Natural organs are spatially heterogeneous, both in material composition and in the cell types within. Engineered tissues, in contrast, remain challenging to create, especially if the goal is to spatially position multiple cell types in a heterogeneous pattern in three dimensions (3D). Here, we describe a simple, inexpensive, yet extremely precise method to create tissue architectures in a digitally specifiable fashion, with morphological and compositional tuning. Specifically, we pattern hydrogel crosslinking via a novel photolithographic process that can trap solutions of extracellular matrix (ECM) components, cells, and diffusible factors in defined 3D shapes, without requiring specialized expensive optics or robotics. By iterating this process with different patterns and different cellular compositions, engineered tissues of varying heterogeneity, size, and complexity can be designed and implemented with microscale precision, in a fashion practical for individual laboratories to perform. We demonstrate the power of this process by engineering tissue building blocks in defined geometries, creating engineered tissues that encapsulate different cells (e.g., primary neurons, embryonic stem cells (ESCs), human umbilical vein endothelial cells (HUVECs), and fibroblasts) at specified locations throughout a single 3D tissue volume. We used the ability to rapidly synthesize tissue volumes of varying scale to examine how tissue volume governs neuron development.
Native tissues consist of multiple cell types and ECM components, which are spatially heterogeneous and organized in a three-dimensional (3D) environment. [1–6] Precision spatial positioning of cells and ECM components is crucial for natural tissue development and function.[3,5,7–12,13] Engineered tissues have been difficult to generate with comparable degrees of material and cell type spatial variability found in natural tissues. Current technologies can pattern cells on a two-dimensional (2D) substrate, providing control over composition and spatial arrangement on a flat surface.[14] However, cells anchored on 2D substrates do not recapitulate important features of cell behavior in vivo, such as cell-cell and cell-microenvironment interactions in 3D.[4–5,9–10,15,16] A current challenge is to develop 3D tissue constructs that replicate the architecture and cellular complexity found in vivo[2,10,15,17] for more accurate platforms for basic science, drug screening and diagnostic platforms, and also for prototyping tissues for clinical replacement. For example, for the brain, which has perhaps thousands of different kinds of cells, arranged in complex 3D architecture, undamentally new kind of tissue engineering techniques might be required.[18–22]
In recent years, many strategies have been proposed for how to define the cellular architectures of engineered tissues in 3D,[5,23–33] but an overall technological pipeline capable of simple, inexpensive, practical, and powerful generation of spatially varying, micron-scale precise, and complex physiological systems, is needed. For example, existing strategies that could be used for the creation of 3D spatially complex engineered tissues (e.g., stereolithography, bioprinting, assembly, and casting) either do not provide degrees of freedom in all three dimensions, essential for defining 3D spatially varying cellular organization, or the cost and time budget rise exponentially as the resolution is pushed to cellular scales (i.e., 10–20 μm), and as the structure dimensions are pushed to centimeter scales–critical for creating 3D spatially varying model tissues of importance for biology and medicine.
To overcome these challenges, thus addressing an unmet need in 3D tissue prototyping with microscale precision,[4,9,10,34] we created an optimized, inexpensive (∼$50 material cost), and easy-to-use version of the powerful method used in semiconductor microfabrication,[35] multilayer photolithography, to build 3D digitally specified tissue constructs (Figure 1). The significance of the presented approach is that it is a scalable and generalizable biomanufacturing method with degrees of freedom in all three dimensions, with multiple-orders-of-magnitude lower cost compared to other specialized systems (e.g., stereolithography devices cost tens of thousands of dollars), and allows ∼10 μm precision within and across layers offering an order-of-magnitude improvement over existing methods without requiring expensive lithographic aligners or computerized-stage microscopes. The key innovations include the strategy for photolithographically crosslinking hydrogels containing cells and ECM components using multiple masks with microscale control, to anchor them at specified coordinates in 3D space (by varying the cell and ECM compositions before the crosslinking step, we can modulate the biological and chemical materials being instantaneously incorporated into the engineered tissue as a function of time and space), as well as the very simple mechanical engineering strategies utilized: since we use simple alignment and layer-by-layer fabrication hardware, no specialized optics or alignment systems are needed for tissue fabrication, making our method eminently practicable in individual laboratories.
