater sample sizes to ensure that relevant subtypes have adequate representation. However, the magnitude of these changes or how to compute them has not been addressed. Moreover, an important question is whether heterogeneous diseases will require different statistical selection methods for identifying biomarkers and if so, which ones? Studies that have examined methods to determine sample size requirements and the relative performance of statistical biomarker selection methods have not considered these questions in the context of heterogeneous diseases(14-21).
A typical biomarker discovery study will screen a large collection of candidate biomarkers, typically genes or proteins, using a set of known disease-positive cases and a set of known disease-negative controls. As the number of candidate biomarkers tested increases, the cost per test increases dramatically. It is often prohibitive to screen all of the available cases and controls against all possible candidate markers. To mitigate this cost, an alternate approach is to conduct screening in two stages(12, 22-24). The first stage, the pre-screen, uses a moderate number of patients and controls to screen the full collection of biomarker candidates, with a goal of eliminating from consideration candidates that show little promise as biomarkers. In the second stage, the remaining candidates are screened using the remaining patients and controls. One advantage of the two-stage design is that in the second stage, the number of biomarker candidates to be screened may be sufficiently small that more detailed studies including additional replicates of measurements may be feasible, all of which can reduce intra-assay variability and variation of measured quantities. While this two-stage screening process is intuitively a cost effective way to screen a large number of candidates when using a large number of patients and controls, few studies have examined the efficiency of this two-stage design or the optimal allocation of cases and controls across the two stages(25-28) and none have examined these issues in the context of a heterogeneous disease.
We used Monte Carlo simulation(29, 30) to examine the relative performance of statistical biomarker selection methods, determine sample size requirements and compare experimental designs One key metric was power, the probability that a true biomarker would be selected in a screening process. We compared the power of several biomarker selection methods with homogeneous and heterogeneous case populations and found dramatic differences in the optimal methods for the two populations. Further, larger sample sizes are required for the heterogeneous disease compared to the homogeneous disease. We report sample size requirements and suggested methods for both homogeneous and heterogeneous diseases. We examined the performance of selection methods in a two-stage screening process and found that for larger studies, two-stage screening can achieve nearly the same power as single-stage screening at significantly reduced cost. We illustrate the impact of using different selection methods by comparing the results from their use in a real screening study for biomarkers for the early detection of breast cancer with a heterogeneous case population.
MATERIALS AND METHODS
Single Stage Screening
We used simulation to estimate and compare the power of biomarker selection methods with homogeneous and heterogeneous case populations using a single stage screening process. We considered a study in which 10,000 candidate biomarkers are to be screened and 50 of these are true biomarkers. In both case populations true biomarkers were set to have only 20% sensitivity at 95% specificity. The choice of 20% sensitivity was based upon consideration of breast cancer, the observation that the best autoantibody biomarkers for breast cancer typically have only 20-30% sensitivity at high specificity(12), and the prevalence of several subtypes of breast cancer, including basal-like, and luminal A and B, are typically reported to be between 10% and 30% depending on the subtype and study population(31, 32). For the homogeneous disease, the biomarker performance arises from a small distributional shift between the control and case populations. For the heterogeneous disease, the performance is due to a large signal observed in a small subpopulation of cases (Figure 1a-c). Importantly, because the particular subpopulation of cases that differ from the remaining cases may depend on the individual biomarker, our heterogeneous disease model does not presuppose how many different subtypes there are for a disease. For each subject and each candidate biomarker we independently simulated a response, which typically would correspond to a normalized and transformed measure of gene expression or protein abundance. We simulated case and control responses to non-biomarkers independently from a standard normal distribution. We also simulated control responses to true biomarkers independently from a standard normal distribution.
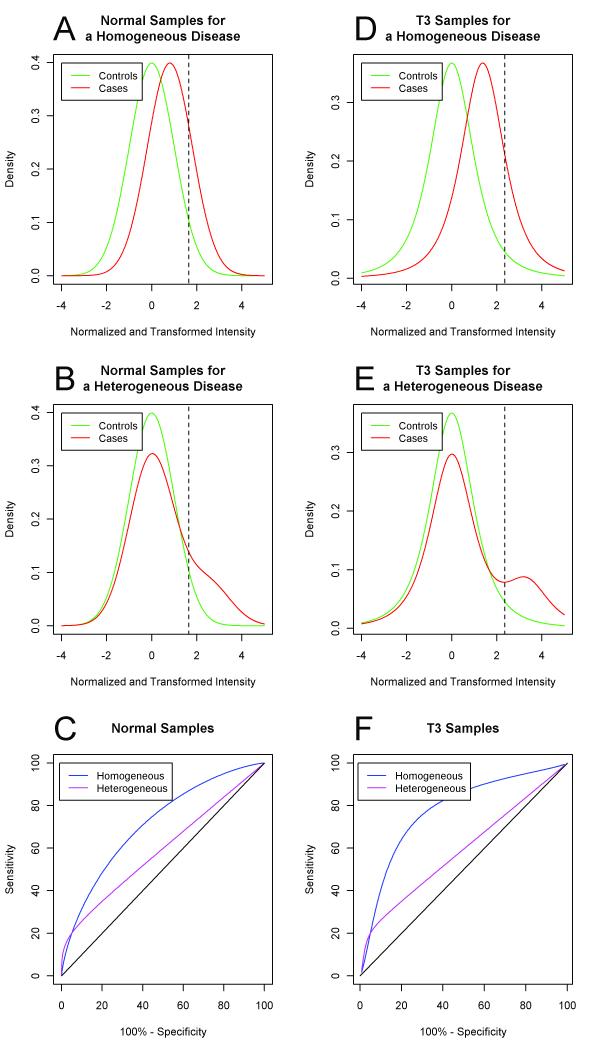
Figure 1.
