Abstract
RNA interference (RNAi) mediated by small interfering RNAs (siRNAs) enables knockdown of a gene of choice, executing the logical operation: silence gene Y. The fact that the siRNA is constitutively active is a significant limitation, making it difficult to confine knockdown to a specific locus and time. To achieve spatiotemporal control over silencing, we seek to engineer small conditional RNAs (scRNAs) that mediate ‘conditional RNAi’ corresponding to the logical operation: if gene X is transcribed, silence independent gene Y. By appropriately selecting gene X, knockdown of gene Y could then be restricted in a tissue- and time-specific manner. To implement the logic of conditional RNAi, our approach is to engineer scRNAs that, upon binding to mRNA ‘detection target’ X, perform shape and sequence transduction to form a Dicer substrate targeting independent mRNA ‘silencing target’ Y, with subsequent Dicer processing yielding an siRNA targeting mRNA Y for destruction. Toward this end, here we design and experimentally validate diverse scRNA mechanisms for conditional Dicer substrate formation. Test tube studies demonstrate strong OFF/ON conditional response, with at least an order of magnitude increase in Dicer substrate production in the presence of the cognate mRNA detection target. By appropriately dimensioning and/or chemically modifying the scRNAs, only the product of signal transduction, and not the reactants or intermediates, is efficiently processed by Dicer, yielding siRNAs. These mechanism studies explore diverse design principles for engineering scRNA signal transduction cascades including reactant stability vs metastability, catalytic vs noncatalytic transduction, pre- vs post-transcriptional transduction, reactant and product molecularity, and modes of molecular self-assembly and disassembly.
Introduction
RNAi enables biologists to knock down expression of a gene of choice in eukaryotes, providing a powerful tool for probing gene function within endogenous biological circuits.1,2 RNAi can be activated by exogenous double-stranded RNAs that are cleaved by the enzyme Dicer to produce siRNAs. One strand of the siRNA duplex (the guide strand) is loaded into the RNA-induced silencing complex (RISC), where it serves as a recognition domain for recruitment of target mRNAs containing the complementary sequence. RISC cleaves and releases the mRNA for subsequent degradation, enabling a single guide strand to mediate destruction of multiple copies of the mRNA silencing target. The conceptual power of RNAi follows from its programmability: by changing the sequence of the siRNA, it is possible to change the identity of the gene that is targeted for knockdown.
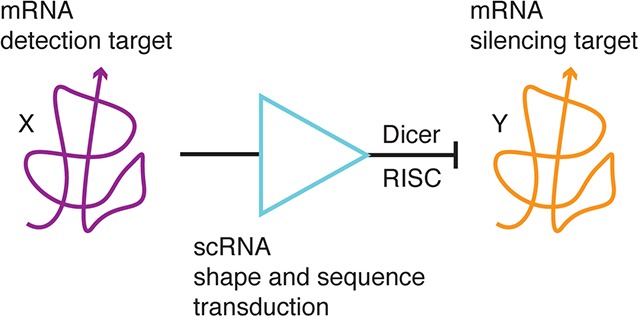
Using an siRNA programmed to silence gene Y, conventional RNAi implements the unconditional molecular logic (inset of Figure 1): silence gene Y. To exert control over the strength and/or timing of gene knockdown, numerous methods have been developed to implement drug-inducible RNAi, where the activation (or inhibition) of knockdown is made dependent on the presence of a small molecule, using either pretranscriptional protein machinery3,4 or post-transcriptional RNA machinery.5−9 To achieve spatiotemporal control over gene knockdown, we seek to engineer scRNAs that mediate conditional RNAi corresponding to the conditional molecular logic (Figure 1): if gene X is transcribed, silence independent gene Y. This logic is programmable at two levels, with input sequence X controlling the scope of silencing and output sequence Y controlling the target of silencing.
Figure 1.
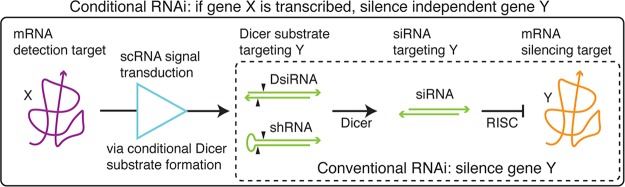
Molecular logic of conditional and conventional RNAi. Conditional RNAi (if gene X is transcribed, silence independent gene Y) provides a conceptual framework for exerting spatiotemporal control over gene knockdown. Toward this end, small conditional RNAs (scRNAs) interact and change conformation to transduce between binding of mRNA ‘detection target’ X and production of a Dicer substrate targeting independent mRNA ‘silencing target’ Y. Inset: Conventional RNAi (silence gene Y) employs constitutively active Dicer substrates, making it difficult to control the locus and time of gene knockdown. We consider conditional formation of Dicer substrates that are either DsiRNAs or shRNAs.
To implement the logic of conditional RNAi, our approach is to engineer scRNA signal transduction cascades in which hybridization of an scRNA to an mRNA ‘detection target’ X initiates downstream conformational changes of one or more scRNAs leading to formation of a Dicer substrate targeting independent mRNA ‘silencing target’ Y. Dicer processing of this substrate then yields an siRNA targeting mRNA Y for destruction. Two types of signal transduction must be performed simultaneously to achieve this goal: conditional shape change is required to produce a molecular geometry that is recognized and processed by Dicer, and conditional sequence change is required to shift from input sequence X to output sequence Y.
Previous studies have shown that suitable Dicer substrates include short hairpin RNAs (shRNAs; 19–29-bp stem with a 2-nt 3′-overhang)1,10,11 and so-called Dicer-substrate RNAs (DsiRNAs; ≈25-bp duplex with a 2-nt 3′-overhang at one end).1,12 Dicer functions as a molecular ruler, measuring from the 2-nt 3′-overhang to cleave ≈21–23-nt siRNA strands that form a duplex with 2-nt 3′-overhangs at both ends.13,14 For this reason, we focus on engineering scRNA transducers that conditionally assemble shRNA or DsiRNA Dicer substrates with a 2-nt 3′-overhang at one end of a minimum 19-bp duplex.