Figure 1.
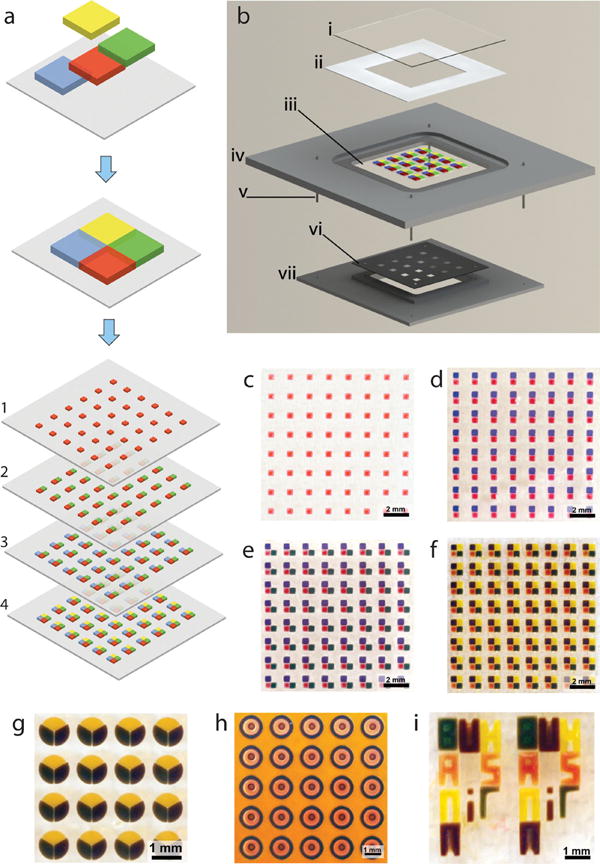
Digitally specified spatially heterogeneous microfabrication and tissue sculpting. (a) In this example, the tissue structure is composed of four different types of building blocks, each with specific cellular and ECM composition shown as different color codes. (b) Fabrication of 3D multilayer tissue prototypes via a layer-by-layer photomasking and alignment approach, with a series of photomasks, and a series of hydrogel solutions to be photo crosslinked to result in digitally sculpted hydrogels (e.g., containing different cell types, ECM components, and diffusible factors). The simple mask alignment and digital sculpting device comprises: (i) top cover slip of fabrication compartment, (ii) thickness control spacer, (iii) a treated glass cover slip on which the sculpted elements are immobilized, (iv) fabrication chamber, (v) alignment pins, (vi) predesigned photomask and (vii) mask holder that aligns on the fabrication chamber. Light is applied through the opening in mask holder. (c-f) Alignment and digital sculpting of individual elements is achieved using a photomask, which has digitally specified, engineered openings allowing photocrosslinking of tissue prototypes in a stepwise manner. Fabrication of cubic digitally sculpted blocks is demonstrated here. Radial circular (g), concentric circular (h) elements can also be sculpted, mimicking cross-section of embedded vasculature in a tissue construct, where yellow hydrogel encapsulates the three types of elements microfabricated as concentric circles. (i) Digital sculpting approach can also be used to generate complex structures such as microscale typescript (e.g., BWH, BAMM, HST, and MIT).
Using this platform, we developed a digitally specifiable 3D environment for brain cells, and explored the effect of 3D spatial confinement on neural cell survival and neurite growth. We further explored microscale engineered tissue complexity by developing prototype structures digitally sculpted with varying cellular composition (i.e., ESCs, HUVECs, and fibroblasts) and geometry (e.g., square, radially fractioned circular, and concentric circular), creating tissue prototypes with a high level of precision, versatility, flexibility, and scalability. Our work will support 3D, hierarchical, modular assembly of heterogeneous engineered tissues, advancing the synthetic biology-driven understanding of how tissues develop and change over time. This approach will also assist in meeting the clinical demand for replacement tissues and organs,[36] by enabling the service of multiscale fabrication techniques in 3D, hierarchical, modular tissue design with broad applications.