(a) Sampling distributions of normalized and transformed intensities for true biomarkers for controls (green) and cases (red) with normal samples for a homogeneous disease. (b) Sampling distribution of normalized and transformed intensities for true biomarkers for controls (green) and cases (red) with normal samples for a heterogeneous disease. (d) ROC curves for true biomarkers with normal samples for homogeneous (blue) and heterogeneous (magenta) diseases. (e) Sampling distributions of normalized and transformed intensities for true biomarkers for controls (green) and cases (red) with a t3 homogeneous disease model. (f) Sampling distribution of normalized and transformed intensities for true biomarkers for controls (green) and cases (red) with a t3 heterogeneous disease model. (g) ROC curves for true biomarkers in the t3 homogeneous (blue) and t3 heterogeneous (magenta) disease models. In panels (a), (b), (d) and (e), the dashed vertical line indicates the threshold for 20% sensitivity and 95% specificity. In panels (c) and (f), the 45 degree line (black) represents the ROC curve when there is no difference between the case and control populations.
For the homogeneous disease, we simulated case responses to true biomarkers independently from a normal distribution with mean 0.80 and standard deviation 1. See Figure 1a. Thus, in this homogeneous disease model, true biomarkers have a sensitivity of 20% at 95% specificity and the area under the receiver operator characteristic (ROC) curve (AUC) is 0.71. For the heterogeneous disease, we simulated responses for each biomarker such that only 20% of cases would have differential response (i.e., the responding subtype) and the remaining 80% of cases would resemble the controls. Specifically, with 20% probability we simulated the response for an individual case from a normal distribution with mean 2.49 and standard deviation of 1; otherwise, we simulated the response from a standard normal distribution. See Figure 1b. Under this mixture model the overall sensitivity is 20% at 95% specificity, which is the same sensitivity as for the homogeneous disease, while the AUC for comparing all cases to controls is only .59. The ROC curves for the homogeneous and heterogeneous diseases are shown in Figure 1c. Note that the heterogeneous model did not presuppose how many different subtypes there are for a disease. Instead, we assumed that each of the multiple and possibly overlapping disease subtypes, which are characterized by a differential response in a biomarker, had a prevalence of 20% in the clinical disease population.
The stochastic models for homogeneous and heterogeneous diseases are simple and transparent. While both models achieve 20% sensitivity at 95% specificity, the heterogeneous disease model uses a mixture distribution for the case responses while a simple normal distribution is used for the homogeneous disease. In other aspects the models are identical. Both models assume that the candidate biomarkers are independent. Although this independence assumption is unlikely to hold in most applications, we make this assumption here to keep the models simple and transparent and because we are focused on selection of individual biomarkers rather than construction of multi-marker panels where the correlation structure between multiple markers could play a significant role. Both models also assume independence across subjects and utilize normal distributions. In a robustness study we replaced the normal distributions in the models with heavier tailed t distributions.
We examined eight selection methods that can be organized into three groups. The first group comprises the t-tests: the ordinary one-sided two-sample t-test, the one-sided Welch’s t-test, and a one-sided empirical Bayes moderated t-test(33). In this study, the moderated t-test represents the best-case scenario for t-tests, as this version exploits the homoscedasticity of the responses across biomarker candidates. These t-tests evaluate whether the mean response for cases is larger than for controls. The second group comprises tests of stochastic dominance: the one-sided Kolmogorov-Smirnov test, the one-sided Mann-Whitney U test, and a permutation test on the AUC. These are nonparametric tests that evaluate whether case responses tend to be larger than control responses. The third group comprises a permutation test on the partial AUC (PAUC) in the region where specificity is greater than 95% and a permutation test on the sensitivity at 95% specificity. These are nonparametric tests that evaluate whether there is sufficient mass in the case response distribution in the right tail of the control response distribution. See Supplementary Data for details on these methods. Most biomarker studies use variants of t-tests, often times through linear models that can easily accommodate covariates into an analysis. The full AUC and Mann-Whitney tests are also commonly used in biomarker studies. While researchers have advocated for the use of the partial AUC and sensitivity tests based upon their focus on clinically-relevant portions of the ROC curve(19), their use in biomarker discovery studies remains limited. For all methods we used the Benjamini-Hochberg procedure to control the false discovery rate (FDR) at 10% for each method based upon their respective p-values(34). Also see Supplementary Data for results under 50% FDR control.