To mediate conditional RNAi via Dicer substrate formation, several design requirements for scRNA function can be identified a priori: First, the sequence of the detection target X must place no restriction on the sequence of the independent silencing target Y. Second, in the absence of detection target X, the scRNAs should not interact to form the Dicer substrate targeting Y. Third, the scRNAs must be capable of detecting a subsequence of a full-length endogenous mRNA detection target X. Fourth, in response to detection of X, the scRNAs must undergo an isothermal hybridization cascade mediating formation of a Dicer substrate targeting Y. Fifth, the Dicer substrate must be efficiently processed by Dicer to produce siRNAs targeting Y. Sixth, the scRNAs should be dimensioned and/or chemically modified appropriately so that only the final Dicer substrate is amenable to Dicer processing. Furthermore, it is likely that other unanticipated design requirements will emerge during the engineering and validation process (e.g., additional constraints imposed by endogenous pathways).
Several groups have achieved subsets of these goals. Masu et al.15 engineered scRNAs that when annealed in a test tube with a short RNA detection target Xs (high temperature followed by slow cooling to room temperature) yielded a Dicer substrate that mediated knockdown of independent silencing target Y upon transfection into mammalian cells. Xie et al.16 engineered scRNAs that detect a 140-nt RNA target X and produce an siRNA that mediates knockdown of a closely related silencing target X′ in Drosophila lysate. Kumar et al.17 express an scRNA in mammalian cells and transfect a short modified-RNA detection target Xs, leading to production of an siRNA that mediates knockdown of independent silencing target Y. Additional work is required to meet all six of the scRNA design requirements.
Previous research in the field of DNA nanotechnology demonstrates that the programmable chemistry of base pairing provides a versatile medium for engineering diverse dynamic functions including catalysis, amplification, logic, and locomotion.18 We seek to exploit mechanism and sequence design principles drawn from this experience to engineer scRNAs (or scDNAs) suitable for interfacing with Dicer and RISC to mediate conditional RNAi in vivo. First, we must address mechanism design: how are the scRNA molecules intended to interact and change conformation in order to effect signal transduction? Second, we must address sequence design: given an envisioned mechanism design and sequences for a pair of independent mRNA detection and silencing targets, X and Y, what scRNA sequences, if any, will encode the intended signal transduction function?
To explore the mechanism and sequence design challenges for conditional Dicer substrate formation, we have engineered five different mechanisms, each satisfying the six design requirements noted above, while examining diverse design alternatives spanning (Table 1): reactant material (scRNA vs scDNA), initial reactant state (metastable vs stable), reactant role (catalytic vs noncatalytic), nucleation mechanism (toehold/toehold vs loop/toehold vs template/toehold), strand displacement mechanism (3-way branch migration vs 4-way branch migration vs spontaneous dissociation), reactant type (hairpin monomer vs duplex dimer), Dicer substrate assembly method (hybridization vs transcription), and Dicer substrate type (DsiRNA vs shRNA). In studying these design alternatives, we sought both to optimize performance in satisfying the six design requirements and to achieve simplicity.
Table 1. Mechanisms and Design Alternatives for Conditional Dicer Substrate Formation.
| mechanism |
|||||
|---|---|---|---|---|---|
| design alternatives | 1 | 2 | 3 | 4 | 5 |
| scRNA reactants | x | x | x | x | |
| scDNA reactants | x | ||||
| metastable reactants | x | x | x | ||
| stable reactants | x | x | |||
| catalytic production | x | x | |||
| noncatalytic production | x | x | x | ||
| toehold/toehold nucleation | x | x | x | x | x |
| loop/toehold nucleation | x | ||||
| template/toehold nucleation | x | ||||
| 3-way branch migration | x | x | x | x | x |
| 4-way branch migration | x | x | |||
| spontaneous dissociation | x | x | |||
| hairpin monomer reactants | 3 | 1 | 0 | 0 | 2 |
| duplex dimer reactants | 0 | 1 | 1 | 2 | 0 |
| Dicer substrate hybridization | x | x | x | x | |
| Dicer substrate transcription | x | ||||
| DsiRNA Dicer substrate | x | x | x | ||
| shRNA Dicer substrate | x | x | |||
For a given scRNA (or scDNA) transduction mechanism, sequence design must be performed subject to the constraints imposed by a given pair of mRNA detection and silencing targets, X and Y (i.e., which subsequences within the full-length mRNA sequences, if any, confer desirable properties on the dynamic conditional response of the scRNA transducers?). These sequence constraints dramatically reduce the size of the design space, increasing the challenge of designing well-behaved sequences. Here, we employ NUPACK to solve a constrained multistate sequence design problem19 based on a set of target secondary structures representing key initial, intermediate, and final states in the intended conditional hybridization cascade. Sequences are optimized with the goal of reducing the ensemble defect for each target structure below a user-specified stop condition.20 For a given target secondary structure and candidate sequence, the ensemble defect is the average number of incorrectly paired nucleotides at equilibrium evaluated over the ensemble of (unpseudoknotted) secondary structures.20,21 Optimization of the ensemble defect encompasses both a positive design paradigm (optimize affinity for the target structure) and a negative design paradigm (optimize selectivity against all other structures in the ensemble).20,21 Hence, multistate ensemble defect optimization provides a framework for designing sequences that execute signal transduction via a prescribed hybridization cascade punctuated by the desired reactant, intermediate, and product secondary structures.
Following mechanism and sequence design, we quantify the OFF/ON response of conditional Dicer substrate formation in test tube studies, introducing either a short RNA or full-length mRNA detection target, and monitoring production of Dicer substrates targeting an independent mRNA silencing target. Studies with recombinant Dicer are used to verify that only the final product of signal transduction, and not the reactants or intermediates, are efficiently processed by Dicer, yielding siRNAs.
