Summary
In animals, many cells reach their destinations by migrating towards higher concentrations of an attractant. However, the nature, generation and interpretation of attractant gradients are poorly understood. Using a GFP fusion and a signaling sensor, we analyzed the distribution of the attractant chemokine Sdf1 during migration of the zebrafish posterior lateral line primordium, a cohort of about 200 cells that migrates over a stripe of cells uniformly expressing sdf1. We find that a small fraction of the total Sdf1 pool is available to signal and induces a linear Sdf1-signaling gradient across the primordium. This signaling gradient is initiated at the rear of the primordium, equilibrates across the primordium within 200 minutes, and operates near steady-state. The rear of the primordium generates this gradient through continuous sequestration of Sdf1 protein by the alternate Sdf1-receptor Cxcr7. Modeling shows that this is a physically plausible scenario.
Introduction
During animal development, homeostasis and disease, cells must move from one location to another to form tissues, assemble into organs, chase a pathogen or – in the case of cancer – populate sites of metastasis. Depending on the process, cells migrate as single cells, chains of cells or as tissue-like collectives of a few cells to hundreds of cells. In order to move in the correct direction, migrating cells need guidance cues. Studies over the last few decades have revealed the identity of many guidance cues. These guidance cues are often secreted from the target tissue and form an attractant gradient, from which migrating cells derive directional information (Parent, 1999; Rørth, 2011; Swaney et al., 2010). There are several ways for how these attractant gradients can guide migrating cells. For example, migrating cells can be guided by long-range attractant gradients emanating from a source at the target tissue (for example (Montell, 2003)), shifting expression domains of the attractant (for example (Affolter and Caussinus, 2008)) or the graded distribution of an immobilized attractant (for example (Weber et al., 2013)). In the simplest model, attractants are secreted from a local source and degraded by a local sink, generating a linear gradient at steady-state. Francis Crick showed in 1970 that this source-sink model can generate stable, linear gradients over several hundreds of micrometers (Crick, 1970).
A classic example for single cell migration is the slime mold Dictyostelium (reviewed in (Parent, 1999)). Dictyostelium cells are attracted by cAMP and move towards higher cAMP concentrations. The cells are about 10 μm in diameter, and while they can sense differences in cAMP concentration of as low as 1% across themselves, they migrate most efficiently when this difference is 3% (Fisher et al., 1989). Intriguingly, Dictyostelium migrate towards higher cAMP concentrations both within pre-steady-state gradients with temporally increasing cAMP concentration as well as within stable, steady-state gradients (Fisher et al., 1989). It is thought that Dictyostelium achieves sensitivity to concentration differences of cAMP and robustness to fluctuations in cAMP concentration by integrating and reinforcing information about local cAMP concentrations sensed by the cAMP receptors on the cell surface (Cai and Devreotes, 2011).
The purpose of this study is to determine the shape (linear or non-linear), dynamics (pre-steady-state or steady-state), and mechanisms of generation and maintenance of an attractant gradient in a living animal. We were motivated by recent studies using overexpressed fluorescently-tagged proteins to describe the distribution of signaling molecules in living animals (Entchev et al., 2000; Kicheva et al., 2007; Muller et al., 2012; Teleman and Cohen, 2000; Yu et al., 2009). These studies reported the distribution of the total population of signaling molecules using overexpressed tagged molecules, but they did not delineate how much of the total signaling molecules are actually involved in signaling events. Therefore, the in vivo distribution of endogenous untagged signaling molecules remains unclear.
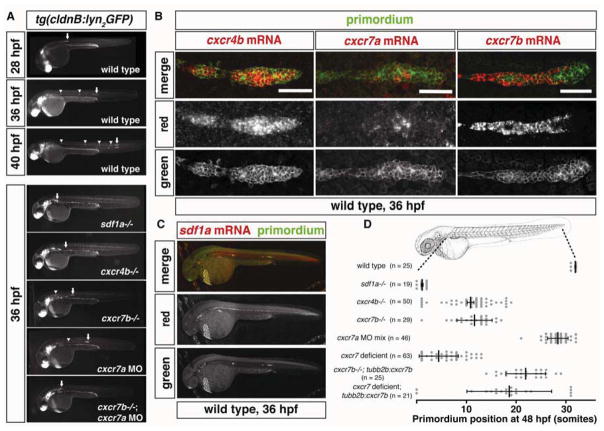
The posterior lateral line primordium in zebrafish is an excellent model for studying how attractants guide migrating cells (Aman and Piotrowski, 2010). The primordium is composed of about 200 epithelial-like cells that are born behind the ear around 19 hours post fertilization (hpf). Over the next 20 hours, these cells migrate collectively along the body of the fish until they reach the tip of the tail around 40 hpf (Figure 1A and Movie S1). During this migration period, the primordium deposits 5 to 7 cell clusters along the trunk and tail of the embryo (Ghysen and Dambly-Chaudière, 2007). Each of these clusters differentiates into a neuromast, a specialized organ that senses water flow around the embryo. The primordium requires the chemokine Sdf1a and its two receptors, Cxcr4b and Cxcr7b, for proper migration (Figure 1A). The cells of the primordium express cxcr4b uniformly starting at 19 hpf when the primordium first forms (Figure 1B). cxcr7b expression turns on specifically in the rear of the primordium (Figure 1B) only once it reaches and starts migrating over a narrow and uniform stripe of sdf1a-expressing cells located along the trunk and tail of the embryo (Figure 1C) (Breau et al., 2012; Dambly-Chaudière et al., 2007; David et al., 2002; Valentin et al., 2007). Although chemokine signaling is required for proper migration, it remains unclear how a stripe of uniform sdf1a can provide directional guidance to the primordium during its journey through the embryo.
Figure 1. Expression and requirement of Sdf1a and its receptors Cxcr4b, Cxcr7a and Cxcr7b during primordium migration.
(A) Live images of embryos of the indicated stage and genotype. Arrow indicates the primordium, arrowheads indicates neuromasts. (B) Fluorescent staining for cxcr4b, cxcr7a or cxcr7b mRNA and GFP protein at 36 hpf. Scale bar 50 μm. (C) Fluorescent staining for sdf1a mRNA and GFP protein in a tg(cldnB:lyn2GFP) embryo at 36 hpf. Anterior is to the left and posterior is to the right. (D) Quantification of primordium migration in 48 hpf embryos of indicated genotypes. The vertical bars represent the average position of the primordium, the error bars represent SD and the circles represent the positions of individual primordia. 48 hpf embryo schematic adapted from (Kimmel et al., 1995). (See also Figure S6).
Here, we developed quantitative reporters for Sdf1a protein and Sdf1-signaling and employed quantitative imaging and mathematical modeling to examine the distribution of total Sdf1a protein and the pool of Sdf1a protein available for signaling through Cxcr4. We find that total Sdf1a protein is distributed uniformly along the stripe of chemokine producing cells underneath the primordium. In contrast, Sdf1-signaling is linearly-graded across the primordium for the duration of its migration, with a slope of 7% per cell. Upon abrogation, this gradient re-emerges and reaches steady-state again within 200 minutes. Mathematical modeling shows that the observed gradient kinetics are inconsistent with freely diffusing Sdf1a protein and suggest that the chemokine is hindered in its diffusivity, probably due to binding to extracellular molecules.
To determine how the primordium converts a uniform source of Sdf1a protein into an Sdf1-signaling gradient, we analyzed the expression of Sdf1a protein within the primordium. We find that the rear of the primordium sequesters 1% of the total Sdf1a protein. Although controversial (Rajagopal et al., 2009), CXCR7 - an alternate receptor for SDF1 - has been proposed to act as a chemokine clearance receptor (Boldajipour et al., 2008; Sánchez-Alcañiz et al., 2011). The two CXCR7 orthologs, Cxcr7a and Cxcr7b, are expressed in the rear of the primordium. We find that the two orthologs are required for Sdf1a protein uptake in the rear of the primordium, Sdf1-signaling gradient formation across the primordium and primordium migration. Additionally, in embryos lacking Cxcr7, both the Sdf1-signaling gradient and primordium migration can be restored by reintroducing Cxcr7b underneath the rear of the primordium. These observations demonstrate that the primordium generates an attractant gradient across itself by sequestering Sdf1a protein in its rear via Cxcr7-mediated chemokine uptake. This self-generated attractant gradient, combined with the route information provided by the stripe of sdf1a-expressing cells, then provides directional guidance to the migrating primordium. Mathematical modeling of a sink moving across a source stripe that provides a constant attractant concentration shows that this a plausible scenario.
