Abstract
Bone marrow-derived macrophages (BMMs) treated with granulocyte-macrophage colony-stimulating factor (GM-CSF) or macrophage colony-stimulating factor (M-CSF), differentiate into GM-CSF-induced mouse bone marrow-derived macrophages (GM-BMMs) or M-CSF-induced mouse bone marrow-derived macrophages (M-BMMs), which have an M1 or M2 profile, respectively. GM-BMMs produce large amounts of proinflammatory cytokines and mediate resistance to pathogens, whereas M-BMMs produce anti-inflammatory cytokines that contribute to tissue repair and remodeling. M-BMMs stimulated with lipopolysaccharide (LPS) are in an antiinflammatory state, with an IL-12low IL-10high phenotype. However, the regulation of this process remains unclear. Klf10 belongs to the family of Krüppel-like transcription factors and was initially described as a TGF-β inducible early gene 1. IL-12p40 is upregulated in LPS-stimulated M-BMMs from Klf10-deficient mice, but downregulated during Klf10 overexpression. Klf11, another member of the Krüppel-like factor family, can also repress the production of IL-12p40. Furthermore, Klf10 binds to the CACCC element of the IL-12p40 promoter and inhibits its transcription. We have therefore identified Klf10 as a transcription factor that regulates the expression of IL-12p40 in M-BMMs.
Keywords: IL-12p40, Inflammatory factor, Klf10, Macrophage, Transcription factor
Introduction
Macrophages are critical in inflammation, tissue regeneration, and tolerance. Macrophages can be generated from bone marrow cells treated with granulocyte-macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) [1, 2] and then induced to become GM-CSF-induced mouse bone marrow-derived macrophage (GM-BMMs) or M-CSF-induced mouse bone marrow-derived macrophages (M-BMMs), which have a M1 (classic activated macrophages) or M2 (alternative activated macrophages) profile. Cytokines are also involved in macrophage polarization. M1 macrophages are induced by IFN-γ, with or without lipopolysaccharides (LPS), whereas M2 macrophages are generated through IL-4 or IL-13 stimulation [1, 3].
GM-BMMs and M-BMMs have different patterns of cytokine expression. GM-BMMs produce large amounts of nitric oxide (NO) and proinflammatory cytokines involved in resistance to pathogens, whereas M-BMMs produce fewer proinflammatory cytokines but more antiinflammatory cytokines responsible for tissue repair and tumor progression [1–3]. However, the transcription factors that regulate macrophage polarization remain largely undefined. IRF5 has been reported to promote the expression of M1-related genes [4], whereas IRF4 and Klf4 can control M2 macrophage polarization by regulating the expression of specific M2 markers [5, 6]. In addition, LPS-stimulated M-BMMs are in an antiinflammatory state with an IL-12lowIL-10high phenotype [7]. Therefore, regulation of inflammatory cytokines such as IL-12 is important in maintaining the steady state of M-BMMs.
IL-12 (IL-12p70), a heterodimeric cytokine comprising the p40 and p35 subunits, is an important cytokine produced mainly by antigen-presenting cells and can regulate innate responses during infection [8]. IL-12 can also induce interferon-γ production and trigger CD4+ T-cell differentiation into type 1 T helper (Th1) cells [9]. Moreover, IL-12 is a phenotypic marker for GM-BMMs [4] and the ratio of IL-12 to IL-10 production is often used to define GM-BMMs and M-BMMs [2]. Macrophages derived from IL-12p40-deficient mice have a bias toward M2 polarization [10]. IL-12p40, a subunit shared by IL-12 and IL-23, is produced predominantly by activated monocytes, macrophages, and dendritic cells. Higher levels of the IL-12p40 subunit is produced than IL-12 and IL-23 heterodimers [11], the production of which is regulated by strict mechanisms. NF-κB family members are activated in the production of IL-12p40 [12]. Several IFN-regulatory factors (IRFs) such as IRF5 and IRF8 are involved in IL-12p40 expression [13, 14]. Moreover, the CCAAT/enhancer-binding protein (C/EBP) family, including C/EBP α and C/EBP β, inhibits the production of IL-12p40 [15, 16].
The Krüppel-like factors (KLFs) are a family of transcriptional regulators with a highly conserved DNA-binding domain that consists of three C2H2-type zinc fingers capable of binding to a CACCC element or GC box consensus sequences [17, 18]. KLFs play different roles in biology through their divergent non-DNA-binding regions that function as trans-activation or trans-repression domains. A total of 17 members of mammalian KLFs have been identified thus far [19], some are found to play important roles in immune and hematopoietic cell biology by regulating gene transcription. For example, Klf1 (erythroid Krüppel-like factor) regulates β-globin expression during erythrocyte development [20, 21] and also affects IL-12p40 production in human macrophages [22]. Klf4 has been reported as a key regulator in monocyte differentiation and macrophage activation [23–25]. Recent studies further demonstrated Klf4 as a novel regulator in M2 macrophage polarization [5].
Klf10 belongs to the KLF family and was initially identified in human osteoblasts as a TGF-β responsive gene [26]. Thus, Klf10 is also called TGF-β inducible early gene 1 (TIEG1) [26]. Osteoblasts from Klf10-deficient mice have been reported as defective in mineralization and in supporting osteoclast differentiation in vitro [27]. Subsequent studies demonstrated that Klf10 is also essential in T-cell biology. Klf10 cooperates with Itch to regulate Foxp3 expression [28] and also regulates CD4+CD25− T cells and Treg cells by targeting TGF-β [29]. TGF-β inhibits several LPS-induced inflammatory cytokines in macrophages [30] and contributes to resolve inflammation. Recent studies revealed that TGF-β also contributes to M2 macrophage polarization [2]. However, as a TGF-β-induced gene, the function of Klf10 in innate immune cells such as macrophages has not been studied thus far.
Here, we demonstrate the role of Klf10 in regulating the production of inflammatory cytokines in M-BMMs. We found that Klf10 expression was downregulated upon TLR activation. The forced expression and loss function assay of Klf10 in M-BMMs revealed a repressive effect on IL-12p40. Moreover, we also observed a similar role for Klf11 as that of Klf10 in regulating IL-12p40 expression. Studies on this mechanism demonstrated that Klf10 inhibits the production of IL-12p40 by binding to the IL-12p40 promoter. Therefore, our observations support the importance of Klf10 as a key transcriptional repressor of inflammatory cytokines in M-CSF-induced macrophages.