Results and Discussion
For our engineering studies, we consider detection target DsRed2 (mRNA X) and silencing target d2EGFP (mRNA Y). Hence, our objective is to design scRNA mechanisms and sequences so that, upon exposure to DsRed2 mRNA, the scRNAs interact and change conformation to form a Dicer substrate targeting d2EGFP mRNA. To focus our attention on scRNA signal transduction and eliminate the confounding effects of native mRNA secondary structure, we also consider short detection targets (Xs) corresponding to the DsRed2 subsequence that is recognized by a given scRNA mechanism. We quantify the relative OFF/ON response of conditional Dicer substrate formation in the absence/presence of the detection target (Xs or X). As a test for off-target effects, we also measure the response to the silencing target Y and to GAPDH (mRNA Z), neither of which should initiate signal transduction. To confirm that scRNA transducers interact with Dicer as intended, we use recombinant Dicer to test for undesired processing of the scRNA reactants and transduction intermediates as well as for efficient processing of the final product (i.e., the cognate Dicer substrate) to produce siRNAs. Experimental characterizations of conditional OFF/ON response (Figures 2–6) are augmented by computational and experimental stepping analyses that characterize the reactants, intermediates, and products for each mechanism (Sections S2–S6).
Figure 2.
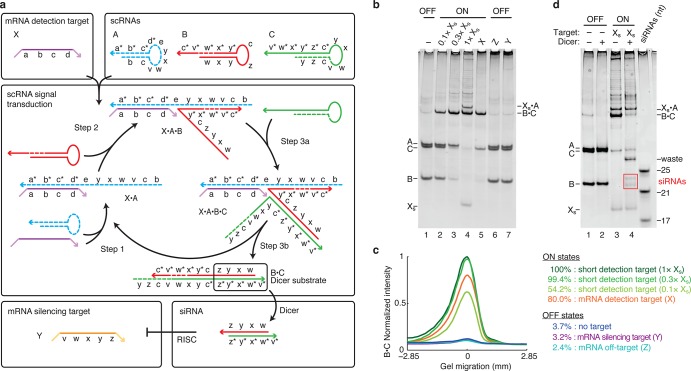
Conditional catalytic DsiRNA formation using metastable scRNAs. (a) Mechanism 1. scRNA A detects mRNA detection target X (containing subsequence ‘a-b-c-d’) to form catalyst X·A, which mediates production of DsiRNA Dicer substrate B·C targeting mRNA silencing target Y (containing independent subsequence ‘v-w-x-y-z’). scRNAs A, B, and C coexist metastably in the absence of X. Successive toehold-mediated 3-way branch migrations enable assembly of X with A (step 1), X·A with B (step 2), X·A·B with C (step 3a), and disassembly of DsiRNA Dicer substrate B·C from catalyst X·A (step 3b). Domain lengths: |a| = 10, |b| = 10, |c| = 5, |d| = 2, |e| = 2, |v| = 2, |w| = 5, |x| = 2, |y| = 6, |z| = 5. Chemical modifications (2′OMe-RNA): A and parts of B and C (dashed backbone). (b) Conditional catalytic Dicer substrate formation. OFF state: minimal production of Dicer substrate B·C in the absence of detection target X, the presence of mRNA silencing target Y, or the presence of mRNA off-target Z. ON state: strong production of B·C in the presence of substoichiometric or stoichiometric short RNA detection target Xs (‘a-b-c-d’) or the presence of full-length mRNA detection target X. (c) Quantification of the Dicer substrate band (B·C) in panel (b). (d) Conditional Dicer processing. OFF state: minimal processing of the reactants (lane 2). ON state: efficient processing of Dicer substrate B·C (lane 4), yielding canonical 21- and 23-nt siRNAs (boxed bands). The non-siRNA remainder of the cleaved substrate is labeled ‘waste’. See Section S2 for additional computational and experimental studies of Mechanism 1.
Figure 6.
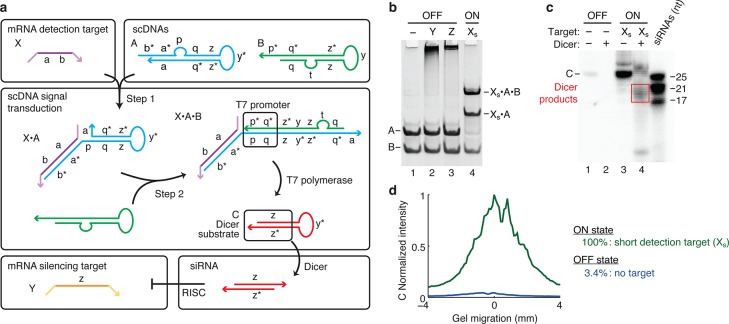
Conditional shRNA transcription using scDNAs. (a) Mechanism 5. scDNA A detects mRNA detection target X (containing subsequence ‘a-b’) and assembles with B to form a transcription template (containing promoter, coding, and termination sequences), leading to transcription of the shRNA Dicer substrate C targeting mRNA silencing target Y (containing independent subsequence ‘z’). scDNAs A and B coexist metastably in the absence of X. X assembles with A via toehold-mediated 3-way branch migration (step 1). Subsequently, X·A assembles with B via toehold-mediated 4-way branch migration to produce a dsDNA transcription template (step 2), mediating transcription of shRNA Dicer substrate C with catalytic turnover. Domain lengths: |a| = 10, |b| = 8, |p| = 8, |q| = 9, |t| = 7, |y| = 6, |z| = 19. (b) Conditional transcription template formation. OFF state: minimal production of transcription template A·B in the absence of short DNA detection target Xs (‘a-b’), the presence of mRNA silencing target Y, or the presence of mRNA off-target Z. ON state: strong production of transcription template Xs·A·B in the presence of Xs. (c) Conditional Dicer substrate transcription and processing. OFF state: minimal transcription of Dicer substrate C in the absence of short DNA detection target Xs (lane 1). ON state: strong transcription of C in the presence of Xs (lane 3) and efficient Dicer processing of shRNA Dicer substrate C (lane 4). (d) Quantification of the Dicer substrate band (C) in lanes 1 and 3 of panel (c). See Section S6 for additional computational and experimental studies of Mechanism 5.