Results
Sdf1a-GFP is distributed evenly along the migratory route of the primordium
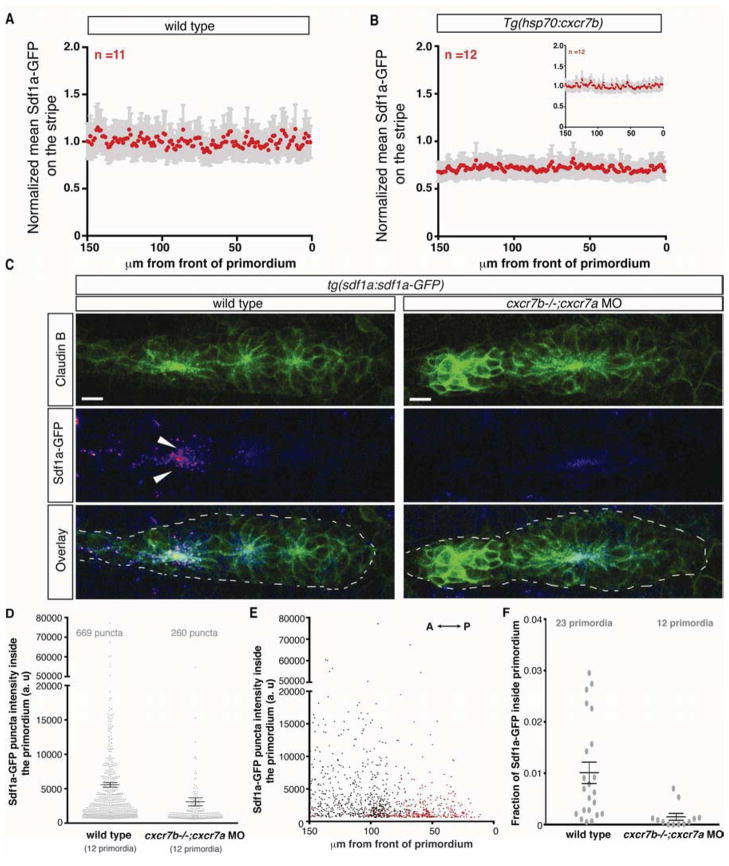
To analyze the distribution of Sdf1a protein along the migratory route of the primordium we generated a transgenic line (tg(sdf1a:sdf1a-GFP)) that expresses Sdf1a fused to green fluorescent protein (GFP) from a bacterial artificial chromosome (BAC). This BAC includes the sdf1a exons and introns, a 55kb sequence upstream of the start codon and a 30kb sequence downstream of the stop codon (Figure S1A). The transgene recapitulates the endogenous sdf1a mRNA expression pattern (Figure S1B and S1C) and restores primordium migration in sdf1a mutant embryos (Figure S1E–S1G), demonstrating that it is functional. We used the tg(sdf1a:sdf1a-GFP) line to examine the distribution of Sdf1a-GFP protein in wild type embryos. Sdf1a-GFP protein is distributed evenly along the migration route of the primordium (Figure S1D) and is confined to the immediate vicinity of the cells that produce it (Movie S2). We quantified the intensity of Sdf1a-GFP on the stripe underneath the primordium and do not detect a difference in the levels of the chemokine between the front and rear of the primordium (Figure 2A). However, close inspection reveals that cells in the rear of the primordium sequester small amounts of Sdf1a-GFP, which appear as discrete intracellular puncta (Figure 2C and Movie S2). Quantification of the number and intensity of Sdf1a-GFP puncta inside the primordia of multiple embryos confirms that cells in the rear of the primordium internalize more Sdf1a-GFP than the cells in the front of the primordium (Figure 2E). This raises the possibility that the rear of the primordium reduces the concentration of Sdf1a beneath it through protein sequestration, suggesting that the primordium is capable of locally modifying the levels of chemokine in its path. However, the Sdf1a-GFP uptake by the rear of the primordium represents only 1% of the total Sdf1a-GFP signal (Figure 2F) and is thus within the noise margin (SEM 18%) of Sdf1a-GFP intensity measurements made from the stripe beneath the primordium. This suggests that the migrating primordium only modifies the chemokine pool in its immediate vicinity, with minimal effects on overall chemokine levels.
Figure 2. Cxcr7 is required for Sdf1a-GFP sequestration by the primordium.
(A, B) Average fluorescence intensity of Sdf1a-GFP protein along the stripe of chemokine producing cells underneath primordia in embryos of indicated genotypes (B, inset, heat-shocked wild-type control embryos). (C) Sum projection of the primordium from 36 hpf embryos of indicated genotypes stained for Claudin-B and GFP. Scale bar corresponds to 10 μm. Direction of migration is to the right. Arrowheads indicate Sdf1a-GFP puncta. (D) Distribution of intensities of Sdf1a-GFP puncta in primordia of indicated genotypes. (E) Distribution of intensities of Sdf1a-GFP puncta in primordia along the anterior-posterior axis. Each dot (D and E) represents an individual punctum (red: cxcr7b−/−; cxcr7a morphant primordia; black: wild type primordia). (F) Fraction of Sdf1a-GFP found inside the primordium of the total Sdf1a-GFP in the indicated genotype. Each dot represents the fraction of Sdf1a-GFP in an individual primordium. Horizontal bars represent the mean +/− SEM. (See also Figure S1)
A novel in vivo Sdf1-signaling sensor
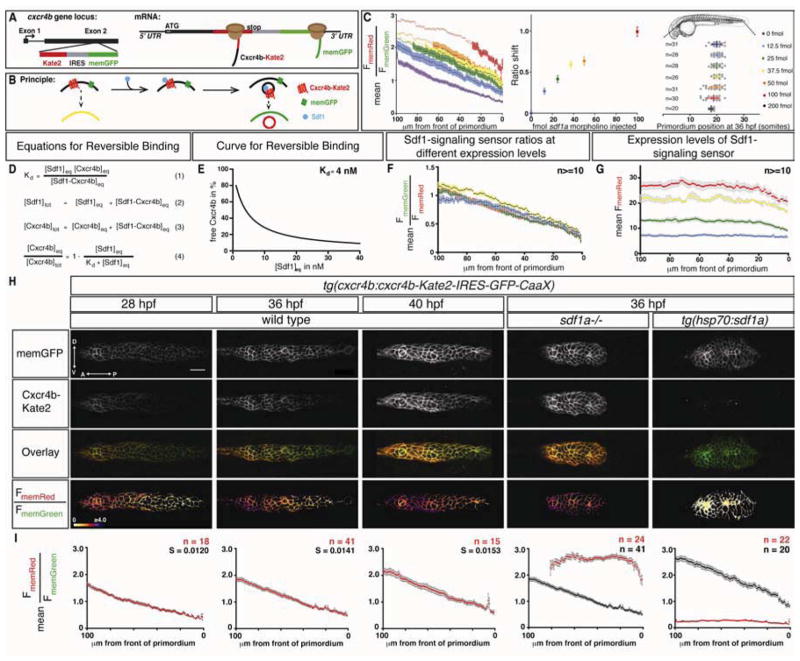
It is possible that the primordium senses and responds to a shallow gradient of Sdf1a that we cannot detect by measuring the total amount of Sdf1a-GFP protein along the stripe. To investigate this possibility, we developed an in vivo Sdf1-signaling sensor designed to measure the levels of Sdf1a that the primordium perceives. Since the binding of SDF1 to CXCR4 triggers rapid internalization of the receptor from the cell membrane and subsequent receptor degradation (Marchese and Benovic, 2001; Marchese et al., 2003; Minina et al., 2007), we reasoned that the levels of Cxcr4b on the cell membrane should correlate inversely to the levels of extracellular Sdf1 protein. To test this idea, we fused the monomeric red fluorescent protein Kate2 to the C-terminus of Cxcr4b (Cxcr4b-Kate2) and expressed this fusion protein from the cxcr4b promoter. As an internal reference, we co-expressed membrane tethered GFP (memGFP) that is co-translated from the same transcript through an internal ribosomal entry site (IRES) (Figure 3A and S1H). This signaling sensor recapitulates the expression of endogenous cxcr4b (Figure S1I–S1K), internalizes with Sdf1a-GFP (Figure S1O and S1P) and rescues primordium migration in cxcr4b mutant embryos (Figure S1L–S1N), demonstrating that it is functional. Since ligand binding causes receptor internalization, the ratio of the red fluorescence from the Cxcr4b-Kate2 fusion protein to the green fluorescence from the memGFP on the membrane of a cell (FmemRed/FmemGreen) should represent a quantitative readout of the amount of Sdf1a protein to which the cell is exposed (Figure 3B). We tested the relationship between Sdf1a protein levels and the Sdf1-signaling sensor in several ways. First, in the absence of Sdf1a protein, the membranes of cells within the primordium exhibit high levels of Cxcr4b-Kate2, resulting in an average FmemRed/FmemGreen ratio of 2.6 (Figure 3H and 3I, column 4). Second, upon global Sdf1a over-expression from an inducible heat-shock promoter (tg(hsp70:sdf1a)), Cxcr4b-Kate2 is found primarily inside the cell rather than on the cell membrane, resulting in an average FmemRed/FmemGreen ratio of 0.2 (Figure 3H and 3I, column 5). Third, injection of increasing amounts of a translation-blocking sdf1a morpholino progressively shifts the FmemRed/FmemGreen ratios across the primordium to higher values in a manner directly proportional to the amount of sdf1a morpholino injected, consistent with progressively decreasing levels of Sdf1a (Figure 3C). Fourth, consistent with theoretical considerations for reversible ligand-receptor binding (Figure 3D and E), we find that altering the expression levels of the Sdf1-signaling sensor does not change the Sdf1-signaling sensor ratios across the primordium (Figure 3F and G). Fifth, in the absence of Sdf1a, Cxcr4b-Kate2 and memGFP are translated at a fairly constant ratio across the primordium (Figure 3H, column 4, and S2F), indicating that memGFP expression from the IRES is uniform across the primordium. Sixth, memGFP is expressed uniformly along the anterior-posterior axis of the primordium (Figure S2A), indicating that the activity of the cxcr4b promoter is fairly constant across the cells of the primordium. Seventh, expression of the Sdf1-signaling sensor in HEK 293T cells indicates that the FmemRed/FmemGreen ratios are directly proportional to the concentration of Sdf1added to the cell media (Figure S3A and S3C). Thus, the ratio of FmemRed/FmemGreen reported by the Sdf1-signaling sensor is linearly related to the levels of Sdf1 protein that the primordium perceives during migration.