We have developed an advanced fabrication process comprising a number of digitally controllable lithography steps (Figure 1a) to generate tissue complexity in 3D (Figure 1b). The geometry of each and every layer is tightly controlled by predesigned photolithography masks (Figure S1) to fabricate architectures and tissue prototypes composed of multiple building blocks (Figure 1c–f). We have shown that complexity and organization of the hydrogel architectures can be tightly controlled for various geometries, i.e., rectangular prism (Figure 1f), radially fractioned circular (radial circular) (Figure 1g), concentric circular (Figure 1h), and typescript (Figure 1i). The method utilizes a mask alignment setup (Figure 1b and S2) to achieve high alignment precision of the building blocks (Figure S3). We defined the alignment precision as the positioning repeatability of the centroid of the individual building blocks during micro-fabrication based on the computer aided design (Figure 1 and Figure S1). Alignment precision was determined (for details, see Supporting Information) to be within: 10.9 μm (±8.2) for square geometry; 11.2 μm (±4.5) for radial circular geometry; and 27.8 μm (±9.8) for concentric circular geometry (mean ± standard deviation (STD), n = 16-24 measurements for each geometry).
To demonstrate the utility of this digital tissue sculpting strategy, we assessed how neuron cell development and neurite growth could be studied in a space-constrained custom-tuned environment. We created digitally specified neural tissues made of lithographically specified hydrogel containing neonatal rat primary cortical neurons. We assessed neuron cell behavior in three different hydrogel sizes, each crafted lithographically as described (100 μm × 100 μm × 150 μm, 200 μm × 200 μm × 150 μm, 500 μm × 500 μm × 150 μm, Figure 2a). To morphologically assess neurons in engineered tissue prototypes, we performed labeling with anti-Tau-1 (axonal stain) and DAPI (nuclear stain), which clearly indicated the morphology of neurites and nuclei of the cells. In each 100 μm size element, only several neurons were encapsulated due to the space constraints, and the few cells exhibited short and undeveloped neurites (Figure 2b–j). We observed that neurons encapsulated in 200 μm (Figure 2k–n) and 500 μm (Figure 2o–q) elements formed more elaborate 3D geometries over comparable timescales (14 days in culture). Neurite (axon and dendrite) lengths were quantified under comparable photopolymerization conditions in different element sizes (Figure 2r). Specifically, neurons displayed longer neurites (p < 0.05, analysis of variance (ANOVA) with Tukey's posthoc test for multiple comparisons) in 200 μm elements compared to 100 μm elements, which demonstrated extremely low growth. Neurons exhibited even longer neurites in 500 μm elements (p < 0.05). Neurite growth was observed in the first three days of culture in 100 μm and 200 μm elements, which reached a stagnant state after three days (Figure 2r). However, in 500 μ m elements, neurite growth continued with a significant increase compared to day 1 and day 3 (p < 0.05) till day 7, after which a significant growth was no longer observed (p > 0.05). Projected neurite length was significantly greater (p < 0.05) in 500 μm elements than other size elements after 3 days of culture, which reached around 800 μm in length over 14 days in culture (Figure 2r). The distribution of neurons and their neurites in 3D was visualized using fluorescent confocal imaging (see video in the Supporting Information). We did not observe any difference in cell survival or morphology in different sections of the engineered constructs in 3D.
Figure 2.
