Abstract
High metabolic activity and low levels of antioxidant enzymes make neurons particularly prone to damage by reactive oxygen species. Thus, repair of oxidative DNA damage is essential for normal brain function. Base excision repair is the major pathway for repair of oxidative DNA damage, and is initiated by DNA glycosylases recognizing and removing the damaged base. In mammalian cells at least five different DNA glycosylases with overlapping substrate specificity, NEIL1, NEIL2, NEIL3, OGG1 and NTH1, remove oxidative DNA base lesions. Here we report mRNA expression and distribution of these five DNA glycosylases in human and rodent brains using in situ hybridization and Northern blotting supported by glycosylase activity assays. NEIL1, NEIL2, OGG1 and NTH1 showed widespread expression at all ages. In situ hybridization studies in mouse brain showed that expression of mNeil1 increased with age. In newborn mouse brain, mNeil3 revealed a discrete expression pattern in brain regions known to harbour stem cell populations, i.e., the subventricular zone, the rostral migratory stream, and the hilar region of the hippocampal formation. Expression of mNeil3 decreased with age, and in old mice brains could be detected only in layer V of neocortex. MNth1 was constitutively expressed during lifespan. In Northern blots, mOgg1 expression showed a transient decrease followed by an increase after 8 weeks of age. Assays for faPy DNA glycosylase activity revealed increased activity level with age in all brain regions analyzed.
The widespread but differential expression of the DNA glycosylases recognizing oxidative base lesions suggests distinct and age dependent roles of these enzymes in genome maintenance in brain. The distribution of mNeil3 is particularly intriguing and points to a specific role of this enzyme in stem cell differentiation.
Keywords: Base excision repair, In situ hybridization, Mice, Human, Aging, BER
1. Introduction
A common feature of neurodegenerative diseases, as well as aging, is elevated levels of DNA damage [1,2]. A major cause of DNA damage is oxidative stress, which contributes to cell death in stroke and chronic conditions such as Alzheimer's and Parkinson's diseases [2–6]. Neurons are continuously challenged by reactive oxygen species (ROS), which may damage nucleic acids and induce cell death. Thus, effective neuronal repair of oxidative damage is critical to genome maintenance and brain function [2].
Base excision repair (BER) is the main pathway for repair of oxidative DNA lesions. BER is initiated by damage-specific DNA glycosylases, which remove the modified base, leaving an abasic site which is acted upon by an intrinsic AP lyase activity (bifunctional glycosylase) or by an AP endonuclease. In the resulting strand break, lyases and/or nucleases remove the sugar-phosphate backbone, and DNA polymerases and ligases fill and reseal the gap [7]. Five mammalian DNA glycosylases excising oxidized DNA bases have been described: NEIL1, NEIL2, NEIL3, OGG1 and NTH1.
HNEIL1 removes a number of oxidative lesions including 8-oxoguanine (8-oxoG), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (faPy) and thymine glycol [8–10]. Embryonic mouse stem cells deficient in mNeil1 are sensitive to low levels of γ-radiation, indicating that mNeil1 is important for the cellular defence against DNA damage [11].
HNEIL2 mainly cleaves oxidized pyrimidine substrates from double stranded DNA, also in bubble-structured DNA [12]. MNeil1 and mNeil2 revealed unique specificity towards spiroiminodihydantoin and guanidinohydantoin lesions, which are more mutagenic, oxidized products of 8-oxoG [13].
HNEIL3 possesses a weak DNA glycosylase activity against faPy in crude insect cell extracts [8]. Moreover, mice deficient in mNeil3 exhibit no apparent phenotype [14].
HOGG1 primarily removes oxidized purines as 8-oxoG and faPyG [15–19], whereas hNTH1 excises oxidized pyrimidines, including thymine glycol, 5-hydroxycytosine, dihydrothymine and dihydrouracil [20,21]. The mild phenotypes associated with targeted disruption of the mouse mOgg1 and mNth1 genes indicate existence of backup enzymes for the repair of these lesions, such as the NEIL enzymes.
There is scant knowledge about oxidative DNA repair in neurons. Several of the BER enzymes have been shown to be expressed in the brain, but distribution of DNA glycosylases in brain has been published only for Ogg1 [22].
We have made extensive tests of commercial and custom-made antibodies to DNA glycosylases and none of these antibodies show sufficient selectivity. Notably, antibodies to OGG1 produce staining even in animals with targeted disruption of this gene. Pending antibodies with adequate selectivity and affinity, localization studies of DNA glycosylases cannot be performed by immunocytochemical approaches. Here we have used Northern analysis and in situ hybridization to describe the distribution of mRNAs of the five different DNA glycosylases, NEIL1, NEIL2, NEIL3, OGG1 and NTH1, in mammalian brains. Finally, we examined the transcript distribution and faPy DNA glycosylase activity during development and aging.
2. Methods
2.1. Northern blot hybridization
Northern blots containing multiple adult human brain tissue samples purchased from Clontech (catalog numbers 636802 and 636838) were probed with full-length DNA probes of hNEIL1, hNEIL2, hNEIL3, hOGG1 and hNTH1 expression. Mouse brain aging blot (MBAB 1009-1) was purchased from Seegene Inc. (Soul, Korea), and probed with full-length mouse Neil1 and Neil3. Ogg1 probe containing exon 4–7, Nth1 probe including exon 4–6 and Neil2 probe of exon 2 was utilized. Northern blot hybridization was carried out using ExpressHyb solution (Clontech) as recommended by the manufacturer. Probes were labelled with (α-32P)-dCTP using the Rediprime DNA labelling system (Amersham Corp.).