Mechanism 1: Conditional Catalytic DsiRNA Formation Using Metastable scRNAs
We begin by exploiting the hairpin motif of Yin et al.,22 which has previously been used to program diverse self-assembly and disassembly hybridization cascades, including catalytic duplex formation. In the present circumstances, the duplex that we wish to form must have the canonical 2-nt 3′-overhang of a DsiRNA, and the catalysis process must also achieve sequence transduction between detection target X and silencing target Y. The transduction mechanism of Figure 2a employs three hairpins (A, B, and C) that coexist metastably in the absence of detection target X (i.e., they are kinetically impeded from assembling into an equilibrium distribution of products). The detection target X opens hairpin A, which in turn opens hairpin B, which in turn opens hairpin C, leading to formation of duplex B·C and regeneration of catalyst X·A. Duplex B·C has a 2-nt 3′-overhang and targets silencing target Y. Chemical modifications (2′OMe-RNA) of A and portions of B and C are employed to prevent Dicer cleavage of scRNA reactants and transduction intermediates, while preserving efficient Dicer processing of the transduction product B·C. In functional terms, A detects X and catalyzes production of DsiRNA B·C targeting Y.
Figure 2b,c examines the conditional OFF/ON response of the transduction mechanism. In the absence of the detection target X, there is minimal production of DsiRNA B·C, corresponding to minimal ‘leakage’ of the kinetically trapped hairpins out of their metastable states. Neither the mRNA silencing target Y (which is necessarily related in sequence to the hairpins) nor the unrelated mRNA off-target Z causes measurable production of B·C above the background leakage. Stoichiometric introduction of the short detection target Xs leads to strong production of B·C, and substoichiometric introduction of Xs demonstrates catalytic turnover in producing B·C. Strong production of B·C is also observed using full-length mRNA detection target X. The OFF/ON conditional response of the transduction mechanism yields more than an order of magnitude increase in production of Dicer substrate above background (Figure 2c).
Figure 2d demonstrates signal transduction in the presence of recombinant Dicer. Only the DsiRNA B·C that is the final product of transduction is recognized and efficiently processed by Dicer, yielding canonical 21- and 23-nt siRNAs (see also Figure S3 and Table S3).
In assessing the advantages and disadvantages of this scRNA transduction mechanism, it is helpful to classify the design features that contribute to conditional shape change and conditional sequence change (Table 1). The hairpin reactants are metastable, and could potentially leak into the DsiRNA product on a biologically relevant time scale, even in the absence of detection target X. Three hairpins are required to effect the necessary shape and sequence transduction: hairpin A achieves partial sequence independence via the loop, hairpin B moves the independent sequence to the end of the strand for presentation to Dicer, and hairpin C contributes the additional independent sequence of the 2-nt 3′-overhang and liberates the fully formed DsiRNA B·C from catalyst X·A. This process requires transient formation of a relatively complex intermediate (tetramer X·A·B·C including mRNA X). Use of monomer hairpins simplifies preparation of purified reactants, but eventual delivery of three scRNA species to cells could prove burdensome. On the other hand, each self-assembly operation (opening of a new A, B, or C hairpin) and disassembly operation (liberation of a new B·C duplex) occurs via toehold-mediated 3-way branch migration,23 providing a robust framework for engineering fast reaction kinetics.18 The mechanism has the potentially useful property that a single detection target X can catalyze production of multiple DsiRNAs targeting silencing target Y, augmenting the catalytic turnover that is already present in the RNAi pathway (via RISC-mediated cleavage of multiple silencing targets using a single guide strand).
Mechanism 2: Conditional DsiRNA Formation Using Stable scRNAs
We wondered whether we could simplify the signal transduction mechanism by exploiting alternative design principles. In particular, it seems intuitively desirable to reduce the number of scRNA reactants, the number of assembly steps in the transduction cascade, and the complexity of the reaction intermediates. These goals are achieved by replacing the A and B hairpins of Mechanism 1 with the A·B duplex of Mechanism 2 (Figure 3a). The detection target X mediates displacement of A from B, which opens C to produce duplex B·C with a 2-nt 3′-overhang. The number of reactants and the number of assembly steps are both reduced from three to two and the largest intermediate is reduced from a tetramer (resulting from three sequential assembly steps) to a trimer (resulting from one assembly step). This simplified signal transduction mechanism dispenses with catalytic turnover, producing one DsiRNA per detected molecule of X. In functional terms, A·B detects X, leading to production of DsiRNA B·C targeting Y.
Figure 3.
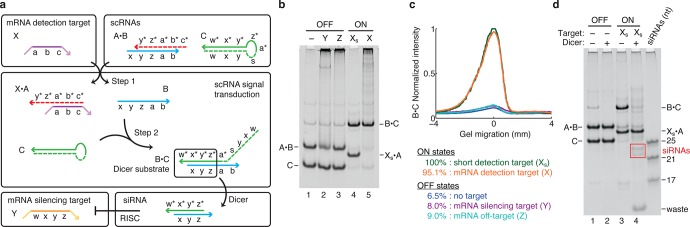
Conditional DsiRNA formation using stable scRNAs. (a) Mechanism 2. scRNA A·B detects mRNA detection target X (containing subsequence ‘a-b-c’), leading to production of DsiRNA Dicer substrate B·C targeting mRNA silencing target Y (containing independent subsequence ‘w-x-y-z’). scRNAs A·B and C are stable in the absence of X. A swaps B for X (step 1) via toehold-mediated 3-way branch migration and spontaneous dissociation. B assembles with C (step 2) via loop/toehold nucleation and 3-way branch migration to form DsiRNA Dicer substrate B·C. Domain lengths: |a| = 6, |b| = 4, |c| = 8, |s| = 5, |w| = 2, |x| = 12, |y| = 4, |z| = 3. Chemical modifications (2′OMe-RNA): A and part of C (dashed backbone). (b) Conditional Dicer substrate formation. OFF state: minimal production of Dicer substrate B·C in the absence of detection target X, the presence of mRNA silencing target Y, or the presence of mRNA off-target Z. ON state: strong production of B·C in the presence of short RNA detection target Xs (‘a-b-c’) or full-length mRNA detection target X. (c) Quantification of the Dicer substrate band (B·C) in panel (b). (d) Conditional Dicer processing. OFF state: minimal Dicer processing of the reactants (lane 2). ON state: efficient Dicer processing of DsiRNA Dicer substrate B·C (lane 4), yielding canonical 21–23-nt siRNAs (boxed bands). The non-siRNA remainder of the cleaved substrate is labeled ‘waste’. See Section S3 for additional computational and experimental studies of Mechanism 2.