Results
Klf10 expression is downregulated upon TLR activation
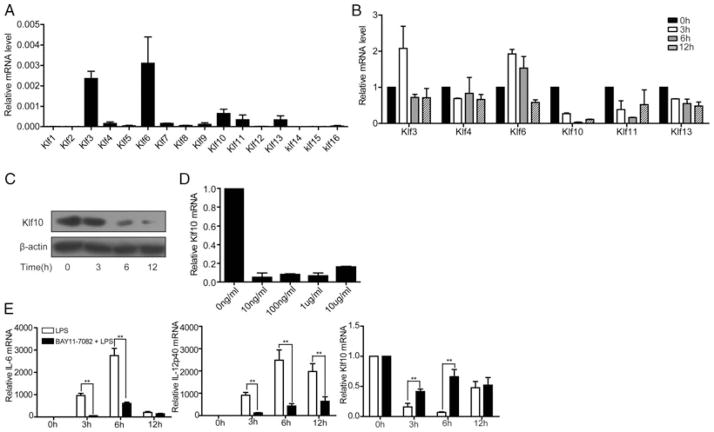
Quantitative PCR (qPCR) analysis for the expression of the KLF family members in M-BMMs was conducted to determine whether the KLF family members can control the inflammatory factors in M-BMMs. The result shows that Klf3, Klf4, Klf6, Klf10, Klf11, and Klf13 have high mRNA level among all family members (Fig. 1A). We performed time course LPS treatment for M-BMMs to verify the response of these KLFs to innate immune stimuli. The qPCR results indicate that Klf3, Klf4, Klf6, and Klf13 exhibited a minor or no increase, whereas Klf10 and Klf11 significantly decreased (Fig. 1B). In addition, KLF expression and response to LPS were investigated in GM-BMMs, and the result was similar to that in M-BMMs (Supporting Information Fig. 1). The decline in Klf10 expression in M-BMMs was further verified by western blot analysis (Fig. 1C). This Klf10 downregulation can be induced by LPS even with a concentration as low as 10 ng/mL (Fig. 1D). LPS is a ligand for TLR4, which localizes on the cell surface. Klf10 expression also decreased when TLR3 and TLR9, located in intracellular vesicles [31], were activated by poly I:C and CpG (Supporting Information Fig. 2). TLR stimulation can result in NF-κB activation, and our observation reveals that Klf10 can respond sensitively to these TLRs. Klf10 is an NF-κB-targeted gene [32]. Thus, we further demonstrate that Klf10 was downregulated in an NF-κB-dependent manner. We pretreated M-BMMs with BAY11–7082, an IκB-α inhibitor, to repress the NF-κB pathway and found that the decrease in Klf10 after LPS challenge can be abrogated (Fig. 1E). Meanwhile, the upregulation of inflammatory cytokines, such as IL-12p40 and IL-6, was abolished (Fig. 1E). These results indicate that klf10 may participate in TLRs and may control the production of inflammatory factors in M-BMMs.
Figure 1.
Downregulation of Klf10 expression upon TLR4 activation by LPS in M-BMMs. (A) The mRNA levels of KLFs in M-BMMs. M-BMMs on day 5 were harvested and the gene expression levels of KLFs were assessed by qPCR. The actin mRNA level in these cells was set as 1. (B) Response of KLFs to LPS stimulation in M-BMMs. M-BMMs on day 5 were treated with 1 μg/mL LPS for the indicated time, then gene expression levels were assessed by qPCR and normalized to those in untreated cells. (C) Western blot analysis of Klf10 in M-BMMs treated with 1 μg/mL LPS for the indicated time. β-Actin was used as a loading control. Data shown are representative of three experiments performed. (D) qPCR analysis of Klf10 in M-BMMs treated with graded concentrations of LPS. (E) The role of the NF-κB pathway in the downregulation of Klf10 in LPS activated M-BMMs. M-BMMs on day 5 were pretreated with 5 μM BAY 11–7082 or without pretreatment, and then stimulated with 100 ng/mL LPS for the indicated time. Cells were harvested for qPCR analysis. (A–E) Data are shown as mean ± SD of three samples pooled from three independent experiments. **p < 0.01, determined by Student’s t-test.
Klf10 overexpression reduces production of IL12p40 in M-BMMs
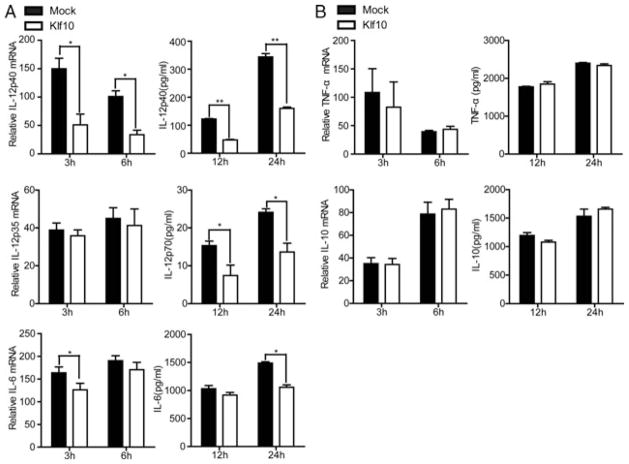
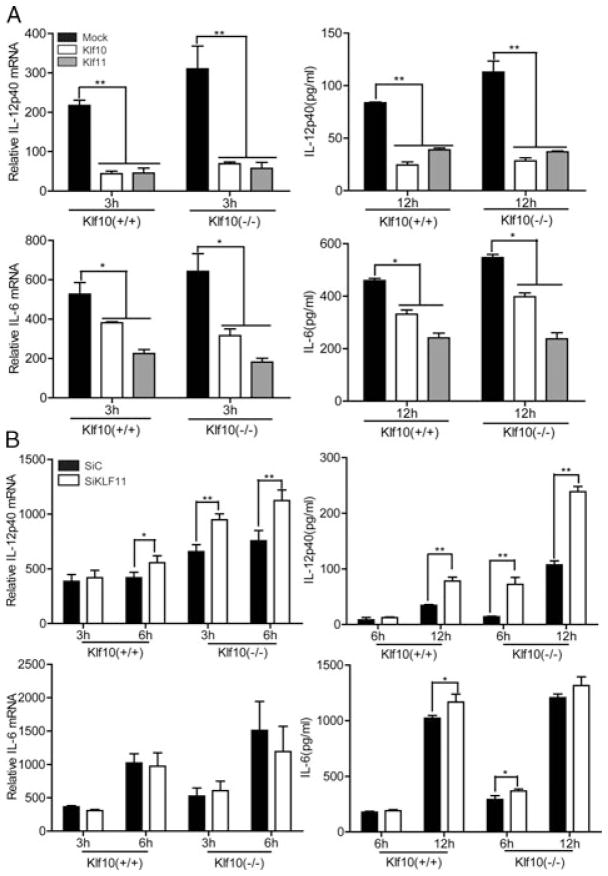
Klf10 was overexpressed in M-BMMs to investigate whether it is involved in the regulation of inflammatory cytokines triggered by TLR4 signaling. The result shows that LPS-induced IL-12p40 was significantly inhibited at both the mRNA and protein levels, which also resulted in a decrease in IL-12p70. However, IL-12p35, the other subunit of IL-12p70, was unaffected (Fig. 2A). Other proinflammatory mediators, such as IL-6 and TNF-α, were slightly affected or unaffected by Klf10 (Fig. 2A and B). IL-10 is a key antiinflammatory factor that can suppress IL-12 and IL-6 expressions in M-BMMs. Thus, we found Klf10 had no effect on IL-10 (Fig. 2B), indicating that the suppression of IL-12p40 and IL-6 was not mediate by IL-10. These observations indicated that Klf10 overexpression inhibited the production of IL-12p40 induced by TLR4 signaling in M-BMMs.