2.2. Animals
Brains from C57Bl/6J mice were used. Mice were housed and handled in accordance with the European Council Directive 86/609/EEC.
2.3. Tissue preparation
Mice at postnatal day 3 (P3), 1 month or 1 year of age were decapitated and the brains carefully removed. The brains were quickly frozen on dry ice and stored at –80 °C. Horizontal or sagittal sections were cut at 15 μm on a cryostat and mounted onto slides (Super frost). The sections were post-fixed in 4% formaldehyde and stored in 96% ethanol at 4 °C until use.
2.4. Probe synthesis
Full-length mouse Neil1 DNA (IMAGE clone 1399170) was cloned into pT7T3D-Pac vector with restriction enzymes Not1 and EcoRI, and linearized with BspHI. MNeil2 exon2, mNeil3 full-length and mNth1 exon 4–6 were cloned into pT7T3-Pac vector with the restriction enzymes Not1 and EcoRI. MNth1 was linearized with FspHI and mNeil2 and mNeil3 were linearized by BspHI. MOgg1 containing exon 4–7 was cloned into Topo Zero blunt vector. All DNA templates were purified by GFX column (Amersham) before transcribing RNA digoxigenin (DIG) probes by DIG-RNA Labelling kit (Roche). Both antisense and sense probes for each gene was generated (for details see [23]). The RNA probes were tested on agarose gel, and the quality of DIG incorporation was tested by dot blot according to DIG nucleic acid detection kit (Roche).
2.5. In situ hybridization
Horizontal and sagittal sections of mouse brain aged P3, 1 month or 1 year were labelled with full-length DIG-RNA anti-sense probes against mNeil1 or mNeil3. For mOgg1 a probe with exon 4–7 was used, for mNeil2 a probe with exon 2 and for mNth1 a probe with exon 4–6. As controls sense DIG-RNA probes were used.
In situ hybridization was performed according to the protocol by Hoover and Goldman (1992) with minor modifications [24]. In brief, post-fixed brain sections were dehydrated in ethanol (from 100% to 50%), rinsed in 2× SSC (150 mM NaCl and 15 mM sodium citrate, pH 7.4) and permeabilized with 10 μg/μl Proteinase K (Roche) in 0.1 M Tris–HCl (pH 7.5) with 50 mM EDTA for 15 min at 37 °C. Sections were then fixed in 4% formaldehyde and treated with 0.2 M HCl for 10 min. Acetylation was done with 0.1 M triethanolamine (TEA), pH 7.5, for 2 min at room temperature, followed by incubation for 10 min in 0.25% of acetic anhydride. Sections were dehydrated in graded alcohol and air dried before incubation with hybridization solution (10 mM Tris–HCl, pH 7.5, 30–50% formamide, 0.3 M NaCl, 1 mM EDTA, 10% dextran sulfate and 1% blocking solution) with 100 ng DIG-RNA probe. The slides were covered with Parafilm and incubated in a humid chamber at 55 °C over night. Then the slides were rinsed in 2× SSC and washed in 2× SSC with 30% formamide at 55 °C for 30 min. Slides were then rinsed in 2× SSC twice for 10 min. To remove unhybridized RNA, the sections were treated with RNase (50 μg/ml) in 10 mM Tris pH 7.5, 0.5 M NaCl and 1 mM EDTA for 30 min at 37 °C. Afterwards the slides were washed in the same buffer without RNase for 30 min at 60 °C. Unspecific staining was blocked by 1% blocking solution (Roche) with 0.05% Triton X-100 in 2× SSC for 2 h before incubation in 1:3000 anti-DIG-alkaline phosphate (Roche) in maleate buffer with 1% blocking solution and 0.3% Triton X-100 over night at 4 °C. The sections were washed in maleate buffer and incubated in 100 mM Tris–HCl, 100 mM NaCl and 50 mM MgCl for 10 min at room temperature before detection with NBT (Nitro blue tetrazolium chloride) and BCIP (5-bromo-4-chloro-3-indolyl phosphate, toluidine salt) (Roche). The sections were analyzed with an Axioplan-2 microscope from Zeiss and pictures were taken with an Olympus camera.
2.6. Preparation of cell extracts from different brain regions
Hippocampus, cortex, cerebellum and caudatoputamen (CP) were isolated from mouse brain at the age of 3–7 (3 animals), 10–15 (5 animals) and 35–49 (4 animals) weeks. The scalpel-macerated brain tissue was homogenized with a pestle in 100 μl PBS, and the cell suspensions were added 100 μl 84% sucrose/40 mM Tris–HCl, pH 8.0/10 mM EDTA (ethylenediamine tetraacetic acid) for plasmolysis. After 10 min incubation on ice, the lysed cells were added 400 μl 50 mM MOPS (morpholinopropanesulfonic acid) pH 7.5/1 mM EDTA/100 mM KCl/1 mM DTT (dithiothreitol) and the nuclei broken by freeze/thawing procedure (ethanol/dry ice bath and 37 °C bath) repeated three times. The debris was spun down at 13.000 rpm for 15 min. The supernatant was aliquoted and frozen at –70 °C.
2.7. Assay for faPy DNA glycosylase activity
FaPy DNA glycosylase activity was assayed in a reaction buffer containing 70 mM MOPS, pH 7.5, 1 mM EDTA, 5% glycerol and 1 mM DTT, and the mixtures were incubated at 37 °C for 40 min. For analyzing faPy removal N-[3H]methyl-N′-nitrosourea (18 Ci/mmol) was used to prepare poly(dG–dC) DNA containing faPy residues (5000 dpm/μg DNA) [25]. FaPy DNA glycosylase activity was measured in a total volume of 50 μl containing 0.4 μg faPy-DNA substrate.