We ran simulations using an equal number (N) of cases and controls, with N = 25, 50, 75, 100, 150, and 200, which encompassed the number of samples commonly used in preliminary biomarker studies. We constructed twenty simulated data sets at each sample size and used the eight statistical methods on each data set to construct lists of significant candidates, which we refer to as hits. We calculated the power for each simulation by computing the proportion of the 50 true biomarkers that appear in the list of hits. We also calculated the actual false discovery rate as the proportion of hits that were not among the 50 true biomarkers.
We estimated the sample size requirements for both homogeneous and heterogeneous diseases for power values of 70%, 80%, 90%, 95%, and 98% by linearly interpolating our power estimates for the best method, excluding the empirical Bayes t-test. We calculated the fold increase in sample size requirements for heterogeneous diseases as the ratio of the number of samples required for heterogeneous diseases to the number of samples required for homogeneous diseases.
Two Stage Screening
Here we consider a common scenario in which the total number of case and control samples is fixed and researchers wish to make the most efficient use of these samples to discover meaningful biomarkers. Specifically, we consider a two-stage process where the first stage uses a subset of the available samples to identify the top 750 candidates out of the 10,000 candidates and the second stage uses the remaining samples to identify the statistically significant hits out of those top 750. Using Monte Carlo simulation, we examined the performance of biomarker selection methods with homogeneous and heterogeneous case populations using a two-stage process, and compared their performance to a single-stage screening process with the same total number of cases and control.
We first conducted a preliminary study to determine the optimal scoring algorithm (out of 8 considered) for the first stage of a two-stage screening study (see Supplementary Data). Based on the results of this study (see Figure S1), for the homogeneous disease model we used empirical Bayes moderated t-statistics when the sample size is at most 40 and the ordinary t-statistic when the sample size is greater than 40. For the heterogeneous disease model we used empirical Bayes moderated t-statistics when N=10 and PAUC otherwise.
We then used the scoring algorithms identified above in the first stage of a two-stage screening process and examined the performance of different second-stage selection methods with the normal homogeneous and heterogeneous disease models. Although greater replication may be possible in the second stage due to the screening of fewer candidates, we here ignored the possible reduced variation that would accompany replication. We fixed the number of total cases and total controls and examined the power across various allocations of those cases and controls to the two stages. With 100 cases and 100 controls, we considered (first stage) : (second stage) allocations ranging from 10:90 up to 50:50. With 200 cases and 200 controls, we considered allocations ranging from 10:190 up to 140:60. We simulated ten data sets for each allocation of 100 cases and 100 controls. With 200 cases and 200 controls, we simulated ten data sets when the number allocated to the first stage was between 30 and 100, inclusive and five data sets when the number allocated was less than 30 or greater than 100.
RESULTS
Single Stage Screening
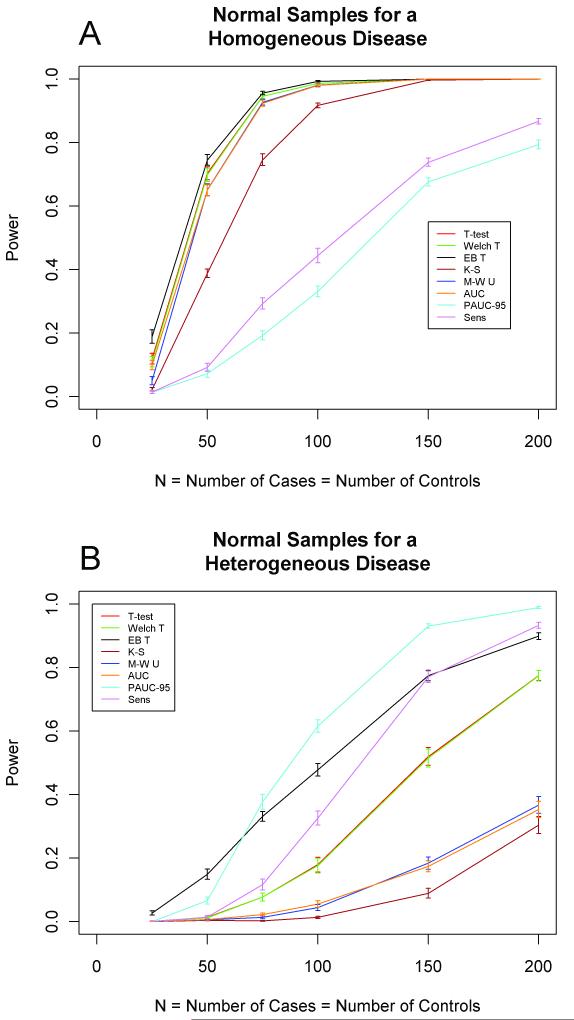
We compared the performance of biomarker selection methods with homogeneous and heterogeneous case populations using Monte Carlo simulation. Power estimates are displayed graphically in Figure 2 for both the homogeneous and heterogeneous disease sampling models. Numerical power estimates are provided in Supplementary Tables S1 and S2. For the homogeneous disease model, the t-tests and the Mann-Whitney and AUC tests identified over 60% of the true biomarkers using only 50 cases and 50 controls. By contrast, Kolmogorov-Smirnov identified less than 40%, and PAUC and the sensitivity test identified fewer than 10% of the true biomarkers with the same sample size.