The mechanism exhibits strong OFF/ON conditional Dicer substrate formation, achieving an order of magnitude increase in DsiRNA production in the presence of either the short detection target Xs or the full-length mRNA detection target X (Figure 3b,c). Chemical modifications of A and portions of C are employed to prevent Dicer processing of the reactants and intermediates. Only the DsiRNA B·C is efficiently processed by Dicer, yielding canonical 21–23-nt siRNAs (Figures 3d and S8 and Table S5).
Compared to Mechanism 1, shape and sequence transduction are achieved based on dramatically altered design principles (Table 1). Mechanism 1 repeatedly exploits toehold/toehold hybridization for nucleation and 3-way branch migration for strand displacement, while Mechanism 2 simplifies the transduction pathway by also exploiting spontaneous dissociation to achieve strand displacement (of B from X·A) and loop/toehold hybridization to nucleate interactions (between B and C). Strikingly, the scRNAs for Mechanism 2 are stable rather than metastable (i.e., if the scRNAs are allowed to equilibrate in the absence of X, they will predominantly remain in the reactant state rather than converting to the product state; see Section S7). This is a major conceptual advantage because it places a thermodynamic rather than a kinetic limit on the amount of spurious DsiRNA that can form in the absence of X. With Mechanism 2, strong production of DsiRNA is only thermodynamically favorable if X is present, whereas with the metastable reactants of Mechanism 1, X catalyzes a reaction that is kinetically impeded but thermodynamically favorable in the absence of X. In our studies, the metastable scRNAs of Mechanism 1 and the stable scRNAs of Mechanism 2 happen to produce comparable amounts of background DsiRNA in the absence of X (both yielding an OFF state that is ≈5% of the ON state achieved using Xs). Nonetheless, stable reactants offer a conceptually appealing framework for engineering robust OFF/ON signal transduction in vivo; if the thermodynamic driving force for spontaneous DsiRNA formation can be further reduced, stable reactants promise a clean and reliable OFF state.
Mechanism 3: Conditional shRNA Formation Using a Single Stable scRNA
Motivated by the simplifications of Mechanism 2, we wished to see if we could push even further to simplify shape and sequence transduction. Mechanism 3 requires only a single duplex scRNA A·B, and in a single step produces a Dicer substrate that is an shRNA monomer instead of a DsiRNA duplex (Figure 4a). The detection target X mediates displacement of A from B to yield a hairpin B with a 2-nt 3′-overhang. The number of reactants and the number of assembly steps are both reduced from two to one. This is the simplest mechanism for conditional Dicer substrate formation that we have devised to date. In functional terms, A·B detects X, leading to production of shRNA B targeting Y.
Figure 4.
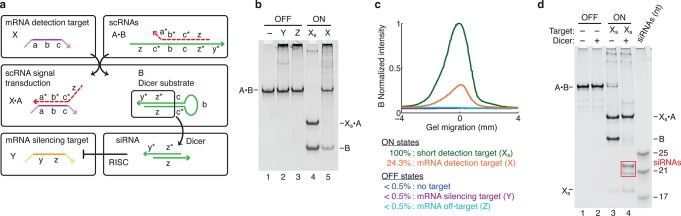
Conditional shRNA formation using a single stable scRNA. (a) Mechanism 3. scRNA A·B detects mRNA detection target X (containing subsequence ‘a-b-c’), leading to production of shRNA Dicer substrate B targeting mRNA silencing target Y (containing independent subsequence ‘y-z’). scRNA A·B is stable in the absence of X. X partially displaces A from B via toehold-mediated 3-way branch migration, exposing a previously sequestered internal toehold, ‘c’, within B, mediating a further 3-way branch migration that disassembles B from X·A to yield shRNA Dicer substrate B. Domain lengths: |a| = 12, |b| = 14, |c| = 3, |y| = 2, |z| = 19. Chemical modifications (2′OMe-RNA): A (dashed backbone). (b) Conditional Dicer substrate formation. OFF state: minimal production of Dicer substrate B in the absence of detection target X, the presence of mRNA silencing target Y, or the presence of mRNA off-target Z. ON state: strong production of B in the presence of short RNA detection target Xs (‘a-b-c’) or full-length mRNA detection target X. (c) Quantification of the Dicer substrate band (B) in panel (b). (d) Conditional Dicer processing. OFF state: minimal processing of the reactants (lane 2). ON state: efficient processing of shRNA Dicer substrate B (lane 4), yielding canonical 21- and 22-nt siRNAs (boxed bands). See Section S4 for additional computational and experimental studies of Mechanism 3.
The mechanism exhibits strong OFF/ON conditional Dicer substrate formation, achieving 2 orders of magnitude increase in shRNA production in the presence of the short detection target Xs and 1 order of magnitude increase for the full-length mRNA target X (Figure 4b,c). It is unclear why the performance is diminished for the full-length target (though still comparable to the performance for other mechanisms); we expect this behavior is specific to this test case and not general to the mechanism. The most striking feature of these data is that the OFF state is undetectable (i.e., smaller than our estimated gel quantification uncertainty).
The clean OFF state follows from the fact that the siRNA reactant, A·B, is highly stable, with very little thermodynamic driving force for production of shRNA B in the absence of X. Hence, this mechanism compellingly exhibits the benefit of using stable rather than metastable reactants.
For Mechanism 3, the design elements underlying sequence and shape transduction are pleasingly simple. X partially displaces A from B via toehold-mediated 3-way branch migration, exposing a previously sequestered internal toehold, which B then uses to nucleate a 3-way branch migration with itself, completing displacement of A. Chemical modifications of A are employed to ensure that only shRNA B is efficiently processed by Dicer, yielding canonical 21- and 22-nt siRNAs (Figures 4d and S12 and Table S7).