3. Results
3.1. Expression of DNA glycosylases removing oxidative DNA lesions in human brain
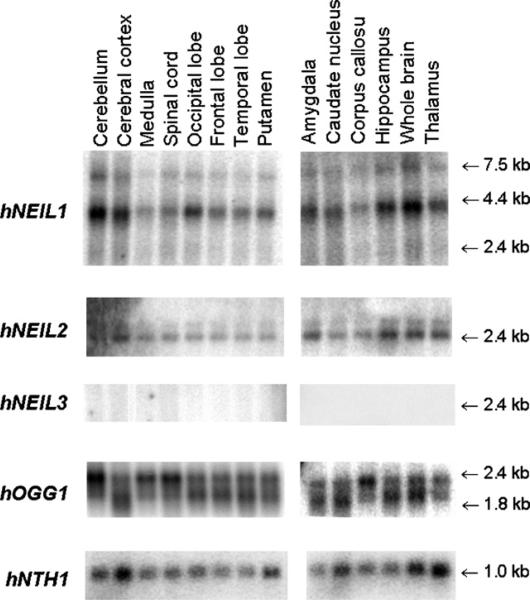
HNEIL1, hNEIL2, hNEIL3, hOGG1 and hNTH1 mRNA expression was investigated in different brain areas of human adults by Northern blot hybridization to full-length cDNA probes (Fig. 1). HOGG1, hNTH1, hNEIL1 and hNEIL2 were expressed in all brain regions analyzed. In contrast, hNEIL3 could not be detected in any brain region of adult humans. HNEIL1 was abundantly expressed in human brain, with highest amounts of transcripts in cerebellum, neocortex and hippocampus. Two strong bands at sizes 4.0 and 7.5 kb were detected with a full-length hNEIL1 probe. The mature mRNA of hNEIL1 is expected to be around 1.8 kb, and such higher bands have previously been observed in addition to the one at 1.8 kb in Northern blots with other human tissues [8,9]. In these studies whole brain was examined and a major band at 4.0 kb was detected in addition to a weak band at 7.5 kb while the 1.8 kb band observed in other tissues was hardly detected. We concluded then that the larger bands were most probably a result of incomplete processing of the transcript. A second possibility would be cross hybridization to an unrelated mRNA transcript. To address this question we reprobed the blots with several smaller probes covering full-length hNEIL1. All probes recognized the 4.0 and 7.5 kb bands (data not shown) indicating that the 4.0 and 7.5 kb transcripts are indeed hNEIL1. HNEIL2 showed high expression in the same three regions as for hNEIL1—cerebellum, neocortex and hippocampus. In addition, transcript level was high in thalamus and amygdala. HOGG1 exhibited highest expression in the cerebellum, brain stem, and spinal cord. Hybridization with the hOGG1 probe revealed two different bands corresponding to the mitochondrial (2.4 kb) and the nuclear transcript (2.0 kb) of the gene [26]. Expression of hNTH1 was highest in cerebral cortex and thalamus among the regions analyzed.
Fig. 1.
Northern blot analysis of hNEIL1, hNEIL2, hNEIL3, hOGG1 and hNTH1 expression in human brain regions. Poly (A)+ mRNA was extracted from different parts of the human brain (4–12 adult individuals) as indicated, and hybridized with full-length cDNA of the five DNA glycosylase genes. All of these genes, except for hNEIL3, were ubiquitously expressed in brain. The absence of hNEIL3 signal in adult brains is consistent with data from mice (Fig. 7), indicating a loss of NEIL3 expression during postnatal development. Duplicate blots were carried out.
3.2. Expression of DNA glycosylases in mouse brain during postnatal development by in situ hybridization
We performed an in situ hybridization analysis to study the expression and distribution pattern of the five DNA glycosylases in mouse brain during postnatal development. As controls, sense riboprobes complementary to the antisense mRNA of the DNA glycosylases were used. No labelling was detected in tissue sections incubated with the respective sense probes (Figs. 2–6, inserts).
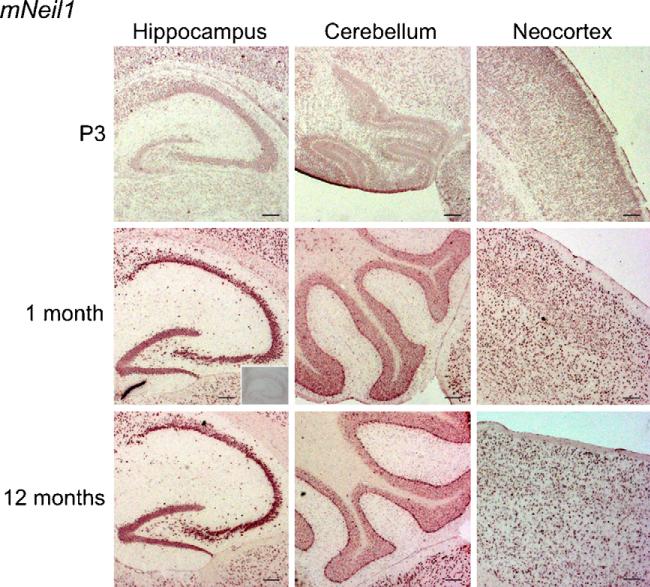
Fig. 2.
In situ hybridization with a DIG-RNA antisense probe to mNeil1 in horizontal brain sections from P3, 1 month and 1-year-old mice. MNeil1 showed strong expression throughout the brain. Particularly intense labelling was found in the cerebellar Purkinje cells, hippocampus, and neocortex. The expression level increased with age. No labelling was seen with the sense probe (insert). Scale bar, 100 μm.