Figure 2.
Estimated power for single stage design using normal samples for a (a) homogeneous disease and a (b) heterogeneous disease. The horizontal axis indicates N, the equal number of cases and controls. The vertical axis indicates power, the proportion of the true biomarkers that are selected by each method. Standard error bars are shown.
For the heterogeneous disease model, the results were strikingly different. At least 100 cases and 100 controls were needed before any method exceeded 60% power, and only the PAUC achieved that level of performance. At that sample size, the empirical Bayes moderated t-test identified nearly 50% of the true biomarkers, the sensitivity test approximately 30%, the other t-tests (ordinary and Welch’s) approximately 20%, and the Mann-Whitney, AUC, and Kolmogorov-Smirnov tests less than 10% of the true biomarkers. In general, among the eight methods the empirical Bayes moderated t-test had the greatest power at small samples sizes (N≤50) whereas PAUC had the greatest power at larger sample sizes (N≥75).
Estimated sample size requirements to achieve a desired level of power are given in Table 1 for both homogeneous and heterogeneous diseases. For each power value, the sample size required for the heterogeneous disease is at least twice that required for the homogeneous disease.
Table 1. Sample Size Requirements for Biomarker Screening Studies.
Suggested methods and sample size requirements for desired power and type of disease. Sample size requirements are given as the equal number of cases and controls in the study and are based upon a linear interpolation of the numerical power estimates, which are provided in Supplementary Tables S1 and S2. Fold increase for heterogeneous disease is the ratio of the number of samples required for a heterogeneous disease to the number of samples required for a homogeneous disease.
| Desired Power | Disease | Suggested Method |
Required Sample Sizes |
Fold Increase in Sample Sizes for Heterogeneous Disease |
|---|---|---|---|---|
| 70% | Homogeneous | T-test | 50 | 2.26 |
| Heterogeneous | PAUC | 113 | ||
| 80% | Homogeneous | T-test | 60 | 2.15 |
| Heterogeneous | PAUC | 129 | ||
| 90% | Homogeneous | T-test | 70 | 2.07 |
| Heterogeneous | PAUC | 145 | ||
| 95% | Homogeneous | T-test | 77 | 2.16 |
| Heterogeneous | PAUC | 166 | ||
| 98% | Homogeneous | T-test | 96 | 2.00 |
| Heterogeneous | PAUC | 192 |
In a separate study (see Supplementary Data), we investigated the robustness of this result by replacing the normal distribution with a t distribution with 3 degrees of freedom (t3). See Figures 1d-f. As with normal samples, the optimal statistical methods differ dramatically depending on whether heterogeneity is present and the sample sizes required under heterogeneity are much greater than under homogeneity. In fact, the difference is more dramatic with t3 samples than with normal samples. See Tables S3 and S4, and Figure S2. Although 90% power can be achieved with 50 cases and 50 controls with homogeneous disease samples, only 5% power can be achieved with heterogeneous disease samples.
Results under 50% FDR control for both normal and t3 samples are similar to those found under 10% FDR control. See Figures S3 and S4 for single stage power curves under 50% FDR control.