Figure 2.
Klf10 overexpression facilitates a reduction in the LPS-induced production of IL-12p40 and IL-6 in M-BMMs. (A and B) M-BMMs on day 5 were transfected with pcDNA3-Klf10 or empty vector by using the Amaxa mouse macrophage Nucleofector kit. The cells were cultured for 24 h, stimulated with 1 μg/mL LPS, and then harvested at the indicated times. Relative mRNA expression of (A) IL-12p40, IL-12p35, IL-6, and (B) TNF-α, and IL-10 was measured by qPCR and normalized to levels in untreated cells (left). Supernatants from the cultures were collected, and the levels of (A) IL-12p40, IL-12p70, IL-6, and (B) TNF-α and IL-10 were evaluated by ELISA (right). Data are shown as mean ± SD of three samples pooled from three independent experiments. *p < 0.05, **p < 0.01; determined by Student’s t-test.
Loss of Klf10 upregulates the production of IL-12p40 in M-BMMs
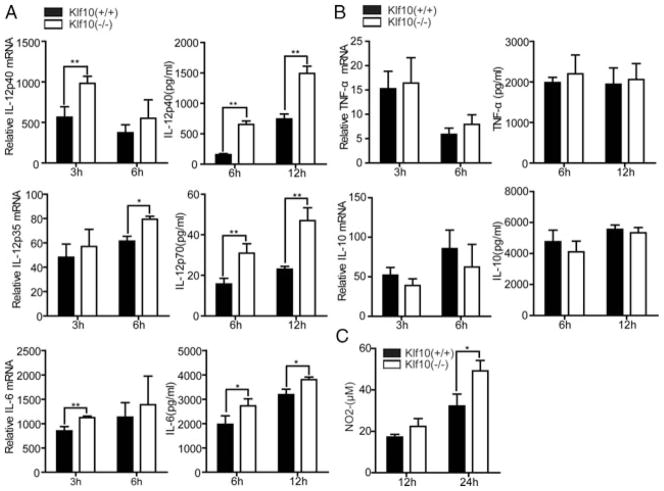
We further performed the loss of function assay with Klf10-deficient mice to verify the aforementioned observation. Surface markers of M-BMMs from WT and Klf10-deficient mice were first characterized by flow cytometry. The result reveal that the proportion of F4/80+CD11b+ mature M-BMMs did not differ between these two markers, indicating that Klf10 was not involved in the differentiation of M-BMMs (Supporting Information Fig. 3A). Moreover, we investigated the markers on M-BMMs such as costimulatory molecules CD80, CD86, TLR4 receptor, and MHC class II, and found that these markers were expressed normally (Supporting Information Fig. 3B). However, when we analyzed the inflammatory factors from these cells after LPS stimulation, IL-12p40 exhibited a significant upregulation at both the mRNA and protein levels in Klf10-deficient cells compared with that in WT cells (Fig. 3A). Meanwhile, the total IL-12p70 upregulated although the IL-12p35 subunit increased slightly, which was consistent with the result obtained from the klf10 over-expression assay (Fig. 3A). IL-12p40 is known to contribute to the production of NO [10] that is an important effect molecule of GM-BMMs, while here we found M-BMMs from klf10-deficient mouse released more NO as well (Fig. 3C), which may indicate a bias toward GM-BMM activation. The production of TNF-α and IL-10 did not exhibit a significant difference in M-BMMs from WT and Klf10-deficient mice, except for the slight increase of IL-6 (Fig. 3A and B). Type I interferon is reported to downregulate IL-12 expression [33,34], while we found that expression of IFN-β was not evidently changed in our assay (Supporting Information Fig. 4). TGF-β is an inhibitor of IL-12 production [35], and its expression in Treg cells is downregulated in Klf10-deficient mice [29]. However, we did not observe a decrease in TGF-β expression in M-BMMs of Klf10-deficient mice. The expression of several other specific markers linked with M-BMMs [36], such as CCL2, CCL5, CCL12, and CXCL10, were also investigated, and we found only a slight increase in CCL2 and CCL5 (Supporting Information Fig. 4). Moreover, we verified several markers in M-BMMs stimulated with IL-4, such as arginase-1 (Arg1), chitinase-like Ym1 (Chi3l3), found in inflammatory zone-1 (Fizz1, also called Retnla) and mannose receptor (Mrc1 encoding MR), and observed that they had similar expression levels in klf10-deficient and WT mice (data not shown). These results indicate that klf10 was not involved in the differentiation of M-BMMs but can repress the expression of IL-12p40 and IL-6 in these cells.
Figure 3.