Figure 3. A quantitative signaling reporter for Sdf1.
(A, B) Schematic of Sdf1-signaling sensor construct (A) and concept (B). (C) Left: Mean FmemRed/FmemGreen ratio along the anterior-posterior axis of n≥20 primordia from tg(cxcr4b:cxcr4b-Kate2-IRES-GFP-CaaX) embryos injected with sdf1a morpholino. Circles are mean ratios and error bars represent SEM. Open circles indicate ratios greater than the standard deviation corrected mean FmemRed/FmemGreen ratio in sdf1a−/− primordia. Middle: Shift of the FmemRed/FmemGreen ratio curves on left. The error bars represent SD. Right: Primordium position in embryos injected with increasing amounts of sdf1a morpholino. The vertical bars represent the average position of the primordium, the error bars represent SD and the circles represent the positions of individual primordia. 35 hpf embryo schematic adapted from reference (Kimmel et al., 1995). (D) Derivation of equation for reversible binding of Sdf1 to Cxcr4b (equation 4) using the definition of the dissociation constant (equation 1) for the reversible reaction where Cxcr4beq is free receptor, Sdf1eq is free ligand, and Cxcr4b-Sdf1eq is receptor-ligand complex, and mass balance for Sdf1 (equation 2) and Cxcr4b (equation 3). (E) Graph of equation 4 for a Kd of 4 nM (Crump et al., 1997). (F) Mean FmemGreen/FmemRed ratio values across 100 μm beginning at the front of the primordium in 36 hpf wild type embryos with increasing levels of expression of the signaling sensor indicated in G. (G) Mean FmemRed intensity values across 100 μm beginning at the front of the primordium in 36 hpf embryos carrying different combinations and copy numbers of the signaling sensor transgenes (blue: Sdf1 sensorGRp1/+, n=14, green: Sdf1 sensorGRp7/+, n=17, yellow: Sdf1 sensorGRp1/Sdf1 sensorGRp7, n=10, red: Sdf1 sensorGRp7/Sdf1 sensorGRp7, n=16). The colored and grey bars indicate SEM in F and G, respectively. (H) Single confocal slices through the primordium in live embryos of the indicated genotypes and stages, all carrying the Sdf1-signaling sensor. The FmemRed/FmemGreen images are inverted heat maps of the ratio. (I) Mean FmemRed/FmemGreen along the anterior-posterior axis of n≥10 primordia with 0 μm representing the front of each primordium. Red circles indicate the mean FmemRed/FmemGreen in embryos of the indicated genotype; black circles, where present, indicate the mean FmemRed/FmemGreen of wild-type embryos or heat-shocked control embryos. Grey bars indicate SEM. Anterior is to the left in H. Scale bar is 20 μm. (See also Figures S1–S5)
A linear Sdf1-signaling gradient across the primordium
Using this sensor, we detected an Sdf1-signaling gradient across the anterior-posterior axis of the migrating primordium in live, 36 hpf wild-type embryos (Figure 3H and 3I, column 2). The gradient begins at the leading edge of the primordium at a mean FmemRed/FmemGreen of below 0.6, increases fairly linearly by 1.2%/μm for the first 100 μm and plateaus at a mean FmemRed/FmemGreen of 2.3 in the rear of the primordium (Figures I column 2 and S2G). The linear gradient moves with the primordium throughout its migration, remaining remarkably constant in shape and amplitude over time (Figure 3H and 3I, columns 1–3 and Movie S3, left). Moreover, the gradient is absent in sdf1a mutant embryos (Figure 3I, column 4) and is rapidly abolished upon global over-expression of Sdf1a from a heat-shock inducible promoter (Figure 3I, column 5 and Movie S3, right), confirming that the signaling gradient depends on Sdf1a protein levels. The Sdf1-signaling gradient is mirrored by an internal gradient of internalized and degraded Cxcr4b. Quantification of the levels of internalized Cxcr4b-Kate2 to memGFP indicates that Cxcr4b-Kate2 is degraded more in the front than in the rear of the primordium and that internalization and degradation of Cxcr4b-Kate2 depends on Sdf1a (Figure S4). To approximate the lower and upper limits of Sdf1-signaling in the primordium, we compared the mean FmemRed/FmemGreen in sdf1a mutant embryos and embryos that globally over-express Sdf1a. The maximal difference in chemokine signaling observed between these two scenarios is 2.4 ratio units (mean FmemRed/FmemGreen of 2.6 and 0.2, respectively, in Figure 3I, columns 4 and 5). When compared to the maximal signaling difference between the front and back of wild-type primordia of 1.7 ratio units (mean FmemRed/FmemGreen of 0.6 and 2.3, respectively, in Figure S1Q), this indicates that 36 hpf wild type primordia use 71% of the Sdf1-signaling dynamic range.
The Sdf1-signaling gradient observed across the primordium - high signaling in the leading cells and low signaling in the trailing cells - suggests that a graded distribution of Sdf1a continuously confers directional information to the migrating primordium. Results from a previous study demonstrated that ectopic sources of the chemokine Sdf1b, the protein encoded by a closely related paralog of sdf1a, can attract the primordium (Li et al., 2004). Since Sdf1b is not expressed along the migratory route of the primordium and dispensable for its migration, we hypothesized that Sdf1a, like Sdf1b, can lure the primordium off course when expressed ectopically, therefore acting as an instructive rather than permissive guidance cue. We tested this hypothesis in two ways. First, we over-expressed Sdf1a from a heat-shock promoter during primordium migration in embryos carrying the cldnB:lyn2GFP transgene or the Sdf1-signaling sensor. In response to global over-expression of Sdf1a, the primordium exhibits uniformly high levels of Cxcr4b-Kate2 internalization, rounds up and ceases to migrate, in contrast to primordia in heat-shocked control embryos that report a steady, linear signaling gradient, maintain an elongated morphology and continue to migrate (Figure S5A–S5C and Movie S3). Second, we generated small, Sdf1a-misexpressing clones along the migratory route of the primordium or within the primordium in tg(cldnB:lyn2GFP) embryos by blastomere transplantation (Figure S5D). Clones positioned dorsal or ventral to the normal migratory route were able to attract the primordium, sending it off course (Figure S5J–S5L), while clones within the primordium caused it to round up and stall (Figure S5F).