Two Stage Screening
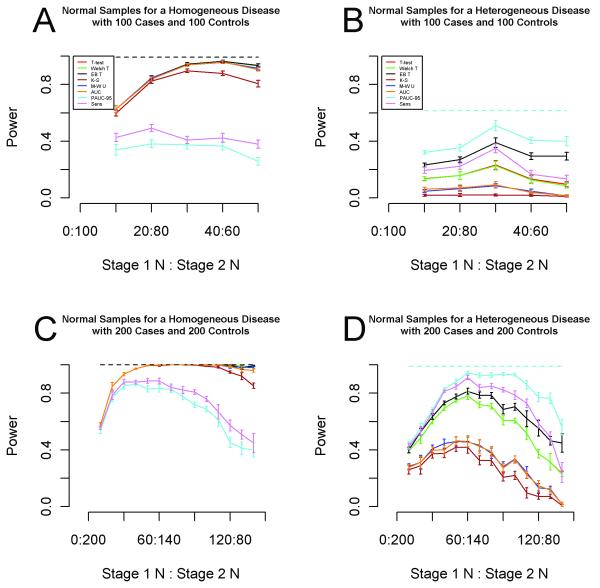
The results for 100 cases and 100 controls are displayed in Figure 3a,b. For a homogeneous disease, a single-stage screening study achieved 98-99% power with 5 of the 8 methods (Kolmogorov-Smirnov, PAUC and the sensitivity tests excluded). The same 5 methods achieved approximately 96% power using a less costly two-stage design with 40 first-stage cases and controls and 60 second-stage cases and controls. For a heterogeneous disease, PAUC achieved nearly 62% power using 100 cases and 100 controls in a single-stage study. The greatest power achieved by any of the eight methods with a total of 100 cases and 100 controls was 51% by PAUC with an allocation of 30:70. Although a loss of 11% power is not insignificant, the single-stage study could easily cost twice that of a 30:70 two-stage study based on our own experience with Nucleic Acid Programmable Protein Arrays (NAPPA)(35). See Figure S5 for similar results under 50% FDR control.
Figure 3.
Estimated power for a two-stage design using 100 total patients and 100 total controls with normal samples for a (a) homogeneous disease and (b) heterogeneous disease. Estimated power for a two-stage design using 200 total patients and 200 total controls with normal samples for a (c) homogeneous disease and (d) heterogeneous disease. The horizontal axis indicates the allocation of the total number of patients and controls across the two stages. The vertical axis indicates power, the proportion of the true biomarkers that are selected by each method. The selection method used for the first stage is the optimal first stage method for the prescribed first stage sample size and is common across the eight methods. The dashed horizontal line indicates the estimated power of the best method in a single-stage design using the same number of total patients and controls. Standard error bars are shown.
Figure 3c,d displays the results for 200 cases and 200 controls. With a homogeneous disease, a single-stage screening study achieved 100% power with 6 of the 8 methods (PAUC and the sensitivity test excluded). The same 6 methods also achieved 100% power using a less costly two-stage design with 50 first-stage cases and controls and 150 second-stage cases and controls. For a heterogeneous disease, PAUC achieved nearly 99% power using 200 cases and 200 controls in a single-stage study. With an allocation of 60:140, PAUC identified 94.4% of the true biomarkers. See Figure S6 for similar results with 50% FDR control.
Actual Breast Cancer Screening Study
To compare the performance of the eight selection methods with a known heterogeneous case population, we analyzed published data from a breast cancer screening study(12). Specifically, 761 antigens were screened against sera from 77 controls and 102 early stage breast cancer patients using NAPPA protein microarrays(35). Normalized and log-transformed intensity data were analyzed using each of the eight selection methods. The numbers of hits with 5% FDR control ranged from 38 with Kolmogorov-Smirnov up to 78 with Welch’s t-test. Summary statistics for these hits are given in Supplementary Table S5. Not surprisingly, the t-tests and tests for stochastic dominance yielded proteins with slightly higher AUC values (e.g., median AUC of 0.63 for t-test versus 0.60 for partial AUC). However, the median sensitivity values at 95% specificity ranged from 14-16% for the t-tests and tests for stochastic dominance, while the median sensitivity improved to 21% for partial AUC and 23% for the sensitivity test. Further, we examined the best hits from across the methods. Supplementary Table S6 lists the 37 proteins that were highly significant (1% FDR control) using any of the eight methods, along with their q-values (the minimum FDR for which the protein would be significant). This list contains many proteins that have previously been identified as having important roles in breast cancer biology and pathogenesis. Some of these have low q-values across all eight methods (EIF3E, PITX1A, PDCD6IP). However, some are identified only using partial AUC or the sensitivity test (RAC3, CTBP1, BMX) and some only by t-tests and tests for stochastic dominance (ARF1).
DISCUSSION
Selecting the best analysis methods
Disease heterogeneity is a well-established phenomenon. However the statistical methodologies most often used to identify pre-diagnostic biomarkers are designed for homogeneous diseases where at most the only case heterogeneity is assumed to be explainable by measured covariates such as age and gender. We have shown here the substantial effect that disease subtype heterogeneity may have on the performance of selection methods. The implications are significant: studies to identify biomarkers for the early detection of heterogeneous diseases require different statistical selection methods and larger sample sizes than if the disease were homogeneous.