Mechanism 4: Conditional DsiRNA Formation via Template-Mediated 4-Way Branch Migration
To date, efforts to engineer conditional hybridization cascades within the field of DNA nanotechnology have focused almost exclusively on strand displacement reactions based on 3-way branch migration, in which an invading strand displaces one strand from a duplex.18 By comparison, there has been very little study of strand displacement reactions based on 4-way branch migration24,25 in which two duplexes exchange partner strands. In the present setting, a DsiRNA signal transduction product is a duplex, so we were curious if 4-way branch migration might prove especially suitable for conditional Dicer substrate formation. Mechanism 4 employs two duplex scRNAs (A·B and C·D of Figure 5a). The detection target X mediates swapping of partner strands, producing duplex B·C with a 2-nt 3′-overhang. Chemical modifications to A and D prevent Dicer cleavage of the reactants and intermediates, while preserving efficient Dicer processing of transduction product B·C. In functional terms, A·B and C·D detect X, leading to production of DsiRNA B·C targeting Y.
Figure 5.
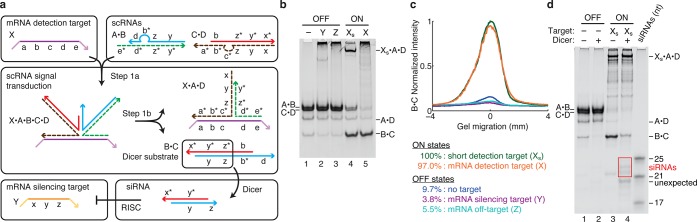
Conditional DsiRNA formation via template-mediated 4-way branch migration. (a) Mechanism 4. scRNAs A·B and C·D detect mRNA detection target X (containing subsequence ‘a-b-c-d-e’), leading to production of DsiRNA Dicer substrate B·C targeting mRNA silencing target Y (containing independent subsequence ‘x-y-z’). scRNAs A·B and C·D coexist metastably in the absence of X. X templates conucleation of A·B and C·D, mediating a short 3-way branch migration that enables toehold/toehold nucleation between B and C to create a 5-way junction (step 1a). Subsequent 4-way branch migration and spontaneous dissociation disassemble DsiRNA Dicer substrate B·C from X·A·D (step 1b). Domain lengths: |a| = 8, |b| = 6, |c| = 6, |d| = 7, |e| = 11, |x| = 2, |y| = 19, |z| = 2. Chemical modifications (2′OMe-RNA): A and D (dashed backbone). (b) Conditional Dicer substrate formation. OFF state: minimal production of Dicer substrate B·C in the absence of detection target X, the presence of mRNA silencing target Y, or the presence of mRNA off-target Z. ON state: strong production of B·C in the presence of short RNA detection target Xs (‘a-b-c-d-e’) or full-length mRNA detection target X. (c) Quantification of the Dicer substrate band (B·C) in panel (b). (d) Conditional Dicer processing. OFF state: minimal processing of the reactants (lane 2). ON state: efficient processing of DsiRNA Dicer substrate B·C (lane 4), yielding canonical 21–24-nt siRNAs (boxed bands). Additional Dicer products are produced by unexpected cleavage of the substrate within domains ‘y’ and ‘y*’. See Section S5 for additional computational and experimental studies of Mechanism 4.
The mechanism exhibits strong OFF/ON conditional Dicer substrate formation, achieving an order of magnitude increase in DsiRNA production in the presence of either the short detection target Xs or the full-length mRNA target X (Figure 5b,c). Only the transduction product B·C is efficiently processed by Dicer, yielding canonical 21–24-nt siRNAs (Figures 5d and S17 and Table S9). Additional Dicer products are produced by unexpected cleavage of the Dicer substrate within domains ‘y’ and ‘y*’, suggesting that for some fraction of the substrates, Dicer is either measuring unusually short siRNAs from the cognate end of the substrate (which has a canonical 2-nt 3′-overhang) or is measuring from the noncognate end of the substrate (which has a 7-nt 3′-overhang). If further studies confirm the latter explanation, Dicer recognition of the noncognate end can be further discouraged by introducing a 5′-overhang.
This mechanism achieves sequence and shape transduction using markedly different design elements than Mechanisms 1–3 (Table 1). Sequence transduction is achieved via the novel approach of templated nucleation, with the two scRNAs A·B and C·D being brought into proximity not via mutual complementarity to each other (as with conventional toehold/toehold or loop/toehold nucleation) but due to complementarity to adjacent segments of another strand—the detection target X, which serves as a template for their nucleation. Templated nucleation provides a simple approach to sequence transduction because by construction, the template sequence (the input) is independent from the sequences of the nucleated duplexes (the output). We are not aware of previous use of nucleic acid templates to mediate conditional strand displacement via either 3- or 4-way branch migration. After the two scRNAs are colocalized via templated nucleation, shape transduction is completed via 4-way branch migration in which the two scRNA duplexes swap base-pairing partners. Initially the scRNAs each undergo short 3-way branch migrations with the template to liberate short mutually complementary toeholds, creating a 5-way junction with the template which resolves into a 4-way branch migration as strand swapping commences. Previous studies demonstrated that 4-way branch migrations are dramatically faster when they are mediated by two toehold/toehold nucleations to create an initial 4-way junction;25 here we adapted this principle to the templated scenario, where each duplex experiences first template/toehold nucleation with X and then toehold/toehold nucleation with each other. By including this auxiliary toehold/toehold nucleation step to enhance branch migration kinetics, we introduced some sequence dependence of duplex B·C on X, which is then removed by Dicer to produce a completely independent siRNA targeting Y. Template-mediated 4-way branch migration provides a simple one-step approach to conditional Dicer substrate formation that provides an intriguing alternative to the more familiar concepts of toehold/toehold nucleation and 3-way branch migration.