Cxcr7 sequesters Sdf1a protein in the rear of the primordium
Our observations that the rear of the primordium sequesters Sdf1a-GFP protein (Figure 2C and Movie S2) and perceives lower levels of Sdf1 than the front (Figure 3H and 3I, columns 1–3) suggest that the rear of the primordium continuously clears Sdf1a protein from the region underneath itself. Previous studies have proposed that the alternate SDF1 receptor CXCR7 can act as a scavenger receptor for chemokines (Boldajipour et al., 2008; Sánchez-Alcañiz et al., 2011). Consistent with this proposition, cxcr7b is expressed in the rear of the primordium (Figure 1B, column 3), and is required for its migration (Figure 1A and 1D) (Dambly-Chaudière et al., 2007; Valentin et al., 2007). Primordia in cxcr7b mutant embryos exhibit slowed migration or stalling (Movie S4, top) (Valentin et al., 2007). Interestingly, we find that cxcr7a - the second ortholog of CXCR7 in zebrafish - is also expressed in the rear of the primordium (Figure 1B, column 2). In embryos injected with morpholinos that block translation of cxcr7a transcripts (Figure S6A and B), the primordium does not always reach the tip of the tail (Figure 1A and 1D). Compromising the function of both CXCR7 orthologs enhances these migration defects (Figure 1A and 1D), often resulting in complete stalling of the primordium (Movie S4, bottom), a defect that is comparable to what we observe in primordia of sdf1a mutant embryos (Figure 1A and Movie S1). However, in contrast to the rounded morphology that the primordium assumes in sdf1a mutant embryos (Figure 1A and Movie S1), primordia deficient in cxcr7a and cxcr7b (collectively referred to as Cxcr7) are more motile and often extend in multiple directions (Movie S4, bottom). Thus, Sdf1a protein sequestration by Cxcr7 is a plausible mechanism for the generation and maintenance of a chemokine attractant gradient across the migrating primordium. To test this, we measured Sdf1a-GFP protein uptake by the primordium in cxcr7 deficient and cxcr7b over-expressing embryos. Consistent with the hypothesis, we find that cxcr7 deficient primordia fail to sequester Sdf1a-GFP protein in the rear of the primordium, in contrast to wild-type primordia that show significant uptake in this region (Figure 2C–2F and Movie S2). The number and intensity of the Sdf1a-GFP puncta are markedly reduced in cxcr7 deficient primordia (Figure 2D and 2E), suggesting that Cxcr7 is required for chemokine sequestration. Conversely, over-expression of Cxcr7b from a heat-shock inducible promoter causes the primordium to assume a rounded morphology similar to that observed in sdf1a mutant embryos and to decelerate (Movies S5 and S1, respectively). Sdf1a-GFP protein levels on the stripe outside the primordium are reduced by 29% in these embryos (Figure 2B), indicating that Cxcr7 activity promotes removal of Sdf1a from the stripe.
Cxcr7 generates an Sdf1-signaling gradient across the primordium
Next, we tested whether Cxcr7-mediated Sdf1a protein sequestration in the rear of the primordium is responsible for generating the Sdf1-signaling gradient across the primordium. Consistent with the variable migration defects observed in embryos deficient for either cxcr7a or cxcr7b, we find that Sdf1-signaling is increased specifically in the rear of the primordium in the absence of cxcr7a or cxcr7b activity compared to wild-type controls (Figure 4A and 4B, columns 1–3), resulting in a 31% or 40% reduction in the steepness of the Sdf1-signaling gradient across the primordium, respectively. These findings indicate that both CXCR7 orthologs contribute to the local clearance of Sdf1a protein and, thus, to the generation of the signaling gradient. Indeed, impairing both cxcr7a and cxcr7b activity in the same embryo increases Sdf1-signaling in the rear to levels that are normally only observed in the front of the primordium (Figure 4A and 4B, column 4). Importantly, this increase of Sdf1-signaling in the rear of the primordium of cxcr7 deficient embryos requires Sdf1a activity since Sdf1-signaling in primordia of embryos lacking cxcr7 and sdf1a resembles Sdf1-signaling in primordia of embryos mutant for sdf1a alone (Figure 4A and 4B, column 5). Conversely, in embryos that over-express Cxcr7b from a heat-shock inducible promoter, Sdf1-signaling is reduced throughout the primordium (Figure 4A and 4B, column 6). The absence of the Sdf1-signaling gradient in cxcr7 deficient primordia resembles the scenario in which Sdf1a is over-expressed globally (Figure 3H and 3I, column 5), while the high signaling levels observed across the primordium upon global Cxcr7b over-expression are similar to what we observed in sdf1a mutant primordia (Figure 3H and 3I, column 4), suggesting that Cxcr7 activity correlates inversely with Sdf1a levels. Furthermore, the abrogation of the Sdf1-signaling gradient in cxcr7 deficient embryos enables the relative quantification of the available Sdf1a levels outside the primordium. In the absence of cxcr7 activity, the mean FmemRed/FmemGreen in the primordium should correspond to the unaltered levels of Sdf1a on the stripe (C0), and the mean FmemRed/FmemGreen in sdf1a mutant embryos should correspond to the absence of Sdf1a. Thus, the combined activities of Cxcr7a and Cxcr7b reduce Sdf1a beneath the rear of the primordium to 0.14×C0, while the front of the primordium perceives C0. In summary, these observations demonstrate that Cxcr7a and Cxcr7b continuously sequester Sdf1a protein in the rear of the primordium. This results in an 86% reduction in the local concentration of Sdf1a in the rear of the primordium, which in turn generates the difference in chemical potential required for the formation of a linear gradient of the attractant along the migration route, which is essential for proper primordium migration.
Figure 4. Cxcr7 generates the Sdf1-signaling gradient across the primordium.
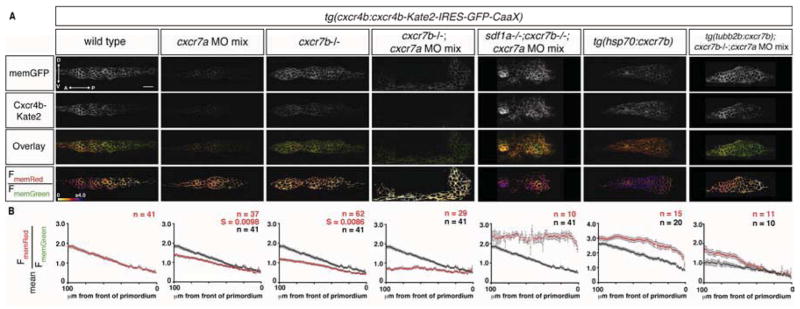
(A) Single confocal slices through the primordium in live 36 hpf embryos of the indicated genotypes, all carrying the Sdf1-signaling sensor. The FmemRed/FmemGreen images are inverted heat maps of the ratio. (B) Mean FmemRed/FmemGreen along the anterior-posterior axis of n≥10 primordia with 0 μm representing the front of each primordium. Red circles indicate the mean FmemRed/FmemGreen in embryos of the indicated genotype; black circles, where present, indicate the mean FmemRed/FmemGreen of wild-type embryos, heat-shocked control embryos or cxcr7 deficient control embryos. Grey bars indicate SEM. Anterior is to the left and dorsal is up in A. Scale bar is 20 μm. Note that cxcr7 deficient; tg(tubb2b:cxcr7b) embryos were injected with a low dose of cxcr7a MO mix. (See also Figures S2, S4 and S6 and supplemental discussion)
Cxcr7 shapes the Sdf1-signaling gradient on the tissue level
Cxcr7 could sculpt the chemokine gradient across the primordium through local competition with Cxcr4b for Sdf1a protein on the cell membranes of individual cells or through global chemokine clearance in the rear of the primordium. To distinguish between these possibilities, we used cell transplantation to generate chimeric primordia composed of wild-type and cxcr7 deficient cells and compared the FmemRed/FmemGreen ratios within and outside the clones. Placement of cxcr7 deficient cells in the rear of a wild-type primordium does not result in increased internalization of Cxcr4b-Kate2 selectively in the cxcr7 deficient clones when compared to adjacent wild-type cells or control chimeras (Figure 5A and 5B). This indicates that - although Cxcr7a and Cxcr7b clear Sdf1a protein locally - the reduction of Sdf1-signaling in the rear of the primordium is generated at the level of the tissue rather than the individual cell.