Regarding the statistical selection methods, we found that the optimal statistical methods (out of the eight considered) differed dramatically between the two disease models. For example, the PAUC was the worst of the eight for the homogeneous disease, but the best for the heterogeneous disease at larger sample sizes. Similarly, the Mann-Whitney and AUC tests were among the best methods for the homogeneous disease and among the worst for the heterogeneous disease. Only the empirical Bayes moderated t-test performed relatively well under both disease models; however, we believe that the performance is artificially inflated due to a limitation of our study in which we used a common variance across the candidate biomarkers. To test this hypothesis, we conducted a small study in which the variances of the candidate biomarkers were simulated from an inverted gamma distribution with mean 1.0 and standard deviations ranging from 0.1 up to 10. We found that while the empirical Bayes moderated t-test produces a real improvement in power when the candidate biomarkers are nearly homoscedastic, this improvement vanishes with greater heteroscedasticity. See Supplementary Data and Figures S7 and S8 for additional details and results.
The explanation for the differences in performance of these tests for the two types of disease lies in the nature of the tests themselves. In the homogeneous disease model the distribution of responses for cases is simply shifted to the right of the distribution of control responses, which results in a moderate difference in the means of the distributions. Therefore t-tests, which test for a difference in the means and tests for stochastic dominance (Kolmogorov-Smirnov, Mann-Whitney, and AUC) perform moderately well at detecting this difference between case and control responses. In the heterogeneous disease model the only difference between the distributions of cases and controls is that there is a small subset of cases with large responses. While these aberrant cases do induce stochastic dominance and do increase the mean of the entire case population, the difference from the control population is less pronounced than in the homogeneous disease model. What better differentiates cases and controls in the heterogeneous disease model are the tails of the distributions. It is therefore not surprising that methods that specifically focus on differences in the tails, such as the partial AUC test and the sensitivity test, outperform tests that focus on differences in means in the heterogeneous disease model.
Although the partial AUC and sensitivity test performed best with a heterogeneous disease, neither of these methods was designed specifically for heterogeneous case populations. It is likely that other methods such as mixture modeling and cluster analysis will perform better with heterogeneous diseases in certain circumstances. Our intent in this study was not to find the optimal method, which will depend on many factors including the distributions of case and control responses; instead we examined the relative performance of methods for homogeneous and heterogeneous diseases in order to highlight the critical role that disease heterogeneity plays in the analysis of biomarker screening studies.
Another valid strategy for biomarker selection is to use multiple selection criteria. The application of the 8 methods on the breast cancer screening data yielded different sets of proteins. RAC3, a protein with an important role in breast cancer biology, was highly significant according to PAUC, but not according to any of the t-tests or the full AUC. Similarly, ARF1 was highly significant according to all tests, except for PAUC and the sensitivity test. The results suggest that the signals from a biomarker, even in a heterogeneous case population, may or may not appear to be heterogeneous, and therefore different testing strategies may be necessary to identify different types of biomarkers.
We have also shown that much larger sample sizes are required when heterogeneity is present. Whereas over 70% power can be achieved using a sample size of 50 cases and 50 controls for the homogeneous disease, only 15% power can be achieved using the same sample sizes for the heterogeneous disease. Further, we showed that at least twice the number of samples is needed for a heterogeneous disease compared to a homogeneous disease in order to obtain the same power. If a power analysis is based upon an assumption that the underlying disease is homogeneous, the study may be severely underpowered if the disease is actually heterogeneous.
Optimizing two stage design
A two-stage design may reduce the cost of screening studies as fewer candidates need to be screened in the second stage. We have shown that for moderately-sized studies, there is some loss in power from the use of a two-stage design, e.g. 62% to 51% using PAUC with 100 cases and 100 controls, but that loss may be justifiable due to the potential cost savings. For larger studies the power of the two-stage design can approach that of the single stage design. We therefore recommend that a two-stage design be considered for moderate to large biomarker discovery studies, where cost is an issue.
Recommendations
We have demonstrated that disease heterogeneity and sample sizes impact both the design and analysis techniques of biomarker screening studies. We strongly recommend that a formal power analysis be conducted for any planned biomarker screening study that accounts for the suspected amount of disease heterogeneity, sample availability, magnitude of differences between cases and controls, sample variability, false discovery rate tolerable and analysis methods. Further, a cost-benefit analysis could be conducted to examine the tradeoffs between using single and two-stage designs. In lieu of such formal analyses, we make the following recommendations for the design and analysis of biomarker screening studies (see Figure 4). For homogeneous case populations, for smaller sample sizes (less than 75 cases or 75 controls) a single stage design should be used, and investigators should seriously consider a two-stage design for larger sample sizes. For heterogeneous case populations, a single stage design should be used if there are fewer than 100 cases or 100 controls, and serious consideration should be given to a two-stage design for larger sample sizes. As for selection methods, the t-tests, Mann-Whitney U test, and AUC test are all viable tests for homogeneous case populations, and analysts should consider both normality of data and inclusion of covariate information when deciding between these approaches. For heterogeneous case populations, we recommend the use of the PAUC or sensitivity test, except for very small studies (fewer than 25 cases or 25 controls). In such small studies with heterogeneous case populations, these nonparametric tests will lack sufficient power. Instead, the best that one can accomplish may be to identify biomarkers that have a similar effect across the disease sub-types. Therefore, for these very small studies, we recommend the use of t-tests, Mann-Whitney U test, or the AUC test.