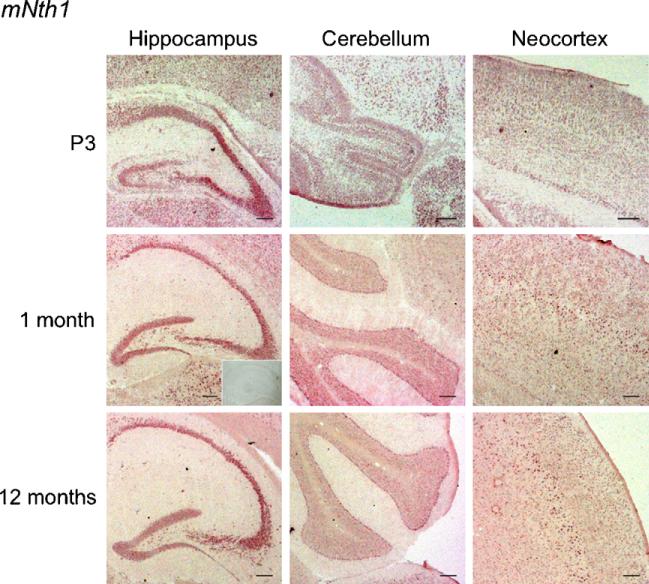
Fig. 6.
In situ hybridization with a DIG-RNA antisense probe to mNth1 in horizontal brain sections from P3, 1-month- and 1-year-old mice. The signal of mNth1 was generally weak, but heterogeneous. High expression was seen in hippocampus and in the cerebellar Purkinje cells. Layer V of neocortex showed a strong signal from 1 month of age. No labelling was detected with the sense probe (insert). Scale bar, 100 μm.
The in situ hybridization analysis indicated that mNeil1 expression increased with age (Fig. 2). However, this was not evident in every experiment. This could be explained by individual variation as the results were reproducible in consecutive experiments using tissue from the same brain. In old animals, transcripts were abundant in most brain regions including CA1, CA3 and dentate gyrus of the hippocampus, the Purkinje cells of cerebellum and neocortex (Fig. 2).
MNeil2 demonstrated a similar expression pattern as mNeil1 in the adult brain, but with an unchanged or slightly increased expression with age (Fig. 3).
Fig. 3.
In situ hybridization with a DIG-RNA antisense probe to mNeil2 in horizontal brain sections from P3, 1 month and 1-year-old mice. The signal detected for mNeil2 was generally weak. However, there was a clear signal in hippocampus and in cerebellar Purkinje cells. In the older animals strong labelling was observed in layer V of neocortex. No labelling was seen with the sense probe (insert). Scale bar, 100 μm.
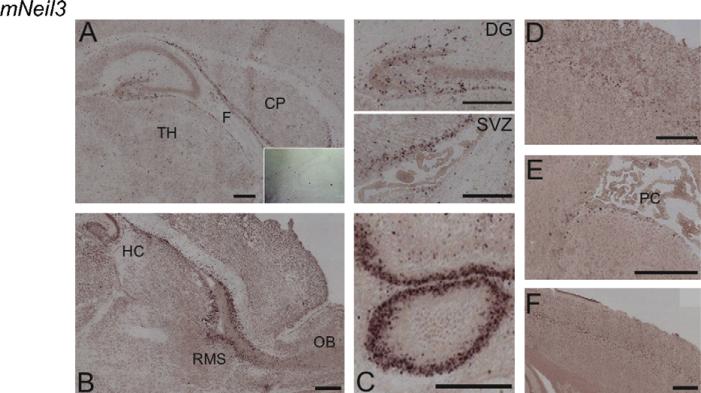
Distribution of mNeil3 differed from the other DNA glycosylases as expression was limited to distinct cell populations. In brains from P3 mice, abundant mNeil3 transcripts were observed in the subventricular zone (SVZ) (Fig. 4A, enlarged), hilus of the hippocampal formation (Fig. 4A, enlarged), the rostral migratory stream (RMS) (Fig. 4B), and the Purkinje cell of the cerebellum (Fig. 4C). The expression level declined with age (compare Fig. 4A–C (P3) with Fig. 4D and E (1 month) and Fig. 4F (1 year)). Thus, in brains from 1-month-old mice mNeil3 was detected in layer V of neocortex (Fig. 4D) and only in a few cells in the SVZ (Fig. 4E), and in brain from 1-year-old mice mNeil3 was observed only in layer V of neocortex (Fig. 4F).
Fig. 4.
In situ hybridization with a DIG-RNA antisense probe to mNeil3 in horizontal and sagittal brain sections from P3, 1-month- and 1-year-old mice. (A) Horizontal section through hippocampus and the subventricular zone (SVZ) of a P3 mouse. Part of the section (DG, dentate gyrus, SVZ, subventricular zone) in A is enlarged in insert. (B) Sagittal section showing the rostral migratory stream (RMS) from olfactory bulb to hippocampus in a P3 mouse. (C) Horizontal section of cerebellum in P3 mouse brain. (D) Horizontal section of 1-month-old mouse brain displaying the layer V labelling in the neocortex. (E) Horizontal section through the SVZ with plexus choroideus (PC) in a 1-month-old mouse brain. (F) Horizontal section of the neocortex in a 1-year-old mouse brain. High expression of mNeil3 was observed in SVZ, RMS, the hilar cells of the hippocampal formation and in cerebellum of P3 mouse brain. Transcripts of mNeil3 in SVZ decreased with age with few labelled cells present in 1-month mice and absence of SVZ transcripts in 1-year-old mice. In 1-month- and 1-year-old brain (but not in P3 brains) mNeil3 was detected in layer V of neocortex. No labelling was detected with the sense probe (insert in A). CP, caudatoputamen; TH, thalamus; F, fornix; PC, plexus choroideus; DG, dentate gyrus; SVZ, subventricular zone; HC, hippocampus, RMS, rostral migratory stream; OB, olfactory bulb. Scale bar, 100 μm.