Figure 5. Cxcr7 modifies the Sdf1-signaling gradient across the primordium at the tissue-level.
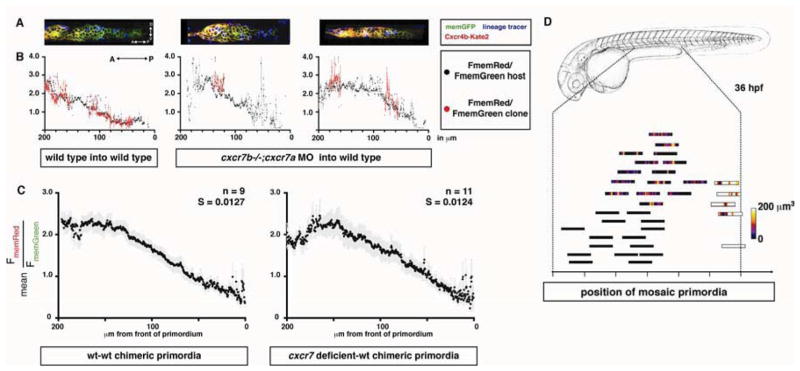
(A) Single confocal slices through mosaic primordia in live 36 hpf embryos of the indicated genotypes. (B) Quantification of mean FmemRed/FmemGreen of host cells (black dots, grey bars SEM) and donor cells (red dots, light red bars SEM) across the anterior-posterior axis of primordia shown in A. (C) FmemRed/FmemGreen ratio on the host cells only across wild-type-wild-type and cxcr7 deficient-wild-type chimeric primordia containing the Sdf1-signaling sensor. The front of the primordium is at 0 μm. The grey bars indicate SEM. (D) Position of mosaic primordia compared to cxcr7b mutant (black rectangles) and wild-type primordia (white rectangle). The amount (heat map in μm3) and position of clonal tissue across 150 μm from the front of the schematized primordia is indicated. (See also Table S1)
Supplying Cxcr7 underneath the rear of the primordium restores the signaling gradient and primordium migration in cxcr7 deficient embryos
If Cxcr7’s sole function is to clear Sdf1a protein from underneath the rear of the primordium, then resupplying Cxcr7 specifically underneath the rear of cxcr7 deficient primordia should restore Sdf1-signaling gradient formation and primordium migration. To test this idea we ectopically expressed cxcr7b in the posterior lateral line nerve - a nerve whose axons closely track the rear of the migrating primordium through GDNF signaling (Schuster et al., 2010) - in cxcr7 deficient embryos. In such embryos, the magnitude of the Sdf1-signaling gradient across the primordium is restored to 85% of the wild-type levels (Figure 4B, column 7), and the primordium migrates on average halfway down the trunk and tail (Figure 1D and S6C). This is in contrast to cxcr7 deficient control embryos in which the Sdf1-signaling gradient is shallower (Figure 4, column 4) and primordium migration is almost completely impaired (Figure 1D and S6C). These observations indicate that Cxcr7 is not necessarily required within the primordium itself and is sufficient for Sdf1-signaling gradient generation and primordium migration when supplied beneath the rear of the primordium.
A steady-state Sdf1-signaling gradient guides the migrating primordium
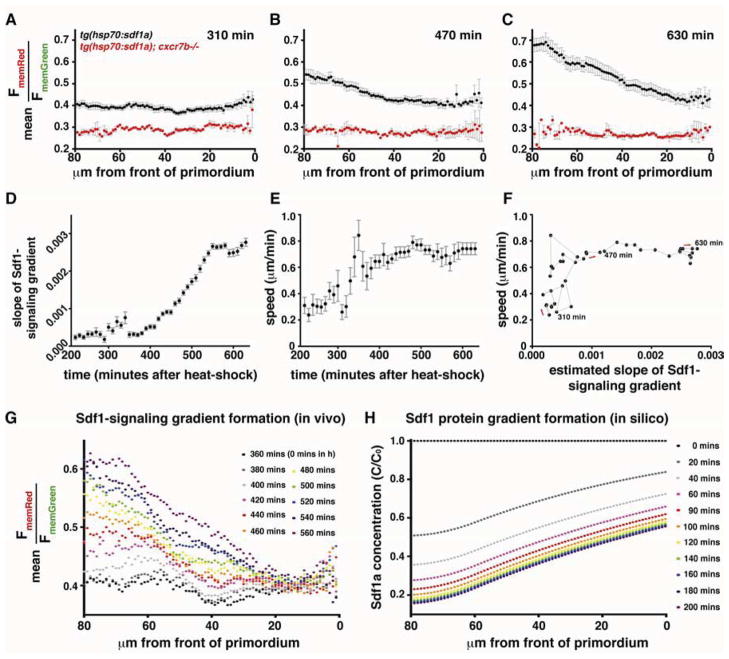
Given sufficient time, the shape and amplitude of signaling molecule gradients will reach steady-state in many scenarios (Muller et al., 2013; Wartlick et al., 2009). However, signaling processes often occur within a few hours, and it is unclear if gradients can reach steady-state within such short time frames, and in turn, if cells interpret pre-steady-state or steady-state signaling gradients in vivo. To address these questions, we analyzed the formation of the Sdf1-signaling gradient over time. A brief heat-shock-induced pulse of global Sdf1a protein expression causes internalization of Cxcr4b-Kate2 throughout the primordium, flattens the Sdf1-signaling gradient and decelerates the primordium (Movie S6, top). The gradient begins to recover ~5–6 hours after the heat-shock and converges to the linear shape that is observed across wild-type primordia (Figure 6A–6C, Figure S7 and Movie S6, top, and Movie S7). Concurrent with recovery of the gradient, the rounded primordium elongates and resumes normal migration (Movie S6, top). Analysis of this recovery reveals a sequence of three distinct states of Sdf1-signaling across the primordium that result in reestablishment of the signaling gradient: At ~5 hours post heat-shock, the Sdf1-signaling gradient across the primordium is absent (Figure 6A, Figure S7A and Movie S7). At ~7–8 hours post heat-shock, Sdf1-signaling is reduced specifically in the rear of the primordium, resulting in a non-linear, sigmoidal Sdf1-signaling gradient (Figure 6B, Figure S7B and Movie S7). Over the next ~2 hours, this sigmoidal gradient equilibrates across the primordium to yield a steeper, linear gradient that resembles the gradient observed in wild-type primordia and remains relatively stable until the end of the imaging period (Figure 6C and 6G, Figure S7C and Movie S7), indicating that it has reached a steady-state. Importantly, the time required for reestablishment of the Sdf1-signaling gradient across the primordium depends on cxcr7 activity. In cxcr7b mutant embryos, a genetic scenario in which the slope of the Sdf1-signaling gradient across the primordium is already reduced by 40% even before heat-shock (Figure 4A and B, column 3), the gradient remains flat ~10 hours following a similar pulse of global Sdf1a protein expression (Figure 6A–6C, Movie S6 bottom and Movie S7).
Figure 6. Kinetics of Sdf1-signaling gradient formation.