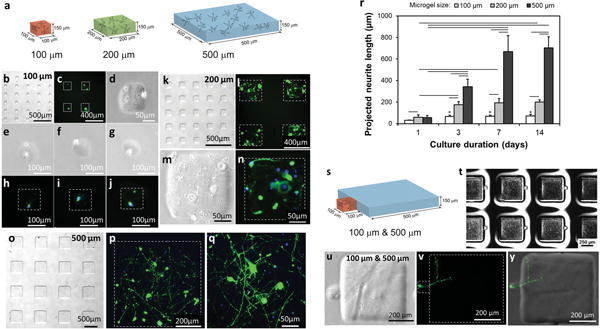
Digitally specified microfabricated neural tissue. (a) Primary neuron cells were inserted into 100 μm, 200 μm and 500 μm elements enabling visualization of neurite growth and development. Neurites (anti-Tau-1 staining in green) and nuclei (DAPI staining in blue) of neurons were observed in neural tissue constructs over 14 days of culture. (b–j) Neurons were digitally sculpted in 100 μm elements, (k–n) in 200 μm elements, and (o–q) in 500 μm elements. (r) Quantitative assessment of projected neurite length in digitally sculpted elements showed different growth dynamics of neurites over time in engineered constructs with different sizes. Horizontal lines connecting individual groups represent statistically significant difference and * represents statistically significant difference compared to 1 day culture (p < 0.05). Error bars represent mean ± STD (n = 3–6). (s) Digital sculpting of 100 μm tissue element next to a 500 μm tissue element, where only the 100 μm element includes neuron cell bodies. (t) Digitally sculpted array of adjacent 100 μm and 500 μm tissue prototypes. (u-y) Neuron cells in digitally sculpted 100 μm elements extended neurites into 500 μm elements. (The video in the Supporting Information shows 3D distribution of neuron cells and the neural circuit formed in digitally sculpted constructs).
The above results suggested that neuronal growth is regulated by available volume. But larger volume tissue cubes also contained greater numbers of neurons. Ideally one would be able to examine effects of volume, and even 3D shape, on neural growth, independent of cell density. Our technology enables such assessments of pure geometrical influences on neuron growth, here demonstrated by our fabrication of complex tissue structures with a 100 μm element touching a 500 μm element (Figure 2s) in an array (Figure 2t). In this configuration, we selectively encapsulated single neuron cells in the 100 μm element, as enabled by our selective photolithography, to investigate the growth of the cell whose body is localized to a confined structure, but for which neurites are allowed to grow into a larger environment. During the 14 day culture period, single neuron cells spatially positioned in 100 μm elements were observed to grow towards the 500 μm element and extend neurites towards the larger gel, resulting in a projected neurite length of 507 μm (± 103, STD, n = 7) (Figure 2u–y). Indeed, such long neurites extending into the larger 3D hydrogel compartment were observed in all the sculpted structures analyzed (n = 7), suggesting that the neurite growth is not only dependent on the number of cells encapsulated in a 100 μm element, but is a function of the complex geometry of spatial confinement. Neurons appeared to be responsive to their surroundings in a novel way, showing how our technology enables the probing of parameters important for developmental as well as tissue engineering studies.
To gain insight into the cellular composition of the 3D neural tissue prototypes, we identified neuronal and glial populations by staining for Tau-1 and GFAP (Figure 3a–i). The numbers of neuronal and glial cells were statistically similar (p > 0.05, non-parametric Mann-Whitney U test) (Figure 3j), displaying a ratio reminiscent of natural brain tissue.[37] We observed both CaMKII- and GAD65-positive neurons indicating that both excitatory and inhibitory neural cells were present in the 3D neural tissue prototypes (Figure 3d–i). Excitatory neurons constituted the majority of the neurons, amounting to 83.9% (±12.6, STD; n = 5) of the total number of neurons, similar to the fraction (i.e., 4:1 ratio of excitatory to inhibitory) of cortical neurons in vivo that are excitatory.[38] Our results suggested that neural networks in elements comprised a complex population of brain cells, including excitatory neurons, inhibitory neurons, and glial cells, appearing at comparable ratios to that in the native rodent cortex, and highlighting the power of our technology to preserve a diversity of brain cell types.