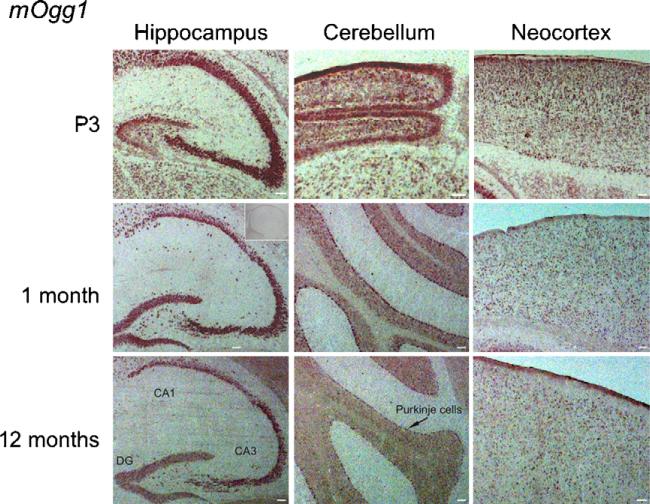
The in situ hybridization signal for mOgg1 was reduced with age (Fig. 5), although the distribution pattern remained the same throughout life with expression in all brain regions.
Fig. 5.
In situ hybridization with a DIG-RNA antisense probe to mOgg1 in horizontal brain sections from P3, 1-month- and 1-year-old mice. MOgg1 showed a heterogeneous expression pattern, with high expression in hippocampus, cerebellum and neocortex. The expression declined with age. No labelling was detected with the sense probe (insert). Scale bar, 100 μm.
MNth1 was also rather ubiquitously expressed, but showed no changes in expression level with development (Fig. 6).
3.3. Expression of mNeil1, mNeil3 and mOgg1 in mouse brain during development by Northern blotting
The in situ hybridization data indicated that three of the glycosylases (mNeil1, mNeil3, and mOgg1) showed distinct changes in expression levels with age. As in situ hybridization has obvious limitations when it comes to quantitation, we decided to perform a correlative Northern blot analysis of the gene transcripts in question (Fig. 7). In accordance with the in situ hybridization data, the Northern blot analysis revealed a modest increase in mNeil1 expression during postnatal development. MNeil3 was clearly downregulated with strong expression before birth, then declining and becoming undetectable from 4 weeks of age. MOgg1 transcripts were abundant until 1 week after birth, then declining and finally increasing again from 8 weeks of age (Fig. 7).
Fig. 7.
Gene expression of mNeil1, mOgg1 and mNeil3 during mouse brain development. The Northern blot contains RNA samples from brain tissue of 17.5-day-old mouse embryos to 12-month-old mice. cDNA from the three mouse genes were utilized as probes, and these were the same sequences as for the RNA probes applied in the in situ hybridization experiments. MNeil1 expression increased with age, in contrast to mOgg1 which was more constantly expressed throughout the life span. MNeil3 could only be detected until 2 weeks of age. The blot was repeated twice.
3.4. DNA glycosylase activity for removal of oxidative DNA damage is increased in aging mouse brain
Formamidopyrimidine (faPy) is a common oxidative DNA base lesion removed by DNA glycosylases such as OGG1, NTH1, NEIL1, NEIL2 and NEIL3. To examine the capacity to initiate BER of oxidative DNA damage, faPy DNA glycosylase activity of cell free protein extracts from four different brain regions of mice, hippocampus, cerebellum, cortex and CP, was measured at the age of 3–7, 10–15 and 35–49 weeks (Fig. 8). Fapy excision increased with age in all regions, consistent with an increased expression of faPy DNA glycosylases. These data indicate that enhanced capacity to initiate repair of oxidative DNA base lesions in aging brain is important for genome stability in neural cells.
Fig. 8.
FaPy DNA glycosylase activity in hippocampus (HIP), cerebral cortex (CTX), cerebellum (CBL) and caudatoputamen (CP) in 3–7, 10–15 and 35–49 weeks old mice. For whole brain FaPy activity was significantly higher in old mice (aged 35–49 weeks) than in young mice (aged 10–15 weeks; Mann–Whitney U-test, p = 0.008/0.016 for one/two-tailed test). FaPy activity was increased with age in all subregions of brain investigated, although statistical significance was not reached for each individual region. Bars represent standard deviation (S.D.). n = 3–5 animals per group.
4. Discussion
It is well established that oxidative stress is involved in several brain disorders, and the mechanisms of oxidative damage have been extensively studied [27–29]. In contrast, little is known about repair of oxidative DNA damage in neurons, and how oxidative stress and DNA damage contribute to neurodegeneration [2,30]. BER is the key pathway for repair of oxidative DNA lesions, and in the present study we have focused on the DNA glycosylases which constitute the damage specific initial step of BER [31]. No information has been available on the distribution in brain of the NEIL and NTH1 DNA glycosylases, but there are some published data for OGG1 [22,32]. We report the expression pattern of five DNA glycosylases involved in excision of oxidative DNA base lesions in mammalian brain. Except for NEIL3, they were all widely distributed in both human and mouse brains, but with a differential expression profile during postnatal development.