(A–C) Mean FmemRed/FmemGreen along the anterior-posterior axis in tg(hsp70:sdf1a) (black dots, n=8) and tg(hsp70:sdf1a);cxcr7b−/− (red dots, n=2) at the indicated number of minutes after induction of a pulse of global Sdf1a expression. The front of the primordium is at 0 μm. Grey bars indicate SEM. (D–F) Relationships of the slope of the gradient, speed of the primordium, and time (n = 8). In D and E, solid black circles indicate mean, grey bars indicate SEM. In F, the grey line connects the data points in chronological order, as indicated by red arrows. (G, H) Sdf1-signaling gradient formation in vivo (G) and Sdf1a protein gradient formation predicted in silico (H). 0 mins (post heat-shock) in H roughly corresponds to 360 min in G. (See also Figure S7)
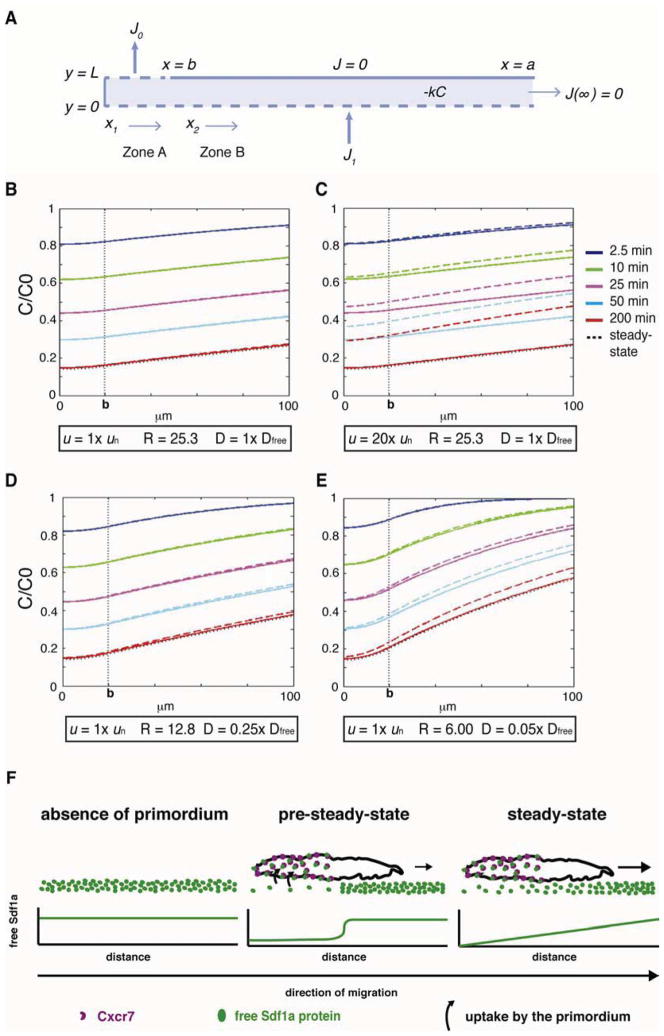
Mathematical modeling of gradient formation by a moving sink
The local sequestration of Sdf1a protein in the rear of the migrating primordium bears a superficial resemblance to the source-sink model described by Crick (1970). Crick showed that a freely diffusing molecule produced by a localized source and degraded by a localized sink should result in a linear gradient at steady-state. Consistent with this prediction, the Sdf1-signaling gradient across wild-type primordia appears linear (Figure 3I, column 1–3) and there is a sink in the rear of the primordium that locally degrades Sdf1a. In contrast to Crick’s localized source model, however, the stripe of sdf1a-expressing cells generates a spatially distributed reservoir of Sdf1a along the migration route. To test whether such a distributed source might provide similar results to Crick’s model, we modeled these dynamics under two assumptions. First, the flux of Sdf1a from a distributed source of chemokine-producing cells is balanced by its degradation to yield a constant reservoir of Sdf1a. Second, the rear of the primordium clears Sdf1a at a constant flow rate (Figure 7A). Initially we used a value of 100 μm2 s−1 for the diffusion coefficient of Sdf1 (i.e. the free diffusion coefficient, see Supplemental Data). Consistent with our analysis of the Sdf1-signaling gradient kinetics and the estimated Peclet number of 0.012 (a measure for whether a system is dominated by diffusion or flow) this model predicts that the primordium migration velocity of ~0.7 μm min−1 does not contribute significantly to the formation of the gradient (Figure 7B and 7C). Moreover, this model shows that a stable, linear, gradient can form in 0.5 to 3 hours and is only slightly perturbed by the motion of the primordium (Figure 7B–7E). However, the model predicts a shallower signaling gradient across the primordium (Figure 7B) than what we observe in vivo (Figure 3I, column 1–3), perhaps reflecting hindered Sdf1a diffusion mediated by molecules in the extracellular matrix that can bind the chemokine, a scenario that is known to reduce the effective diffusion coefficient (Crank 1975, Chapter 14). Three of our observations are consistent with this idea. First, Sdf1a protein is produced by the stripe of chemokine expressing cells throughout the 20-hour migration period but does not diffuse to detectable levels into adjacent tissues (Movie S2), suggesting that Sdf1a protein is retained close to its source. Second, only 1% of the total Sdf1a-GFP protein on the stripe is sequestered through Cxcr7 in the rear of the primordium (Figure 2F). This suggests that a large fraction of the chemokine is bound to the extracellular matrix, a proposition that has also been put forward for other secreted signaling molecules (Muller et al., 2013). Importantly, prolonged global over-expression of Cxcr7b results in removal of 29% of the total Sdf1a-GFP protein from the stripe (Figure 2B), indicating that a larger fraction of Sdf1a protein than the 1% sequestered by the primordium is present but not accessible to Cxcr7 in the rear of the primordium. Third, the kinetics of Sdf1 gradient formation are approximated by the model if the free diffusion coefficient of Sdf1 is reduced by a factor of between 4 and 20 (Figure 7D and 7E). Note that even when the diffusion coefficient is reduced by 20, the Peclet number is still 0.24. In summary, this modeling analysis supports the plausibility of a scenario in which a localized Cxcr7-mediated sink activity combined with hindered Sdf1a protein diffusion (i.e. reduced diffusion coefficient) from a distributed source and degradation mechanism generates a quasi-linear and stable Sdf1-signaling gradient across the primordium (Supplemental Data).
Figure 7. Model for the evolution of chemokine gradient with different values of velocity (u) and effective diffusion coefficient (D).
(A) Model. L = thickness of chemokine reservoir, k = Sdf1 degradation rate in reservoir, J0 flux into the region of primordium absorbing chemokine (b>x> 0, y = L with b = 20 μm), J1 flux of Sdf1 from producing cells (x> 0, y = 0), C concentration of chemokine, C0 initial value of C at time t = 0. The gradient is sensed over the surface of a>x>b, y = L where a is 100 μm and there is no flux over this region. Two zones are defined: Zone A represents b>x1> 0 and Zone B represents a>x2>b. In panels (B–E) the colored solid lines correspond to the gradients calculated with velocity of primordium u = 0 and the colored dashed lines are calculated with the specified u. Each gradient is calculated at the indicated time, assuming that C/C0 = 1 at t = 0. The dotted black line indicates steady-state. k is fixed at 0.0003 s−1. R = J0/J1 is the ratio of flux values chosen so that the steady-state baseline value of C under the sink (x = 0) corresponds to the measured value of 0.14 C0. (B–E) Simulations with different u and D expressed as multiples of measured un (0.7 μm min−1) and Dfree (100 μm2 s−1, Veldkamp 2005). (F) Model for chemokine-signaling gradient by the primordium. In the pre-steady-state, sequestration of Sdf1a protein by Cxcr7 decreases Sdf1a protein beneath the trailing half of the primordium resulting in reduced chemokine signaling in the rear. Diffusion from areas of higher Sdf1a protein concentration equilibrates the chemokine distribution across the primordium, resulting in a linear, stable signaling gradient.
Steepness of Sdf1-signaling gradient correlates with efficient primordium migration
Theoretical considerations and in vitro experiments have suggested that increasing the steepness of an attractant gradient can promote directionality and motility (Fisher et al., 1989; Hatzikirou and Deutsch, 2008; Keller and Segel, 1971; Parent, 1999). To test this model in vivo, we followed the recovery of the Sdf1-signaling gradient and primordium migration speed following exposure to a global pulse of Sdf1a. By comparing the average slope of the signaling gradient and the average speed of the primordium (Figure 6D–6F), we found that when the slope of the Sdf1-signaling gradient is at or above ~46% of its steady-state value (470 min in Figure 6F), the speed of the primordium stabilizes at ~0.7 μm/min (Figure 6F), which is similar to the speed observed in wild-type primordia (Haas and Gilmour, 2006). During this recovery period, the Sdf1-signaling gradient increases fairly linearly (Figure 6D) until it stabilizes at steady-state. When the gradient is less than ~46% of the steady-state value, however, both the speed and directionality of the primordium are unpredictable (Figure 6F), with primordia either stalling or exhibiting serpentine movement rather than straight migration. Importantly, shifting the gradient to higher ratios by low dose Sdf1a morpholino injections without changing its slope (Figure 3C left) does not affect primordium migration (Figure 3C right). These observations are consistent with the idea that it is the steepness of an attractant gradient rather than the absolute amount of signaling that instructs both speed and directionality of migration in vivo.