Loss of Klf10 promotes the production of IL-12p40, IL-6, and NO in M-BMMs. (A and B) M-BMMs on day 5 from WT or Klf10-deficient mouse were stimulated with 1 μg/mL LPS. Cells were harvested at the indicated times. Relative mRNA expression of (A) IL-12p40, IL-12p35, IL-6, and (B) TNF-α and IL-10 were measured by qPCR and normalized to levels in untreated cells (left). Supernatants from the cultures were collected, and the levels of (A) IL-12p40, IL-12p70, IL-6, and (B) TNF-α and IL-10 were evaluated by ELISA (right). (C) NO levels in the supernatants were determined via the Griess reaction. Data are shown as mean ± SD of three samples pooled from three independent experiments. *p < 0.05, **p < 0.01; determined by Student’s t-test.
Loss of Klf10 does not affect the production of IL-12p40 in GM-BMMs
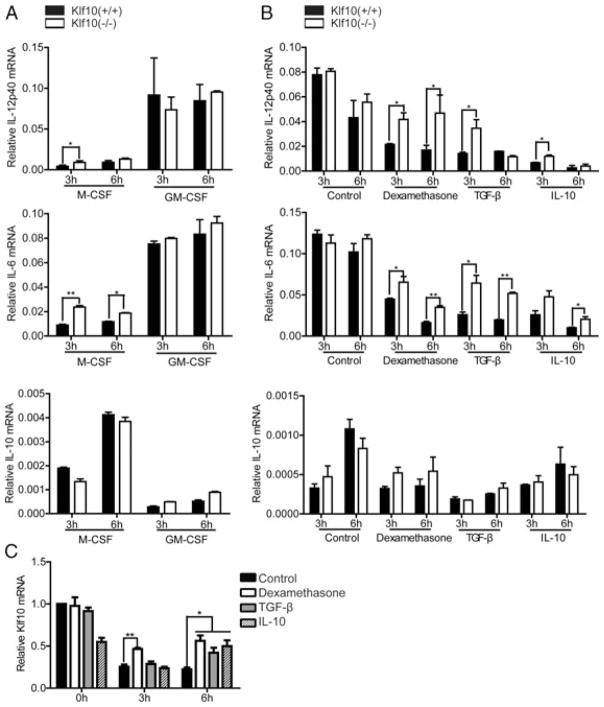
GM-BMMs can make more inflammatory factors than M-BMMs. Similar to other studies [7, 37, 38], we found that GM-BMMs produces more IL-12p40 and IL-6, but less IL-10 than M-BMMs (Fig. 4A). However, no differences were observed in the production of IL-12p40 and IL-6 in LPS-stimulated GM-BMMs between WT and Klf10-deficient mice compared with M-BMMs (Fig. 4A). Meanwhile, IFN-γ is reported as a priming agent for the enhancement of IL-12p40 production [14, 39]. The upregulation of IL-12p40 in Klf10-deficient mice compared with WT mice was also not found in GM-BMMs under the IFN-γ priming conditions (data not shown).
Figure 4.
Different functions of Klf10 in M-BMMs and GM-BMMs. (A) Different expression patterns of IL-12p40, IL-6, and IL-10 in GM-BMMs and M-BMMs. Bone marrow cells from WT or Klf10-deficient mouse were cultured with either 20 ng/mL GM-CSF or 10 ng/mL M-CSF for macrophage polarization. On day 5, cells were stimulated with 1 μg/mL LPS. Cells were harvested at the indicated time, and the relative mRNA expression levels of IL-12p40, IL-6, and IL-10 were measured by qPCR and normalized to levels in untreated cells. (B) GM-BMMs on day 5 were pretreated with 2.5 × 10−7 M dexamethasone, 2 ng/mL TGF-β, or 10 ng/mL IL-10 for 16 h for deactivation, and then stimulated with 1 μg/mL LPS. Cells were harvested at the indicated time, and the relative mRNA expression levels of IL-12p40, IL-6, and IL-10 were measured by qPCR and normalized to those in untreated cells. (C) The expression of Klf10 in GM-BMMs from WT mouse in (B) was measured by qPCR. Data are shown as mean + SD of three samples pooled from three independent experiments. *p < 0.05, **p < 0.01; determined by Student’s t-test.
The expression of Klf10 in M-BMMs and GM-BMMs was first analyzed to investigate the reason for the aforementioned observations. However, no obvious difference was observed between them (Supporting Information Fig. 5). As shown above, GM-BMMs produce more inflammatory cytokines than M-BMMs. Deactivation of GM-BMMs was performed by using several inflammation inhibitors, such as dexamethasone, TGF-β, and IL-10, as previously described [40, 41], to investigate whether Klf10 functions in intense inflammation. The result shows that the expression of relevant cytokines decreased after deactivation. In addition, the expression of IL-12p40 and IL-6 was higher in GM-BMMs from Klf10-deficient mice than that from WT mice after deactivation (Fig. 4B). Moreover, the downregulation of Klf10 was abolished to some extent (Fig. 4C), which may enhance its inhibitive function on the cytokines. These data may indicate that Klf10 alone is insufficient to inhibit the inflammatory factors in GM-BMMs; other factors are possibly involved in the suppression of inflammation in deactivated GM-BMMs.
Klf11 plays similar roles as Klf10 in repressing IL-12p40 production
Klf11, another member of the KLF family, was also identified as a downregulated gene in LPS-stimulated M-BMMs. Klf11 is described as a TGF-β inducible early gene 2 and shares a highly conserved C-terminal DNA-binding domain with Klf10 [18]. In addition, Klf10 and Klf11 have three repression domains in the N-terminal, which define them as a subfamily of repressor. We supposed that Klf11 may have a similar role in the regulation of inflammatory factors in M-BMMs. First, we found that overexpression of both Klf10 and Klf11 can inhibit the production of IL-12p40 and IL-6 in M-BMMs from WT mice and can rescue their overproduction in M-BMMs from Klf10-deficient mice (Fig. 5A). Therefore, we knocked down Klf11 through RNA-mediated interference. The efficiency of RNAi was confirmed by qPCR (Supporting Information Fig. 6A). The inhibition of Klf11 resulted in an increased production of IL-12p40 in WT cells, and this phenomenon was more notable in Klf10-deficient cells (Fig. 5B). Therefore, Klf10 and Klf11 may have a similar function in the regulation of IL-12p40. However, interference of Klf11 in the same conditions did not result in a significant change of IL-6 as that in the overexpression assay. Moreover, we used another SiRNA, as previously reported [42], to confirm the inhibitory function of Klf11 in the regulation of IL-12p40 (Supporting Information Fig. 6B and C). These data indicated that Klf11 can act similarly to Klf10 in the inhibition of IL-12p40 production.