Due to lack of appropriate and specific antibodies the present study was restricted to analyses of mRNA expression and enzyme activity assays. The in situ hybridization data and Northern data mutually reinforced each other to show that the glycosylases fall into two distinct categories: NEIL1, NEIL2, OGG1 and NTH1, which are rather ubiquitously expressed across neuronal populations and brain regions, and NEIL3, whose expression is limited to a few cell populations and to early stages of postnatal development. The in situ hybridization and Northern data also concurred in showing postnatal changes in the expression levels of OGG1 and NEIL1. In contrast to NEIL3, mRNA for the latter two enzymes remained strongly expressed through adulthood. Our data thus suggest that neurons are endowed with several different glycosylases throughout postnatal development, and that DNA glycosylase expression is characterized by overlap rather than segregation. Finally, DNA glycosylase assays suggest that the capacity to initiate repair of oxidative base lesions in brain regions such as hippocampus, cerebellum, cortex and caudatoputamen increases through adulthood.
4.1. NEIL1 AND NEIL2: expression and functional roles
Neil1 and Neil2 are widely distributed in human and murine brain. NEIL1 has been suggested to contribute to repair during replication, while NEIL2 has been linked to repair during transcription [9,33]. DNA repair on the transcribed strand during transcription is termed transcription-coupled repair (TCR), whereas overall repair in the genome is referred to as global genomic repair (GGR). There are indications that terminally differentiated cells, like neurons, display proficient TCR but attenuated GGR.
Neuronal cell lines and human primary neurons have been found to exhibit modest GGR, but effective transcription-coupled nucleotide excision repair in their postmitotic state [34,35]. However, neither the neuron-like cell lines nor the primary cultures showed the strand bias that is typical of TCR repair [36]. A new repair pathway in neurons termed “differentiation-associated repair (DAR)” was therefore suggested, as efficient repair both in the non-transcribed strand as well as the transcribed strand of active genes was demonstrated [35,37]. These findings disclose that repair during transcription is essential for non-dividing cells like neurons. This is also indicated by Cockayne's syndrome (CS), which is a disease caused by mutations in genes involved in TCR of the nucleotide excision repair (NER) pathway [38]. CS patients display severe neurodegeneration [39–41]. The CSB gene of CS appears to be involved in repair of 8-oxoG, which is an important substrate for OGG1 initiated BER [42]. Thus, mice deficient in both mOgg1 and Csb were expected to show a more distinct phenotype than the mild phenotype of the mOgg1–/– mice. However, mOgg1–/–/mCsb–/– double knockouts were viable with only a weak elevation of cancer incidence in liver [43]. The lack of a neurological phenotype of these mice could be explained by the widespread expression of NEIL1 and NEIL2 presently observed. The high level of NEIL1 in brain indicates that the main function of the protein cannot be coupled to replication as neurons in general do not divide. NEIL1 and NEIL2 has been found to be involved in repair of several lesions in double stranded, single stranded and looped DNA [12]. Further, intranuclear localization showed that NEIL1 is concentrated in nucleolus [8]. Thus, the role of NEIL1 and NEIL2 in repair of rDNA and DAR should be further investigated.
4.2. NEIL1, NEIL2, and OGG1: postnatal changes in expression levels
In support of our findings in mouse brain, Englander and Ma [33] reported an upregulation of rNeil1 and rNeil2 mRNA transcripts during aging in rat brain. However, they found no increase in incision of 5-hydroxyuracil in bubble-structured DNA, which is a major substrate for hNEIL1 and hNEIL2. The same work also reported downregulation of Ogg1 during ontogeny in rat brain. The level of mRNA transcripts does not necessarily indicate the protein level, but a downregulation of OGG1 with age has also been shown by other methods [44]. Protein level of mOgg1 was found by immunocytochemistry to decline with age in brain, but not in the liver [45]. Reverse transcriptase PCR (RT-PCR) studies did not detect any modulation of rOgg1 in rat brain during aging [22]. Another study revealed maximum 8-oxoG activity in rat brain 5 days after birth, then a reduction until 12 months of age, followed by an increased excision activity up to 20 months [46]. Thus, the literature data are not entirely consistent when it comes to the expression and activity of OGG1 in the rat brain.
There are several ways to regulate the enzyme activity other than to modulate transcription of the gene. Post-translational modifications are widely known to alter proteins and enzyme activity. Recently, it was published that 8-oxoG removal by hOGG1 was stimulated by phosphorylation [47] and acetylation [48]. In contrast, acetylation of hNEIL2 reduced the excision activity [49]. The possible role of post-translational modifications should be taken into account in the interpretation of the present data on gene transcription levels. Moreover, protein–protein interactions can modulate enzymatic activities such as the XPG and WRN proteins that stimulate the NTH1 and OGG1 glycosylases, respectively [50,51].
4.3. NEIL3: expression in brain regions with progenitor cells
HNEIL3 was not detected in adult human brain by Northern blot. However, transcripts were observed in mouse brain up to 2 weeks after birth. In our previous Northern analysis, we detected hNEIL3 transcript only in adult human testis and thymus [8], but hNEIL3 was observed in several human cell lines by RT-PCR (own unpublished results). Thus, it is possible that Northern blotting is not sensitive enough to detect hNEIL3 transcripts in adult human brain. Analysis of mNeil3 by in situ hybridization disclosed a distinct expression pattern in brains from P3 mice. MNeil3 was expressed in the SVZ, the RMS, the dentate gyrus of the hippocampal formation, and the Purkinje cells of cerebellum. The expression in the former three locations indicates that mNeil3 is present in progenitor cells. This has also been observed in our laboratory by examining Neil3 in foetal mice brain, and a clear reduction of mNeil3 from foetal neurospheres to adult neurospheres has been revealed (Hildrestrand et al., in preparation), indicating a role of mNeil3 in development. This role cannot be essential for viability as Neil3 knock-out mice were healthy and fertile for at last 24 weeks after birth [14]. The mouse transcript of the mNeil3 gene was found in thymus, spleen and bone marrow, and also in several B cell lines. Additionally, mitogen stimulation of mouse splenocytes stimulated expression of mNeil3 [14].