Discussion
Guidance of migrating cells by shallow attractant gradients
In vitro studies using Dictyostelium and neutrophils have shown that individually migrating cells require at least a 3% difference in concentration between the front and the back of the cell for efficient directional migration (Fisher et al., 1989; Mato et al., 1975). This is similar to the 7% difference in Sdf1-signaling observed across the front to the back of a cell in the lateral line primordium, suggesting that shallow gradients are sufficient for efficient directional migration both in vitro and in vivo. Since most scenarios involving a local attractant source yield non-linear gradients whose slope is shallow far from the source and steeper closer to the source (Wartlick et al., 2009), the ability of cells to detect small differences in attractant concentration is essential for migration towards the attractant source from a distance.
Collectively migrating cells can potentially compare differences in attractant concentration sensed by cells at the front and at the rear of the collective to polarize the tissue towards higher attractant concentrations (Rørth, 2007). The induction of polarity across collectively migrating border cells in flies by local activation of Rac (Wang et al., 2010) and the promotion of migration in primordia with a few wild type cells in an otherwise cxcr4b mutant primordium (Haas and Gilmour, 2006) support this idea. However, reducing the difference in Sdf1-signaling across a cell within the primordium to 3% results in inefficient migration and stalling of the primordium, even though there still exists a 40% difference in Sdf1-signaling from the front to back across the primordium in cxcr7b mutant embryos. These observations suggest that either a 3% difference in signaling across cells might be too low to induce polarity across the primordium. Alternatively, the primordium might not compare concentrations of the attractant across the collective to enhance its ability to detect attractant gradients.
Kinetics and dynamics of signaling gradients
Signaling molecules disperse away from their source through a complex environment to pattern a field of cells or to provide guidance to migrating cells. The signaling range depends on the time the signaling molecules have to disperse and the ability of the signaling molecules to move through the tissue (Müller and Schier, 2011). For many scenarios with constant production, diffusion and clearance rates, the distribution of signaling molecules will converge towards a stable gradient (constant amplitude and shape) over time (Muller et al., 2013; Wartlick et al., 2009). Measurements of the total pool of fluorescently tagged and overexpressed signaling molecules indicate that it takes 30 minutes (in the case of nodal (Muller et al., 2012)) to 3 to 4 hours (in the cases of FGF (Yu et al., 2009) and dpp (Entchev et al., 2000; Teleman and Cohen, 2000)) for the signaling gradient to reach steady state. This is similar to the time it takes for the signaling gradient of untagged, endogenous Sdf1 to converge towards steady state, given that the distribution of the pool of total signaling molecules and the pool of actively signaling molecules do not necessarily need to display similar gradient kinetics.
The movement of signaling molecules through tissues is impeded by obstacles that increase the path length of the moving molecule and by transient binding to the extracellular matrix. This reduces the global diffusivity of the signaling molecule and increases the time it takes for the gradient to converge towards steady state (Muller et al., 2013; Crank, 1975). The FGF gradient in the early zebrafish embryo, for example, approaches steady state over a period of 3 to 4 hours instead of less than an hour as predicted for freely diffusing FGF, suggesting that the movement of FGF is hindered by transient binding to extracellular molecules, such as proteoglycans (Duchesne et al., 2012; Muller et al., 2013). Similarly, the shape of the gradient of Sdf1 suggests that the chemokine is hindered in its diffusivity, a supposition supported by the observation that only a small fraction of the total Sdf1a protein pool actively participates in signaling. Although this might depend on the signaling molecule and the tissue context, these observations are consistent with the idea that a large fraction of the signaling molecules are bound to extracellular molecules at any given time and only a small fraction is locally available to engage in signaling.
Self-generated attractant gradients
The primordium is born as a polarized tissue with a rosette in its rear that will be deposited later as the first neuromast (Nechiporuk and Raible, 2008). Shortly afterwards, the rear of the primordium starts expressing cxcr7b (Breau et al., 2012; Dambly-Chaudière et al., 2007). Although the molecular mechanism leading to the expression of cxcr7b in the rear is unknown, this restricted expression polarizes the primordium molecularly. Three conflicting models have been proposed about how Cxcr7b activity in the rear could provide directionality to the migrating primordium. In the first model, Cxcr7b activity is thought to repress the expression of cxcr4b in the rear and Cxcr4b activity in the front is thought to repress cxcr7b expression in the front of the primordium. This cross-repression would cause Cxcr4b activity to become graded across the primordium, with more Cxcr4b available for signaling in the front than in the back (Dambly-Chaudière et al., 2007). In the second model, Cxcr7b is thought not to regulate cxcr4b expression but instead to elicit a response that is distinct from Cxcr4b upon Sdf1a activation. This response is thought to endow cells in the rear of the primordium with a different migratory behavior than cells in the front (Valentin et al., 2007). In the third model, Cxcr7b activity was proposed to internalize Sdf1 protein in the rear of the primordium, which in turn was postulated to generate a gradient of Sdf1 protein across the primordium, which the primordium would follow (Raz and Mahabaleshwar, 2009; Weijer, 2009). Our expression analysis of the transgenic reporters are consistent with the second model in which Cxcr4b and Cxcr7b do not regulate each other’s expression (Figure 1B). However, the findings that Cxcr7 expression in the rear primordium or near the rear of the primordium both similarly lead to the formation of a Sdf1-signaling gradient and primordium migration (Figure 1D, 4B, 5D and S6C) indicate that Cxcr7 acts not as a signaling receptor but rather as a chemokine clearance receptor during primordium migration. This is consistent with the third model, in which Cxcr7 acts as a sink for Sdf1 in the rear of the primordium, generates an Sdf1-signaling gradient across the primordium and propels its migration along a uniform source of attractant (Figure 7F) but inconsistent with a role of Cxcr7 in signaling in the rear of the primordium as postulated in the second model.
It is conceivable that ligand sequestration by a subset of cells in a migrating cluster represents a more general mechanism of generating, maintaining or enhancing a gradient of an attractant – or any signaling molecule – in order to provide directional and/or positional information to cells and tissues. Key to this mechanism is the ability to change the availability of an extracellular signaling molecule, which depends on the signaling molecule’s concentration, the number of sink cells and the rate of ligand sequestration. Many other tissues such as sprouting blood vessels, epithelia and metastasizing tumors exhibit collective migration (Friedl and Gilmour, 2009; Montell, 2008; Rørth, 2009). Thus, a migrating collective modulating the availability of its own guidance cue may represent an elegant mechanism of cell guidance.
Experimental Procedures
Sdf1a-GFP and Sdf1-signaling sensor transgenics
The Sdf1-signaling sensor and the Sdf1a-GFP fusion constructs were generated using recombineering of a bacterial artificial chromosome (BAC) spanning the cxcr4b and sdf1a genomic loci, respectively, and transgenic zebrafish were obtained by co-injecting BAC DNA and tol2 transposase mRNA into one-cell-stage embryos. See also Extended Experimental Procedures.
Immunohistochemistry, embryonic manipulations and transgenesis
In situ hybridization was conducted as previously described (Thisse and Thisse, 2008). Antibody stainings were detected chromogenically with DAB or fluorescently with Cy3, Alexa488 or Alexa 647 conjugated secondary antibodies. Mosaic embryos containing either the Sdf1-signaling sensor or tg(hsp70:sdf1a) and tg(cldnB:lyn2GFP) were generated through cell transplantation. See also Extended Experimental Procedures.
Microscopy and image processing
For live imaging, the primordium was imaged using a Leica SP5 II confocal laser scanning microscope. Quantification of Sdf1a-GFP on the stripe and FmemRed/FmemGreen ratios for each voxel representing part of a cell membrane in the primordium were calculated using ImageJ software. See also Extended Experimental Procedures.