Figure 4.
Informal recommendations for the design and analysis of biomarker screening studies in lieu of formal power and cost-benefit analyses. The sample size (N) is the minimum of the number of cases and number of controls available for screening. In the diagram, ‘T’ refers to t-tests and many other linear modeling approaches, ‘M-W U’ is the Mann-Whitney U test, ‘AUC’ is a test for a large area under the ROC curve, ‘PAUC’ is a test for a large partial area under the ROC curve, and ‘Sensitivity’ is a test for high sensitivity at a given level of specificity.
The results of this study and these recommendations are based upon a Monte Carlo analysis of biomarker screening and are therefore dependent on the underlying stochastic model. Here we chose a simple mixture model to introduce disease heterogeneity in an intuitive and transparent fashion. When planning a biomarker screening study, we recommend that a formal power analysis be conducted with careful consideration given to modeling assumptions. In particular, our recommendations are based on simulating biomarker responses using normal distributions. If after normalization and transformation data do not follow normal distributions, sample size requirements may be greater. We have shown that replacing normal distributions with heavier-tailed t3 distributions leads to substantial changes in both power estimates and preferred selection methods. If data from a pilot study are available, bootstrap methods(36) can be used for power analyses of screening studies without requiring such distributional assumptions.
This study has shown that disease heterogeneity plays a significant role in the statistical selection of biomarkers. Disease heterogeneity may similarly impact drug discovery studies and drug trials. Examples abound of drugs for which only a subset of patients respond: trastuzumab (Herceptin) in HER2-overexpressed breast cancer, gefitinib (Iressa) for EGFR-overexpressed lung cancer, and cetuximab (Erbitux) for EGFR-overexpressed colorectal cancer. Accounting for disease heterogeneity in the design and analysis of drug studies may improve the efficiency and success rate of these studies.
Supplementary Material
Acknowledgements
We gratefully acknowledge financial support from the Early Detection Research Network (NIH/NCI 7U01CA117374), the Juvenile Diabetes Research Foundation (17-2007-1045) and the Virginia G. Piper Foundation. We thank Dr. Valentin Dinu for providing helpful feedback on this manuscript and the ASU Advanced Computing Center for computational resources.
Financial Support: All three authors were supported by the Early Detection Research Network (NIH/NCI 7U01CA117374). Wallstrom and LaBaer were also supported by the Juvenile Diabetes Research Foundation (17-2007-1045) and the Virginia G. Piper Foundation.
Footnotes
Conflicts of Interest: None
References
- 1.Lapointe J, Li C, Higgins JP, Rijn Mvd, Bair E, Montgomery K, et al. Gene Expression Profiling Identifies Clinically Relevant Subtypes of Prostate Cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi D, Spina V, Deambrogi C, Rasi S, Laurenti L, Stamatopoulos K, et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117(12):3391–401. doi: 10.1182/blood-2010-09-302174. [DOI] [PubMed] [Google Scholar]
- 3.Köbel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian Carcinoma Subtypes Are Different Diseases: Implications for Biomarker Studies. PLoS Med. 2008;5(12):e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nacht M, Dracheva T, Gao Y, Fujii T, Chen Y, Player A, et al. Molecular characteristics of non-small cell lung cancer. Proceedings of the National Academy of Sciences. 2001;98(26):15203–8. doi: 10.1073/pnas.261414598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular Definition of Breast Tumor Heterogeneity. Cancer Cell. 2007;11(3):259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Cervera N, Tarpin C, et al. Gene Expression Profiling Identifies Molecular Subtypes of Inflammatory Breast Cancer. Cancer Res. 2005;65(6):2170–8. doi: 10.1158/0008-5472.CAN-04-4115. [DOI] [PubMed] [Google Scholar]