A high expression of mNeil3 in brains of young mice is in line with our finding that mNeil3 is expressed in progenitor cells and hence implicated in development. Nevertheless, mNeil3 expression in layer V of cortex was increased with age. Layer V of the neocortex is the primary source of projections to subcortical sites. Layer V neurons of the motor cortex are of particular interest in the context of DNA repair as they constitute the pool of upper motor neurons that degenerate in amyotrophic lateral sclerosis (ALS) [52]. The presence of mNeil3 in neocortical neurons warrants further analyses.
4.4. NTH1
No distribution data on mammalian NTH1 in brain are available, but activity studies have been performed [44,53]. However, both NEIL1 and NEIL2 exhibit overlapping substrate specificities with NTH1, confounding the interpretation of activity assays. Our study indicates a stable function of NTH1 throughout life in mouse brain.
5. Conclusion
To our knowledge this is the first report on distribution in brain of mRNAs encoding mammalian NEILs and NTH1 DNA glycosylases. As specific antibodies to these enzymes are not available it cannot be resolved whether the distribution of mRNAs matches the distribution of the respective proteins. This limitation must be taken into account in the interpretation of the present data. Our findings indicate a widespread distribution of NEIL and NTH1 DNA glycosylases, except NEIL3, suggesting that BER is important for protecting the genetic material in neurons against oxidative DNA damage. The subtle differences in expression pattern suggest distinct functions in particular brain regions. Given their role in removal of aberrant bases in loop structured or single stranded DNA [12], hNEIL1 and hNEIL2 in particular may be essential for maintaining integrity of transcribed DNA in postmitotic neurons. Our observation of an upregulation of the transcripts of these genes during aging is in line with this idea. Of special interest is the distinct expression pattern of the scantly studied Neil3 gene. The strong expression of this gene in select populations of progenitor cells indicates a role for NEIL3 in brain development and calls for further investigations.
Acknowledgements
The authors wish to thank Jorunn Knutsen and Carina Knudsen for excellent technical assistance and Reidun Torp for fruitful discussions. This research was supported by the Norwegian Cancer Society, the European Union Biomed Project GRIPANNT 005320, The Norwegian Research Council (FUGE and Storforsk), Rikshospitalet-Radiumhospitalet HF, the University of Oslo and R01 GM 066359.
Abbreviations
- 8-oxoG
8-oxoguanine
- AP
apurinic/apyrimidinic
- BER
base excision repair
- CP
caudatoputamen
- CS
Cockayne syndrome
- DAR
differentiation-associated repair
- faPy
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- GGR
global genomic repair
- NEIL
Escherichia coli endonuclease VIII-like
- NTH
Endonuclease III
- OGG
8-oxoG-DNA glycosylase
- P3
postnatal day 3
- RMS
rostral migratory stream
- ROS
reactive oxygen species
- SVZ
subventricular zone
- TCR
transcription-coupled repair
Footnotes
Conflict of interest
None.
REFERENCES
- 1.Rao KS. DNA repair in aging rat neurons. Neuroscience. 2007;145:1330–1340. doi: 10.1016/j.neuroscience.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 2.Bohr VA, Ottersen OP, Tonjum T. Genome instability and DNA repair in brain, ageing and neurological disease. Neuroscience. 2007;145:1183–1186. doi: 10.1016/j.neuroscience.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Gabbita SP, Lovell MA, Markesbery WR. Increased nuclear DNA oxidation in the brain in alzheimer's disease. J. Neurochem. 1998;71:2034–2040. doi: 10.1046/j.1471-4159.1998.71052034.x. [DOI] [PubMed] [Google Scholar]
- 4.Lovell MA, Gabbita SP, Markesbery WR. Increased DNA oxidation and decreased levels of repair products in Alzheimer's disease ventricular CSF. J. Neurochem. 1999;72:771–776. doi: 10.1046/j.1471-4159.1999.0720771.x. [DOI] [PubMed] [Google Scholar]
- 5.Cui K, Luo X, Xu K, Ven Murthy MR. Role of oxidative stress in neurodegeneration: recent developments in assay methods for oxidative stress and nutraceutical antioxidants, Prog. Neuropsychopharmacol. Biol. Psychiatry. 2004;28:771–799. doi: 10.1016/j.pnpbp.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Alam ZI, Jenner A, Daniel SE, Lees AJ, Cairns N, Marsden CD, Jenner P, Halliwell B. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J. Neurochem. 1997;69:1196–1203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DM, III, McNeill DR. Base excision repair and the central nervous system. Neuroscience. 2007;145:1187–1200. doi: 10.1016/j.neuroscience.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Morland I, Rolseth V, Luna L, Rognes T, Bjørås M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99(6):3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 11.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Rep. 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 12.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 13.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Rep. 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J. Biochem. (Tokyo) 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- 15.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 18.Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16(20):6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of lys-212 as the active nucleophilic residue. J. Biol. Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 21.Eide L, Luna L, Gustad EC, Henderson PT, Essigmann JM, Demple B, Seeberg E. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry (Mosc.) 2001;40:6653–6659. doi: 10.1021/bi0028901. [DOI] [PubMed] [Google Scholar]