Supplementary Material
Highlights.
Development of a quantitative Sdf1-signaling sensor for live imaging
Only a small fraction of the total Sdf1 protein pool is free to signal through Cxcr4
Collectively migrating cells follow a linear and stable Sdf1-signaling gradient
The Sdf1-signaling gradient is self-generated by the migrating cells
Acknowledgments
We thank J. Torres-Vazquez, F. Schnorrer, J. Nance, R. Lehmann, R. Cinalli, J. Hubbard and J. Treisman for critical comments; D. Gilmour, P. Krieg and A.J. Hudspeth for reagents; F. Fuentes for excellent fish care; K. Cadwell and G. David for cell lines; M. Stadtfeld for recombinant LIF; and A. Sfeir and M. Tahiliani for advice and reagents for tissue culture experiments. For providing the zebrafish knockout allele cxcr7bsa16 we thank the Sanger Institute Zebrafish Mutation Resource, sponsored by the Wellcome Trust [grant number WT 077047/Z/05/Z]. This work was supported by NIH grants NS069839 (H.K.), NS28642 (C.N.) and HD007520 (S.W.L.) and an American Heart Association fellowship 12POST12060278 (G.V.).
Footnotes
Supplemental Data includes seven Supplemental Figures and legends, seven Supplemental Movie legends, one Supplemental Table, Extended Experimental Procedures and Supplemental Equations and Discussion.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affolter M, Caussinus E. Tracheal branching morphogenesis in Drosophila: new insights into cell behaviour and organ architecture. Development. 2008;135:2055–2064. doi: 10.1242/dev.014498. [DOI] [PubMed] [Google Scholar]
- Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of Chemokine-Guided Cell Migration by Ligand Sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Breau MA, Wilson D, Wilkinson DG, Xu Q. Chemokine and Fgf signalling act as opposing guidance cues in formation of the lateral line primordium. Development. 2012;139:2246–2253. doi: 10.1242/dev.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Devreotes PN. Moving in the right direction: How eukaryotic cells migrate along chemical gradients. Semin Cell Dev Biol. 2011:1–8. doi: 10.1016/j.semcdb.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Diffusion in embryogenesis. Nature. 1970;225:420–422. doi: 10.1038/225420a0. [DOI] [PubMed] [Google Scholar]
- Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. Embo J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambly-Chaudière C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David NB, Sapède D, Saint-Etienne L, Thisse C, Thisse B, Dambly-Chaudière C, Rosa FM, Ghysen A. Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc Natl Acad Sci USA. 2002;99:16297–16302. doi: 10.1073/pnas.252339399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne L, Octeau V, Bearon RN, Beckett A, Prior IA, Lounis B, Fernig DG. Transport of Fibroblast Growth Factor 2 in the Pericellular Matrix Is Controlled by the Spatial Distribution of Its Binding Sites in Heparan Sulfate. PLoS Biol. 2012;10:e1001361. doi: 10.1371/journal.pbio.1001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev EV, Schwabedissen A, Gonzalez-Gaitan M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–991. doi: 10.1016/s0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- Fisher PR, Merkl R, Gerisch G. Quantitative analysis of cell motility and chemotaxis in Dictyostelium discoideum by using an image processing system and a novel chemotaxis chamber providing stationary chemical gradients. The Journal of Cell Biology. 1989;108:973–984. doi: 10.1083/jcb.108.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudière C. The lateral line microcosmos. Genes Dev. 2007;21:2118–2130. doi: 10.1101/gad.1568407. [DOI] [PubMed] [Google Scholar]
- Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell. 2006;10:673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Hatzikirou H, Deutsch A. Cellular automata as microscopic models of cell migration in heterogeneous environments. Curr Top Dev Biol. 2008;81:401–434. doi: 10.1016/S0070-2153(07)81014-3. [DOI] [PubMed] [Google Scholar]
- Keller EF, Segel LA. Model for chemotaxis. Journal of Theoretical Biology. 1971;30:225–234. doi: 10.1016/0022-5193(71)90050-6. [DOI] [PubMed] [Google Scholar]
- Kicheva A, Pantazis P, Bollenbach T, Kalaidzidis Y, Bittig T, Jülicher F, González-Gaitán M. Kinetics of morphogen gradient formation. Science. 2007;315:521–525. doi: 10.1126/science.1135774. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Li Q, Shirabe K, Kuwada J. Chemokine signaling regulates sensory cell migration in zebrafish. Dev Biol. 2004;269:123–136. doi: 10.1016/j.ydbio.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Marchese A, Benovic JL. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J Biol Chem. 2001;276:45509–45512. doi: 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003;5:709–722. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- Mato JM, Losada A, Nanjundiah V, Konijn TM. Signal input for a chemotactic response in the cellular slime mold Dictyostelium discoideum. Proc Natl Acad Sci USA. 1975;72:4991–4993. doi: 10.1073/pnas.72.12.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina S, Reichman-Fried M, Raz E. Control of receptor internalization, signaling level, and precise arrival at the target in guided cell migration. Curr Biol. 2007;17:1164–1172. doi: 10.1016/j.cub.2007.05.073. [DOI] [PubMed] [Google Scholar]
- Montell DJ. Morphogenetic Cell Movements: Diversity from Modular Mechanical Properties. Science. 2008;322:1502–1505. doi: 10.1126/science.1164073. [DOI] [PubMed] [Google Scholar]
- Montell DJ. Border-cell migration: the race is on. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- Muller P, Rogers KW, Jordan BM, Lee JS, Robson D, Ramanathan S, Schier AF. Differential Diffusivity of Nodal and Lefty Underlies a Reaction-Diffusion Patterning System. Science. 2012 doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Rogers KW, Yu SR, Brand M, Schier AF. Morphogen transport. Development. 2013;140:1621–1638. doi: 10.1242/dev.083519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Schier AF. Extracellular movement of signaling molecules. Dev Cell. 2011;21:145–158. doi: 10.1016/j.devcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Raible DW. FGF-dependent mechanosensory organ patterning in zebrafish. Science. 2008;320:1774–1777. doi: 10.1126/science.1156547. [DOI] [PubMed] [Google Scholar]
- Parent CA. A Cell’s Sense of Direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP, Gerard C, Lefkowitz RJ. -arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proceedings of the National Academy of Sciences. 2009:1–5. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: a fisheye view. Development. 2009;136:1223–1229. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- Rørth P. Collective guidance of collective cell migration. Trends in Cell Biology. 2007;17:575–579. doi: 10.1016/j.tcb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Rørth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- Rørth P. Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev Cell. 2011;20:9–18. doi: 10.1016/j.devcel.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Sánchez-Alcañiz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, López-Bendito G, Stumm R, Marín O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Schuster K, Dambly-Chaudiere C, Ghysen A. Glial cell line-derived neurotrophic factor defines the path of developing and regenerating axons in the lateral line system of zebrafish. Proceedings of the National Academy of Sciences. 2010:1–6. doi: 10.1073/pnas.1002171107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Huang C-H, Devreotes PN. Eukaryotic Chemotaxis: A Network of Signaling Pathways Controls Motility, Directional Sensing, and Polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Cohen SM. Dpp gradient formation in the Drosophila wing imaginal disc. Cell. 2000;103:971–980. doi: 10.1016/s0092-8674(00)00199-9. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17:1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartlick O, Kicheva A, Gonzalez-Gaitan M. Morphogen Gradient Formation. Cold Spring Harbor Perspectives in Biology. 2009;1:a001255–a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hauschild R, Schwarz J, Moussion C, de Vries I, Legler DF, Luther SA, Bollenbach T, Sixt M. Interstitial Dendritic Cell Guidance by Haptotactic Chemokine Gradients. Science. 2013;339:328–332. doi: 10.1126/science.1228456. [DOI] [PubMed] [Google Scholar]
- Weijer CJ. Collective cell migration in development. J Cell Sci. 2009;122:3215–3223. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- Yu SR, Burkhardt M, Nowak M, Ries J, Petrášek Z, Scholpp S, Schwille P, Brand M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009:1–5. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]
- Crank . Mathematics of diffusion. Oxford University Press; 1975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.