- 8.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast Cancer Molecular Subtypes Respond Differently to Preoperative Chemotherapy. Clinical Cancer Research. 2005;11(16):5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 9.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA: The Journal of the American Medical Association. 2006;295(21):2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 10.Calza S, Hall P, Auer G, Bjohle J, Klaar S, Kronenwett U, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Research. 2006;8(4):R34. doi: 10.1186/bcr1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, et al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Annals of Oncology. 2007;18(5):868–73. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- 12.Anderson KS, Sibani S, Wallstrom G, Qiu J, Mendoza EA, Raphael J, et al. Protein Microarray Signature of Autoantibody Biomarkers for the Early Detection of Breast Cancer. Journal of Proteome Research. 2010;10(1):85–96. doi: 10.1021/pr100686b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson KS, LaBaer J. The Sentinel Within: Exploiting the Immune System for Cancer Biomarkers†. Journal of Proteome Research. 2005;4(4):1123–33. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobbin K, Simon R. Sample size determination in microarray experiments for class comparison and prognostic classification. Biostatistics. 2005;6:27–38. doi: 10.1093/biostatistics/kxh015. [DOI] [PubMed] [Google Scholar]
- 15.Gary L, Gadbury QX, Jode W, Edwards, Grier P. Page, David B. Allison. The Role of Sample Size on Measures of Uncertainty and Power. In: Allison GPP David B., Beasley T. Mark, Edwards Jode W., editors. DNA Microarrays and Related Genomic Techniques: Design, Analysis, and Interpretation of Experiments. Chapman & Hall / CRC; Boca Raton: 2006. [Google Scholar]
- 16.Jeffery I, Higgins D, Culhane A. Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinformatics. 2006;7(1):359. doi: 10.1186/1471-2105-7-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kooperberg C, Sipione S, LeBlanc M, Strand AD, Cattaneo E, Olson JM. Evaluating test statistics to select interesting genes in microarray experiments. Human Molecular Genetics. 2002;11(19):2223–32. doi: 10.1093/hmg/11.19.2223. [DOI] [PubMed] [Google Scholar]
- 18.Lin W-J, Hsueh H-M, Chen J. Power and sample size estimation in microarray studies. BMC Bioinformatics. 2010;11(1):48. doi: 10.1186/1471-2105-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pepe M, Longton G, Anderson G, Schummer M. Selecting differentially expressed genes from microarray experiments. Biometrics. 2003;59(1):133–42. doi: 10.1111/1541-0420.00016. [DOI] [PubMed] [Google Scholar]
- 20.Shao Y, Tseng C-H. Sample size calculation with dependence adjustment for FDR-control in microarray studies. Statist Med. 2007;26:4219–37. doi: 10.1002/sim.2862. [DOI] [PubMed] [Google Scholar]
- 21.Tibshirani R. A simple method for assessing sample sizes in microarray experiments. BMC Bioinformatics. 2006;7:106. doi: 10.1186/1471-2105-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu C-J, Song G, Huang W, Liu G-Z, Deng C-W, Zeng H-P, et al. Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Molecular & Cellular Proteomics. 2012 doi: 10.1074/mcp.M111.015529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Q, Liu G, Hu S, Zhang Y, Tao Y, Han Y, et al. Novel Autoimmune Hepatitis-Specific Autoantigens Identified Using Protein Microarray Technology. Journal of Proteome Research. 2009;9(1):30–9. doi: 10.1021/pr900131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cima I, Schiess R, Wild P, Kaelin M, Schüffler P, Lange V, et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong T-T, Hsu C-H. Two-stage classification methods for microarray data. Expert Systems with Applications. 2008;34(1):375–83. [Google Scholar]
- 26.Satagopan JM, Verbel DA, Venkatraman ES, Offit KE, Begg CB. Two-Stage Designs for Gene–Disease Association Studies. Biometrics. 2002;58(1):163–70. doi: 10.1111/j.0006-341x.2002.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goll A. Bauer P. Two-stage designs applying methods differing in costs. Bioinformatics. 2007;23(12):1519–26. doi: 10.1093/bioinformatics/btm140. [DOI] [PubMed] [Google Scholar]
- 28.Zehetmayer S, Bauer P, Posch M. Two-stage designs for experiments with a large number of hypotheses. Bioinformatics. 21(19):3771–7. doi: 10.1093/bioinformatics/bti604. [DOI] [PubMed] [Google Scholar]
- 29.Metropolis N, Ulam S. The Monte Carlo Method. Journal of the American Statistical Association. 1949;44(247):335–41. doi: 10.1080/01621459.1949.10483310. [DOI] [PubMed] [Google Scholar]
- 30.Robert CP, Casella G. Monte Carlo Statistical Methods. Second ed Springer; New York: 2004. [Google Scholar]
- 31.O’Brien KM, Cole SR, Tse C-K, Perou CM, Carey LA, Foulkes WD, et al. Intrinsic Breast Tumor Subtypes, Race, and Long-Term Survival in the Carolina Breast Cancer Study. Clinical Cancer Research. 2010;16(24):6100–10. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth G. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical applications in genetics and molecular biology. 2004;3(1) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 35.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, et al. Self-Assembling Protein Microarrays. Science. 2004;305(5680):86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 36.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman and Hall; New York, NY: 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.