Figure 5.
Similar function of Klf11 and Klf10 in the regulation of IL-12p40 expression.(A) Overexpression of Klf10 or Klf11 reduced the LPS-induced production of IL-12 p40 and IL-6 in M-BMMs from WT and Klf10-deficient mice. M-BMMs on day 5 were transfected with pcDNA3-Klf10, pcDNA3-Klf11, or empty vector (Mock), cultured for 24 h and then stimulated with 1 μg/mL LPS. Cells were harvested at the indicated times, and relative mRNA expressions of IL-12p40 and IL-6 was measured by qPCR and normalized to levels in untreated cells (left). Supernatants from the cultures were collected and the levels of IL-12p40 and IL-6 were evaluated by ELISA (right). (B) Silencing of Klf11 expression promoted the production of LPS-induced IL-12p40 in M-BMMs from WT and Klf10-deficient mice. On day 5, M-BMMs were transfected with Klf11 SiRNA or control SiRNA, cultured for 36 h, and then stimulated with 1 μg/mL LPS. Cells were harvested at the indicated times, and relative mRNA expression of IL-12p40 and IL-6 was measured by qPCR and normalized to levels in untreated cells (left). Supernatants from the cultures were collected, and the levels of IL-12p40 and IL-6 were evaluated by ELISA (right). Data are shown as mean ± SD of three samples pooled from three independent experiments. *p < 0.05, **p < 0.01, determined by Student’s t-test.
Klf10 binds to the IL-12p40 promoter to inhibit its transcriptional activity
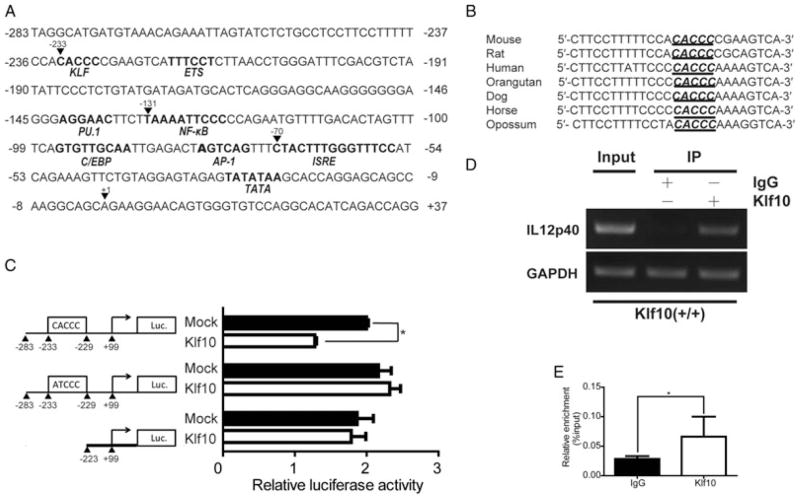
The KLF family members are characterized by a DNA-binding domain capable of binding to target genes to regulate their transcriptional activities and gene expressions. IL-12p40 promoter was sequenced to determine whether Klf10 can regulate the expression of IL-12p40 in a direct manner and a CACCC site was found therein (Fig. 6A). The binding site was highly conserved in mammals (Fig. 6B), similar to the CACCC-binding site of erythroid Krüppel-like factor in human macrophages. Subsequently, a series of luciferase reporter construct that can encode a WT IL-12p40 promoter (−283 to +99 bp), a mutant with 2-bp mutations in the CACCC site (at −233 bp), or a 5′ deletion promoter construct (−223 bp) were constructed to investigate whether Klf10 can repress the transcription of IL-12p40. We found that Klf10 can significantly inhibit the transcriptional activity of the IL-12p40 promoter, but was unable to suppress the luciferase activity of these two mutants (Fig. 6C), suggesting that Klf10 may inhibit IL-12p40 by binding directly to the CACCC site of the promoter. ChIP assays were performed to determine whether Klf10 was recruited to the CACCC element of IL-12p40 promoter. Semi-qPCR and qPCR results verify that Klf10 can bind to the CACCC site of the IL-12p40 promoter (Fig. 6D and E). Therefore, we demonstrate that Klf10 inhibited the transcriptional activity of IL-12p40 by binding directly to the CACCC site of the IL12p40 promoter.
Figure 6.
Klf10 binds to the promoter of IL12p40 and inhibits its transcriptional activity. (A) Putative and verified transcription factor-binding sites in the mouse IL-12p40 promoter region. The core-binding sequences are shown in bold. The positions are relative to the transcription initiation site (TIS) that is defined as +1. (B) High conservation of the CACCC-binding site in IL-12p40 promoter in mammals. (C) Klf10 inhibits the luciferase activity of the IL-12p40 promoter. HEK 293 cells were infected for 48 h with a WT IL-12p40 luciferase reporter (top), an IL-12p40 luciferase reporter with 2 bp mutations in the CACCC site at positions −233 to −229 (middle), or an IL-12p40 luciferase reporter that lack the CACCC-binding domain (bottom), plus a construct that encodes Klf10 or empty vector (mock), and then cells were harvested and analyzed for luciferase activity. (D–E) PCR and qPCR assays of the recruitment of Klf10 proteins to the IL-12p40 promoter. On day 5, M-BMMs were prepared and subjected to ChIP with antibody to IgG or Klf10. The immunoprecipitated DNA were analyzed by PCR and qPCR. The qPCR results were presented relative to those obtained with genomic DNA (input). Results shown are (D) representative of three independent experiments or (C, E) presented as mean + SD of three samples pooled from three independent experiments. *p < 0.05; determined by Student’s t-test.
Discussion
Macrophages are important mediators in immune responses to inflammation. The remarkable plasticity of macrophages has recently been the subject of intense investigation. M-CSF and GM-CSF are mediators involved in the regulation of macrophage heterogeneity. Macrophages induced by GM-CSF and stimulated with IFN-γ and LPS are characterized by a high expression of inflammatory cytokines and iNOS. By contrast, macrophages induced by M-CSF and then stimulated with IL-4 are responsible for the resolution of inflammation. Controlling the expression of inflammatory factors is critical in maintaining the antiinflammatory state in M-CSF-induced macrophages.