Mechanism 5: Conditional shRNA Transcription Using scDNAs
The previous mechanisms explored design alternatives for conditional Dicer substrate hybridization using scRNAs. Here, we consider the alternative strategy of conditional Dicer substrate transcription based on signal transduction with scDNAs. Kim et al.26 have previously demonstrated conditional in vitro transcription mediated by conditional hybridization of a double-stranded DNA promoter sequence. Here, we combine conditional promoter assembly with sequence transduction to implement conditional Dicer substrate transcription. For this test tube design study, T7 RNA polymerase is employed for in vitro transcription, taking advantage of well-characterized promoter and termination sequences.27 Future applications in eukaryotic cells would require use of eukaryotic promoter and termination sequences (e.g., the H1 promoter and poly-T termination sequences for RNA polymerase III, which are commonly used for shRNA transcription).3,28 Mechanism 5 employs two metastable DNA hairpins (A and B of Figure 6a). The detection target X opens hairpin A, which in turn opens hairpin B via a 4-way branch migration to assemble a dsDNA template for transcription of RNA hairpin C (including promoter, coding, and termination sequences). This signal transduction approach incorporates the catalytic turnover inherent in repeated transcription of the template. In functional terms, A detects X leading to transcription of shRNA C targeting Y.
Here, we characterize both the OFF/ON response of conditional transcription template formation and the OFF/ON response of conditional Dicer substrate transcription. For this mechanism, we engineered scDNAs to detect a random short DNA target Xs so we have not characterized performance for a full-length mRNA detection target X. We were nonetheless able to characterize spurious transcription template formation using the full-length mRNA silencing target Y and off-target mRNA Z.
In the absence/presence of short DNA detection target Xs the mechanism demonstrates strong OFF/ON conditional transcription template formation (Figure 6b) and transcription of shRNA Dicer substrate C (Figure 6c), yielding more than an order of magnitude increase in shRNA production (Figure 6d). The transcription product C is efficiently processed by Dicer (Figure 6c and Figure S21), but the poly-U 3′-overhang and short 19-bp stem (consistent with functional shRNAs transcribed with RNA polymerase III in vivo)3,11,28 lead to noncanonical Dicer products (see Section S6.6 for details).
Conclusions
We have engineered five nucleic acid mechanisms for executing the molecular logic: if mRNA detection target X is present, form a Dicer substrate targeting independent mRNA silencing target Y. scRNA sequences encoding the desired shape and sequence transduction properties were designed using the multistate sequence design feature of the NUPACK web application,19 supplying mRNA X (DsRed2) and mRNA Y (d2EGFP) as external sequence constraints. For each transduction mechanism, equilibrium test tube calculations were used to characterize the stability of the designed reactants and to step through the intended molecular assembly and disassembly operations to verify that the targets, reactants, intermediates, and products were predicted to be well-formed with high yield (see Section S1.8). Reflecting the challenge of designing scRNA sequences that are highly constrained by the sequences of mRNAs X and Y, sequence domains that were intended to be perfectly unstructured were often predicted to have some base pairing on average at equilibrium. Experiments confirmed the predicted reactant stability properties, and mechanism stepping experiments confirmed that the intended assembly and disassembly operations occurred with high yield (see Section S1.8).
For each of the five mechanisms, test tube experiments demonstrated a strong OFF/ON conditional response, with at least an order of magnitude increase in Dicer substrate formation in the presence of the cognate full-length mRNA detection target X (for the scDNAs of Mechanism 5, the detection target was not constrained to be an mRNA sequence, so only the designed short DNA detection target Xs was tested experimentally). Reactant structural domains were dimensioned and/or chemically modified to ensure that only the cognate Dicer substrates that were the final products of signal transduction were efficiently processed by Dicer, yielding canonical siRNAs for Mechanisms 1–4 as well as noncanonical Dicer products for Mechanisms 4 and 5.
These mechanism studies explored diverse design principles for shape and sequence transduction via conditional assembly and disassembly of scRNA and scDNA complexes (summarized in Table 1). In broad terms, it appears that varied design concepts that have paced progress in the field of dynamic DNA nanotechnology (including mechanisms for strand nucleation, strand displacement, catalytic hybridization, and motif metastability)18 are equally applicable to dynamic RNA nanotechnology, which is relatively unexplored, yet holds great potential for synthetic regulation in the context of biology; biological RNAs interface with diverse endogenous pathways, and hence synthetic RNA signal transducers that accept RNA inputs and produce RNA outputs represent a particularly appealing framework for engineering conditional regulation in vivo.
The considerable challenge remains of demonstrating robust scRNA signal transduction within living cells. The cellular setting introduces additional uncertainties beyond those addressed here, including the need to deliver the scRNAs and the potential for off-pathway interactions (including with endogenous pathways that are as yet undiscovered). The design versatility demonstrated in the present work will facilitate a flexible approach in engineering around additional design constraints that must be imposed going forward. If scRNA-mediated conditional RNAi eventually performs robustly in vivo, it will provide biologists with a powerful tool for the study of genetic necessity via tissue- and time-specific gene knockdown. The same molecular logic would also have significant medical potential, where transcript X could be chosen to be a diagnostic target and transcript Y could be chosen to be an independent therapeutic target. More generally, if the challenges of operating in vivo can be surmounted, programmable signal transduction with small conditional RNAs will provide an enticing conceptual framework for implementing diverse modes of conditional regulation.
Methods Summary
Sequences were designed and analyzed using the NUPACK web application,19 and oligonucleotides were synthesized by IDT. Target mRNAs were transcribed in vitro. Duplex scRNAs were purified by native PAGE prior to use. To quantify conditional OFF/ON signal transduction response, reactions were run in 100 mM potassium acetate, 20 mM HEPES, pH 7.5 (Mechanisms 1–4), or 50 mM Na2HPO4, 0.5 M NaCl, pH 7.5 (Mechanism 5) at 37 °C for 2 h (0.5 μM per strand, with concentrations adjusted relative to Xs to ensure proper stoichiometry). Reactions were then characterized using native PAGE with SYBR Gold poststaining. Dicer processing was characterized using the Recombinant Human Turbo Dicer Enzyme kit (Genlantis), native PAGE, and mass spectrometry. For Mechanisms 1–4, scRNA transduction and Dicer processing were performed simultaneously (0.5 μM per strand, incubated with 0.5–1 units of recombinant Dicer at 37 °C for 2 h) and then characterized by native PAGE poststained with SYBR Gold. For Mechanism 5, scDNA transduction and T7 in vitro transcription were performed simultaneously (0.1 μM per strand, 37 °C for 3 h in the presence of radioactive UTP). The transcription product was purified, incubated with 1 unit of recombinant Dicer per 20,000 cpm at 37 °C for 2 h and then characterized by native PAGE visualized by phosphorimaging. See Section S1 for full details.