- 22.Verjat T, Dhenaut A, Radicella JP, Araneda S. Detection of 8-oxoG DNA glycosylase activity and OGG1 transcripts in the rat CNS. Mut. Res./DNA Rep. 2000;460:127–138. doi: 10.1016/s0921-8777(00)00022-7. [DOI] [PubMed] [Google Scholar]
- 23.Torp R, Hoover F, Danbolt NC, Storm-Mathisen J, Ottersen OP. Differential distribution of the glutamate transporters GLT1 and rEAAC1 in rat cerebral cortex and thalamus: an in situ hybridization analysis. Anat. Embryol. (Berl.) 1997;195:317–326. doi: 10.1007/s004290050051. [DOI] [PubMed] [Google Scholar]
- 24.Hoover F, Goldman D. Temporally correlated expression of nAChR genes during development of the mammalian retina. Exp. Eye Res. 1992;54(4):561–571. doi: 10.1016/0014-4835(92)90135-f. [DOI] [PubMed] [Google Scholar]
- 25.Boiteux S, Belleney J, Roques BP, Laval J. Two rotameric forms of open ring 7-methylguanine are present in alkylated polnucleotides. Nucleic Acids Res. 1984;12:5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayre LM, Smith MA, Perry G. Chemistry and biochemistry of oxidative stress in neurodegenerative disease. Curr. Med. Chem. 2001;8(7):721–738. doi: 10.2174/0929867013372922. [DOI] [PubMed] [Google Scholar]
- 28.Sayre LM, Perry G, Smith MA. Oxidative stress and neurotoxicity. Chem. Res. Toxicol. 2008;21:172–188. doi: 10.1021/tx700210j. [DOI] [PubMed] [Google Scholar]
- 29.Halliwell B. Oxidative stress and neurodegeneration: where are we now? J. Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 30.Rolseth V, Rundén-Pran E, Neurauter CG, Yndestad A, Luna L, Aukrust P, Ottersen OP, Bjørås M. Base excision repair activities in organotypic hippocampal slice cultures exposed to oxygen and glucose deprivation. DNA Rep. 2008;7:869–878. doi: 10.1016/j.dnarep.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145:1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 32.Mosquera DI, Stedeford T, Cardozo-Pelaez F, Sanchez-Ramos J. Strain-specific differences in the expression and activity of Ogg1 in the CNS. Gene Exp. 2003;11(1):47–53. doi: 10.3727/000000003783992333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Englander EW, Ma H. Differential modulation of base excision repair activities during brain ontogeny: implications for repair of transcribed DNA. Mech. Ageing Dev. 2006;127:64–69. doi: 10.1016/j.mad.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Hanawalt PC, Gee P, Ho L, Hsu RK, Kane CJ. Genomic heterogeneity of DNA repair. Role in aging? Ann. N. Y. Acad. Sci. 1992;663:17–25. doi: 10.1111/j.1749-6632.1992.tb38644.x. [DOI] [PubMed] [Google Scholar]
- 35.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol. Cell. Biol. 2000:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Rep. 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- 37.Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145:1213–1221. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype–phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frosina G. The current evidence for defective repair of oxidatively damaged DNA in Cockayne syndrome. Free Radic. Biol. Med. 2007;43:165–177. doi: 10.1016/j.freeradbiomed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Perry JJP, Fan L, Tainer JA. Developing master keys to brain pathology, cancer and aging from the structural biology of proteins controlling reactive oxygen species and DNA repair. Neuroscience. 2007;145:1280–1299. doi: 10.1016/j.neuroscience.2006.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 1992;42(1):68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 42.Osterod M, Larsen E, Le PF, Hengstler JG, Van Der Horst GT, Boiteux S, Klungland A, Epe B. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene. 2002;21(54):8232–8239. doi: 10.1038/sj.onc.1206027. [DOI] [PubMed] [Google Scholar]
- 43.van der Horst GT, van SH, Berg RJ, van Gool AJ, de WJ, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR, Bootsma D, Hoeijmakers JH. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 44.Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol. Aging. 2006;27:1129–1136. doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Larsen E, Reite K, Nesse G, Gran C, Seeberg E, Klungland A. Repair and mutagenesis at oxidized DNA lesions in the developing brain of wild-type and Ogg1–/– mice. Oncogene. 2006;25(17):2425–2432. doi: 10.1038/sj.onc.1209284. [DOI] [PubMed] [Google Scholar]
- 46.Bolin CM, Basha R, Cox D, Zawia NH, Maloney B, Lahiri DK, Cardozo-Pelaez F. Exposure to lead (Pb) and the developmental origin of oxidative DNA damage in the aging brain. FASEB J. 2006;6:788–790. doi: 10.1096/fj.05-5091fje. [DOI] [PubMed] [Google Scholar]
- 47.Hu J, Imam SZ, Hashiguchi K, de Souza-Pinto NC, Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase ({alpha}-OGG1) modulates its function. Nucleic Acids Res. 2005;33:3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhakat KK, Hazra TK, Mitra S. Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 2004;32:3033–3039. doi: 10.1093/nar/gkh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klungland A, H÷ss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, Bolton PH, Wood RD, Lindahl T. Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 51.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 52.Aguilar J.-L.G.d., Echaniz-Laguna A, Fergani A, Rene F, Meininger V, Loeffler JP, Dupuis L. Amyotrophic lateral sclerosis: all roads lead to Rome. J. Neurochem. 2007;101:1153–1160. doi: 10.1111/j.1471-4159.2006.04408.x. [DOI] [PubMed] [Google Scholar]
- 53.Karahalil BENS, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]