KLFs are important zinc finger transcription factors that can regulate the transcriptional activity of target genes, thereby affecting their expression. So far, Klf4 has been demonstrated to be critical during macrophage differentiation. Klf4 is expressed in a monocyte-restricted and stage-specific pattern during myelopoiesis [23]. Recent studies identified Klf4 as a key regulator in M2 macrophage polarization [5]. Klf4 is also related to macrophage activation. Klf4 overexpression can induce macrophage activation marker iNOS and inhibit TGF-β1 and Smad3 signaling [25].
Klf10, initially identified and named as TGF-β inducible early gene 1 in human osteoblasts [26], has been reported to have a critical role in T-cell biology [28,29]. In this study, we demonstrated that Klf10 functions as a specific repressor to IL-12p40 in M-BMMs, whereas the expression of other cytokines, such as TNF-α and IL-10, were not obviously affected. IL-12p40 is a subunit shared by IL-12p70 and IL-23, and its regulation is important for both innate and adaptive immunity. IL-10 can suppress IL-12 by inhibiting the transcription of its encoding genes [43]. TGF-β is also an inhibitor of IL-12 production through the reduction of the stability of IL-12 p40 mRNA [35]. Type I interferons, such as IFN-α and IFN-β, can also inhibit the production of IL-12 [33]. However, the aforementioned cytokines that regulate IL-12p40 were unaffected by Klf10 in our results. In addition, some transcription factors, such as IRF5, IRF8, C/EBP α, and C/EBP β, regulate the expression of IL-12p40. We found that the expression of these factors was not obviously affected in Klf10-deficient mice (data not shown). Therefore, Klf10 may directly regulate the expression of IL-12p40 in transcriptional levels. Most KLFs can bind to a CACCC element or a GC box sequence to regulate the transcription of target genes. We found a highly conserved CACCC element in the promoter of IL-12p40. Further studies through ChIP and luciferase reporter assays showed that Klf10 can bind to the CACCC site and inhibit the transcription of IL-12p40.
Klf10 is initially identified as a TGF-β responsive gene, and previous studies focused mainly on its roles in the TGF-β signaling pathway. Our study was the first to demonstrate the function of Klf10 in repressing IL-12p40 in M-BMMs upon TLR activation. TLRs can trigger intracellular signaling pathways, upregulate the expression of inflammatory factors and further contribute to the killing of microorganisms [31]. Meanwhile, the TGF-β pathway is chiefly responsible for repressing the levels of inflammatory cytokines to maintain tolerance and to resolve inflammation [44]. Although a deficiency in the expression of TGF-β in Treg cells from Klf10-deficient mice was reported, we did not observe a decrease in the expression of TGF-β in M-BMMs from Klf10-deficient mice (Supporting Information Fig. 4). This result indicates that Klf10 is unimportant in maintaining the expression of TGF-β in M-BMMs. TGF-β1 is a key factor involved in endotoxin tolerance, whereas smad3 and smad4 are also required in endotoxin tolerance [45,46]. However, no obvious difference was observed between WT and Klf10-deficient cells in LPS-mediated endotoxin tolerance (Supporting Information Fig. 7). Therefore, Klf10 can inhibit the production of IL-12p40 in M-BMMs, which may not rely on the TGF-β pathway to some extent.
In conclusion, we demonstrate that Klf10 can repress the expression of IL-12p40 in M-CSF-induced macrophages and may help maintain the steady antiinflammatory state of such macrophages.
Materials and methods
Mice and reagents
C57BL/6 mice were purchased from Shanghai Slac Animal Inc. (Shanghai, China). Klf10-deficient mouse were originally from the laboratory of Dr. Thomas Spelsberg (Mayo Clinic, MN, USA). Mice were maintained in Experimental Animal Center of Zhejiang University. Experiments and animal care were performed in accordance with the guidelines of Zhejiang University. LPS (Escherichia coli 055:B5) and Poly I:C (P1038) were obtained from Sigma (St. Louis, MO, USA). Phosphorothioate-CpG ODN (5′-TCC ATG ACG TTC CTG ACG TT-3′) was synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Antibodies against Klf10 (sc-130408, sc-34544 X) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HRP-conjugated anti-mouse IgG (7076) were from Cell Signaling Technology. Anti-mouse CD11b (M1/70), anti-mouse F4/80 (BM8), anti-mouse MHC Class II (M5/114.15.2), anti-mouse TLR4 (UT41), anti-mouse CD80 (16–10A1), and anti-mouse CD86 (GL1) antibodies were purchased from eBioscience (San Diego, CA, USA). The pGL-3 luciferase and pRL-TK-Renilla luciferase plasmids were from Promega (Fitchburg, WI, USA).
Plasmid constructs
Recombinant vector encoding mouse Klf10 (mKlf10, GenBank Accession number NM 013692.2) and mouse Klf11 (mKlf11, Gen-Bank Accession number NM 178357.3) were constructed by PCR-based amplification and subcloned into the pcDNA3 eukaryotic expression vector (Invitrogen, Carlsbad, CA, USA).
The primers were as followed:
Klf10-pcDNA3: GAATTCGCAGCCAGGCAGCTCGCGAC, GCGGCCGCTCACTGTGCGGAAGCAGGGGT
Klf11-pcDNA3: GAATTCCTCCTGCCTCGCAGCATTGCT, GCGGCCGCTCAGCCAGAGGCCGGCAAGG
Cell preparation and culture
Bone marrow cells were isolated from the tibia and femur and cultured in RPMI 1640 medium with 10% FBS (Hyclone, UT, USA), 2 mM glutamine, 100 units/mL penicillin-streptomycin, 10 ng/mL M-CSF (PeproTech, NJ, USA), or 20 ng/mL murine GM-CSF (R&D systems, MN, USA) at 37°C with 5% CO2 for 5 days to harvest M-BMMs or GM-BMMs, respectively. HEK293 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM supplemented with 2 mM glutamine, 100 units/mL penicillin and streptomycin, and 10% FBS at 37°C in the presence of 5% CO2.
Transfection
Transient transfection into primary mouse bone marrow derived macrophage using Amaxa Mouse Macrophage Nucleofection kit (Cat. No. VPA-1009) was performed according to manufacturer’s instruction. Transient transfection into HEK293 cells was performed by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction.