Acknowledgments
We thank J. J. Rossi for helpful discussions, C. R. Calvert for performing preliminary studies, and B. R. Wolfe, J. N. Zadeh, and R. M. Dirks for the use of unpublished multistate sequence design software. We thank M. Kirk for assistance with figure preparation. This work draws on design principles developed within the NSF Molecular Programming Project (NSF-CCF-0832824) and was funded by the National Institutes of Health (NIH 5R01CA140759), the Gordon and Betty Moore Foundation (GBMF2809), and the Elsa U. Pardee Foundation.
Supporting Information Available
Methods and materials as well as additional studies for each mechanism: computational stepping analysis, mechanism stepping gel, Dicer processing stepping gel, quantification of Dicer substrate production, mass spectrometry of Dicer products. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
∇ Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, United States; Department of Systems Biology, Harvard Medical School, Boston, MA 02115, United States.
Author Contributions
# These authors contributed equally.
The authors declare the following competing financial interest(s): US patents and pending US patents.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kim D. H.; Rossi J. J. Nat. Rev. Genet. 2007, 8, 173–184. [DOI] [PubMed] [Google Scholar]
- Kurreck J. Angew. Chem., Int. Ed. 2009, 48, 1378–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M.; Szulc J.; Trono D. Nat. Methods 2006, 3, 682–688. [DOI] [PubMed] [Google Scholar]
- Lee S. K.; Kumar P. Adv. Drug Delivery Rev. 2009, 61, 650–664. [DOI] [PubMed] [Google Scholar]
- An C. I.; Trinh V. B.; Yokobayashi Y. RNA 2006, 12, 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C. L.; Bayer T. S.; Hoff K. G.; Smolke C. D. Mol. Syst. Biol. 2008, 4, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuleuova N.; An C. I.; Ramanculov E.; Revzin A.; Yokobayashi Y. Biochem. Biophys. Res. Commun. 2008, 376, 169–173. [DOI] [PubMed] [Google Scholar]
- Kumar D.; An C. I.; Yokobayashi Y. J. Am. Chem. Soc. 2009, 131, 13906–13907. [DOI] [PubMed] [Google Scholar]
- Beisel C. L.; Chen Y. Y.; Culler S. J.; Hoff K. G.; Smolke C. D. Nucleic Acids Res. 2011, 39, 2981–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D.; Lerner C.; Burchard J.; Ge W.; Linsley P. S.; Paddison P. J.; Hannon G. J.; Cleary M. A. Nat. Biotechnol. 2005, 23, 227–231. [DOI] [PubMed] [Google Scholar]
- Mcintyre G. J.; Yu Y. H.; Lomas M.; Fanning G. C. BMC Mol. Biol. 2011, 12, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H.; Behlke M. A.; Rose S. D.; Chang M.-S.; Choi S.; Rossi J. J. Nat. Biotechnol. 2005, 23, 222–226. [DOI] [PubMed] [Google Scholar]
- Provost P.; Dishart D.; Doucet J.; Frendewey D.; Samuelsson B.; Radmark O. EMBO J. 2002, 21, 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M.; Doudna J. A. Nature 2009, 457, 405–412. [DOI] [PubMed] [Google Scholar]
- Masu H.; Narita A.; Tokunaga T.; Ohashi M.; Aoyama Y.; Sando S. Angew. Chem., Int. Ed. 2009, 48, 9481–9483. [DOI] [PubMed] [Google Scholar]
- Xie Z.; Liu S. J.; Bleris L.; Benenson Y. Nucleic Acids Res. 2010, 38, 2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D.; Kim S. H.; Yokobayashi Y. J. Am. Chem. Soc. 2011, 133, 2783–2788. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y.; Seelig G. Nat. Chem. 2011, 3, 103–113. [DOI] [PubMed] [Google Scholar]
- Zadeh J. N.; Steenberg C. D.; Bois J. S.; Wolfe B. R.; Pierce M. B.; Khan A. R.; Dirks R. M.; Pierce N. A. J. Comput. Chem. 2011, 32, 170–173. [DOI] [PubMed] [Google Scholar]
- Zadeh J. N.; Wolfe B. R.; Pierce N. A. J. Comput. Chem. 2011, 32, 439–452. [DOI] [PubMed] [Google Scholar]
- Dirks R. M.; Lin M.; Winfree E.; Pierce N. A. Nucleic Acids Res. 2004, 32, 1392–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P.; Choi H. M. T.; Calvert C. R.; Pierce N. A. Nature 2008, 451, 318–322. [DOI] [PubMed] [Google Scholar]
- Yurke B.; Turberfield A. J.; Mills A. P. Jr.; Simmel F. C.; Neumann J. L. Nature 2000, 406, 605–608. [DOI] [PubMed] [Google Scholar]
- Venkataraman S.; Dirks R. M.; Rothemund P. W. K.; Winfree E.; Pierce N. A. Nat. Nanotechnol. 2007, 2, 490–494. [DOI] [PubMed] [Google Scholar]
- Dabby N. L., Synthetic molecular machines for active self-assembly: prototype algorithms, designs, and experimental study, Ph. D. Thesis, California Institute of Technology, 2013. [Google Scholar]
- Kim J.; White K. S.; Winfree E. Mol. Syst. Biol. 2006, 2, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J.; Studier F. W. J. Mol. Biol. 1983, 166, 477–535. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T. R.; Bernards R.; Agami R. Science 2002, 296, 550–553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.