RNA-mediated interference
To knock down Klf11 in M-BMMs from WT or klf10-deficient mouse, the ON-TARGET plus SMART pool mouse-TIEG3 (194655) or the negative control siRNA (Thermo Scientific Dharmacon, Lafayette, CO, USA) were transfected into M-BMMs using INTERFERin™ (Polyplus, Graffenstaden, France) according to the manufacturer’s instruction. Another siRNA against Klf11 (5′-UGCAUGUGGACCUUUCGCUGUCAUG-3′) and control siRNA were synthesized by Shanghai GenePharma Co., Ltd. and were transfected using INTERFERin™ (Polyplus, Graffenstaden, France) according to the manufacturer’s instruction.
RNA extraction and real-time qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Cat. No.15596026). The cDNA was synthesized from total RNA using PrimeScript Reverse Transcriptase (Takara, Cat. No. DRR063A). Real-time PCR was accomplished with the ABI Prism 7500 analyzer (Applied Biosystems, Carlsbad, CA) using SYBR Premix Ex TaqTM (Takara, Cat. No. DRR041A). The corresponding primers were as follows:
Klf10: GGGTGTGGCAAGACTTACTT, AATGGTCGCTCCTCATAAAC
Klf11: GTGTTCCTTGCCTCCAGC, TCTTGTCACAGCCGTCCC
IL-12p40: AGGTGCGTTCCTCGTAGAGA, AAAGCCAACCAAGCAGAAGA
IL-12p35: CTGTGCCTTGGTAGCATCTA, TTTCACTCTGTAAGGGTCTG
TNF-α: CGGTGCCTATGTCTCAGCCT, GAGGGTCTGGGCCATAGAAC
IL-6: AGTTGCCTTCTTGGGACTGA, TCCACGATTTCCCAGAGAAC
IL-10: CCAAGCCTTATCGGAAATGA, TTTTCACAGGGGAGAAATCG
IFN-β: CCCTATGGAGATGACGGAGA, CTGTCTGCTGGTGGAGTTCA
TGF-β: GGCGGTGCTCGCTTTGTA, CCCGAATGTCTGACGTATTGA
CCL2: CAGCAAGATGATCCCAATGA, GTAGGTTCTGATCTCATTTG
CCL5: ACCACTCCCTGCTGCTTT, ACACTTGGCGGTTCCTTC
CCL12: ACTTCTATGCCTCCTGCTC, CACTGGCTGCTTGTGATTC
CXCL10: CGGAATCTAAGACCATCAA, TCACCTTTCAGAAGACCAA
Measurement of cytokines
Mouse IL-12p40, IL-12p70, TNF-α, IL-6, and IL-10 concentrations in cell culture supernatants were quantified by ELISA kits (eBio-science: IL-12/IL-23 total p40, Cat. No. 88–7100-22; IL-12p70, Cat. No. 88–7121-22; TNF-α, Cat. No. 88–7324-22; IL-6, Cat. No. 88–7064-22; IL-10, Cat. No. 88–7104-22) according to the manufacturer’s instruction.
Detection of NO in culture supernatants
M-BMMs on day 5 from WT and Klf10-deficient mouse were stimulated with 1 μg/mL LPS for 12 and 24 h. Culture supernatants were analyzed for NO by the Griess reaction. Briefly, 50 μL supernatant was incubated with 50 μL Griess reagent for 5 min at room temperature, and NO2 level was determined by measuring the absorbance at 540 nm relative to the reference sample.
Western blot
Whole cell lysates were prepared by complete Lysis-M kit (Roche; Cat. No. 04719956001) and the concentration was determined by the bicinchoninic acid protein assay (Thermo Scientific; Lot # MC 155209). The same amounts of protein were resolved on SDS-PAGE gels, transferred to polyvinylidene fluoride membrane. After blocking with 5% nonfat dry milk/PBS, the membranes were further incubated with the indicated primary antibodies overnight, reacted with a secondary antibody, and then protein bands were visualized by ECL.
Flow cytometry
Cells were harvested and incubated with relative antibodies for 30 min on ice, washed, and analyzed in a FACS calibur flow cytometer (Becton Dickinson).
Luciferase reporter assay
The promoter of IL-12p40 and its mutants were produced by PCR-based amplification and subcloned into the pGL3-Enhancer Vector to forming luciferase report plasmid.
Human embryonic kidney (HEK293) cells were cotransfected with 100 ng luciferase reporter plasmid, 10 ng thymidine kinase promoter-Renilla luciferase reporter plasmid, plus the pCDNA3-Klf10, or control vector. After 48 h, luciferase activities were determined by the Dual-Luciferase Reporter Assay System (Promega, Cat. No. E10910) according to the manufacturer’s instructions.
The primers were as followed:
P40-promoter-WT: CTCGAGTAGGCATGATGTAAACAGAAAT, AAGCTTCTAGATGCAGGGAGTTAGC
P40-promoter-Δ: CTCGAGTCATTTCCTCTTAACCTGGG, AAGCTTCTAGATGCAGGGAGTTAGC
P40-promoter-mut: CTCGAGTAGGCATGATGTAAACAGAAATTA GTATCTCTGCCTCCTTCCTTTTTCCAATCCCCGA, AAGCTTCTAGATGCAGGGAGTTAGC
Chromatin immunoprecipitation
Chromatin-immunoprecipitation assays were done essentially as the manufacturer’s protocol (Active motif, CHIP-IT™ Express). The immunoprecipitated DNA fragments were then analyzed by semi-qPCR and qPCR.
The primers used were as followed:
GAPDH: TTACTTTCGCGCCCTGAG, GCGGTTCATTCATTTCCTTC
IL-12p40: TGCCGCCTCTATTCACCTTA, CTGACTAGTCTCAATTGCAACA
Statistical analysis
Data are presented as the mean ± SD. Statistical significance was determined by Student’s t-test. A value of p < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank L. Lu for discussions; F. Xing for assistance with manuscript editing. This work was supported by grants from the National Basic Research Program of China (973 Program) (2012CB966603), the National Natural Science Foundation of China (J20111945), Zhejiang Provincial Natural Science Foundation of China (R20110298), Doctoral Fund of Ministry of Education of China (20110101120102), the Fundamental Research Funds for the Central Universities (2011QNA7001), and the National Technology Key Projects of China (008ZX1002-008).
Abbreviations
- C/EBP
CCAAT/enhancer-binding protein
- GM-BMM
GM-CSF-induced mouse bone marrow-derived macrophage
- IRF
interferon regulatory factor
- KLF
Krüppel-like factor
- M-BMM
M-CSF-induced mouse bone marrow-derived macrophage
- qPCR
quantitative PCR
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s website
References
- 1.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 4.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 5.Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, et al. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 7.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 8.Trinchieri G. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu Rev Immunol. 1995;13:251–276. doi: 10.1146/annurev.iy.13.040195.001343. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 10.Bastos KR, Alvarez JM, Marinho CR, Rizzo LV, Lima MR. Macrophages from IL-12p40-deficient mice have a bias toward the M2 activation profile. J Leukoc Biol. 2002;71:271–278. [PubMed] [Google Scholar]
- 11.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 12.Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci USA. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 14.Wang IM, Contursi C, Masumi A, Ma X, Trinchieri G, Ozato K. An IFN-gamma-inducible transcription factor, IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J Immunol. 2000;165:271–279. doi: 10.4049/jimmunol.165.1.271. [DOI] [PubMed] [Google Scholar]
- 15.Gorgoni B, Maritano D, Marthyn P, Righi M, Poli V. C/EBP beta gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J Immunol. 2002;168:4055–4062. doi: 10.4049/jimmunol.168.8.4055. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Ma X. Triptolide inhibits IL-12/IL-23 expression in APCs via CCAAT/enhancer-binding protein alpha. J Immunol. 2010;184:3866–3877. doi: 10.4049/jimmunol.0903417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood. 2010;116:4404–4414. doi: 10.1182/blood-2010-05-285353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Bieker JJ. Unanticipated repression function linked to erythroid Kruppel-like factor. Mol Cell Biol. 2001;21:3118–3125. doi: 10.1128/MCB.21.9.3118-3125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Q. Activation and repression of interleukin-12 p40 transcription by erythroid Kruppel-like factor in macrophages. J Biol Chem. 2004;279:18451–18456. doi: 10.1074/jbc.M400320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, et al. Kruppel-like factor 4 is essential for inflamatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 26.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, et al. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, Su J, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Z, Wara AK, Icli B, Sun X, Packard RRS, Esen F, Stapleton CJ, et al. Kruppel-like factor KLF10 targets transforming growth factor-1 to regulate CD4+CD25− T cells and T regulatory cells. J Biol Chem. 2009;284:24914–24924. doi: 10.1074/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner F, Jain MK, Feinberg MW, Sibinga NE, Pellacani A, Wiesel P, Chin MT, et al. Transforming growth factor-beta 1 inhibition of macrophage activation is mediated via Smad3. J Biol Chem. 2000;275:36653–36658. doi: 10.1074/jbc.M004536200. [DOI] [PubMed] [Google Scholar]
- 31.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 32.Zhou A, Scoggin S, Gaynor RB, Williams NS. Identification of NF-κB-regulated genes induced by TNFα utilizing expression profiling and RNA interference. Oncogene. 2003;22:2054–2064. doi: 10.1038/sj.onc.1206262. [DOI] [PubMed] [Google Scholar]
- 33.Cousens LP, Peterson R, Hsu S, Dorner A, Altman JD, Ahmed R, Biron CA. Two roads diverged: interferon alpha/beta-and interleukin 12-mediated pathways in promoting T-cell interferon gamma responses during viral infection. J Exp Med. 1999;189:1315–1328. doi: 10.1084/jem.189.8.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu BS, Janssen HL, Boonstra A. Type I and III interferons enhance IL-10R expression on human monocytes and macrophages, resulting in IL-10-mediated suppression of TLR-induced IL-12. Eur J Immunol. 2012;42:2431–2440. doi: 10.1002/eji.201142360. [DOI] [PubMed] [Google Scholar]
- 35.Du C, Sriram S. Mechanism of inhibition of LPS-induced IL-12p40 production by IL-10 and TGF-beta in ANA-1 cells. J Leukoc Biol. 1998;64:92–97. doi: 10.1002/jlb.64.1.92. [DOI] [PubMed] [Google Scholar]
- 36.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, et al. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- 37.Tadokoro CE, de Almeida Abrahamsohn I. Bone marrow-derived macrophages grown in GM-CSF or M-CSF differ in their ability to produce IL-12 and to induce IFN-gamma production after stimulation with Trypanosoma cruzi antigens. Immunol Lett. 2001;77:31–38. doi: 10.1016/s0165-2478(01)00197-3. [DOI] [PubMed] [Google Scholar]
- 38.Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ, Hamilton JA. GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J Leukoc Biol. 2009;86:411–421. doi: 10.1189/jlb.1108702. [DOI] [PubMed] [Google Scholar]
- 39.Ma X, Chow JM, Gri G, Carra G, Gerosa F, Wolf SF, Dzialo R, et al. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier-Osusky I, Schoedon G, Blauer F, Schneemann M, Schaffner A. Comparison of the antimicrobial activity of deactivated human macrophages challenged with Aspergillus fumigatus and Listeria monocytogenes. J Infect Dis. 1996;174:651–654. doi: 10.1093/infdis/174.3.651. [DOI] [PubMed] [Google Scholar]
- 41.Blauer F, Groscurth P, Schneemann M, Schoedon G, Schaffner A. Modulation of the antilisterial activity of human blood-derived macrophages by activating and deactivating cytokines. J Interferon Cytokine Res. 1995;15:105–114. doi: 10.1089/jir.1995.15.105. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto K, Sakaguchi M, Medina RJ, Niida A, Sakaguchi Y, Miyazaki M, Kataoka K, et al. Transcriptional regulation of a brown adipocyte-specific gene, UCP1, by KLF11 and KLF15. Biochem Biophys Res Commun. 2010;400:175–180. doi: 10.1016/j.bbrc.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 43.Aste-Amezaga M, Ma X, Sartori A, Trinchieri G. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J Immunol. 1998;160:5936–5944. [PubMed] [Google Scholar]
- 44.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 45.McCartney-Francis N, Jin W, Wahl SM. Aberrant Toll receptor expression and endotoxin hypersensitivity in mice lacking a functional TGF-beta 1 signaling pathway. J Immunol. 2004;172:3814–3821. doi: 10.4049/jimmunol.172.6.3814. [DOI] [PubMed] [Google Scholar]
- 46.Pan H, Ding E, Hu M, Lagoo AS, Datto MB, Lagoo-Deenadayalan SA. SMAD4 is required for development of maximal endotoxin tolerance. J Immunol. 2010;184:5502–5509. doi: 10.4049/jimmunol.0901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.