Figure 3.
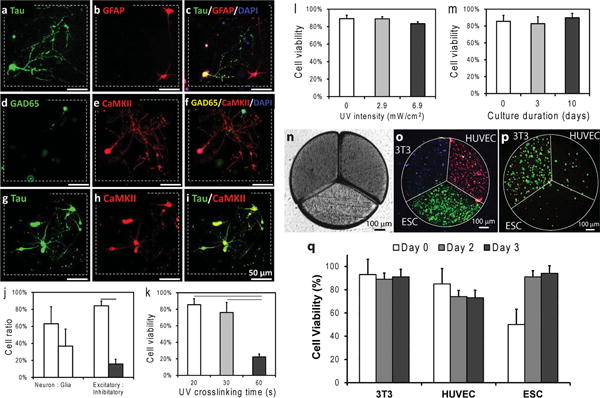
Immunofluorescent labeling of neurons, glia, excitatory, and inhibitory cells and multicell-type tissue prototypes generated using digitally specified tissue engineering. (a–c) Anti-Tau-1 labeled neurons (green), anti-GFAP labeled glial cells (red), and merged image is shown (DAPI in blue). (d–f) Anti-GAD65 labeled inhibitory neurons (green), anti-CaMKII labeled excitatory neurons (red), and merged image is shown (DAPI in blue). (g,h) Tau-1-positive label (green) and CaMKII-positive label (red), and merged image. Scale bars represent 50 μm of length. Dashed squares represent the borders of digitally specified 3D neural tissue constructs. (j) Quantitative plot showing the cell type ratio in hydrogels after 3 weeks of culture. Anti-Tau-1 for neurons (63.16 ± 19.92%), anti-GFAP for glia (36.84 ± 19.92%); anti-CaMKII for excitatory neurons (84.20 ± 10.96%), anti-GAD65 for inhibitory neurons (15.80 ± 10.96%) (n = 4-5). (k) UV light exposure of up to 30 seconds did not significantly affect neuron cell viability (p > 0.05). (l) Light intensity did not have a statistically significant effect on neuron cell viability. (m) Neuron viability was observed in tissue prototypes up to 10 days in culture n = 3–8). (n) Light microscope image of digitally sculpted radially fractioned circle with three different cell types. (o) Encapsulation of three cell types (ESCs in green, 3T3 cells in blue, HUVECs in red) in a single tissue prototype (106 cells/mL for all cell types). Cells were stained using cell tracker dyes to display their distribution after microfabrication. (p) Encapsulation of three cell types in varying concentrations using digital sculpting method: ESCs (0.5 × 106 cells/mL), HUVEC (0.75 × 106 cells/mL) and 3T3 (106 cells/mL). Live/dead stained cells after digital sculpting (green: live cells, red: nonviable cells). (q) Cells remained highly viable at the end of three day in vitro culture in digitally specified tissue constructs: 92% (± 5%), 74% (±8%), and 95% (± 3%) for ESCs, and 3T3s, and HUVECs, respectively (n = 3–7). Error bars represent mean ± STD of the mean. Horizontal lines in the plots (j and k) connecting individual groups represent statistically significant difference (p < 0.05).
Photopolymerizable polymers and hydrogels (e.g., poly(ethylene glycol)/poly(L-lysine), chitosan) have been used in combination with primary neurons, neural stem and progenitor cells for neural tissue engineering and neural cell delivery applications.[30,39–41] In the current study, in which we extended the power of photopolymerizable hydrogels by 3D digital tissue sculpting, we accordingly validated the method with neuron cell viability studies. We evaluated the effects of the duration and intensity of ultraviolet (UV) light exposure to create photo polymerizable gelatin methacrylate hydrogels encapsulating primary neuron cells. We first assessed the effect of light exposure duration (20, 30, and 60 s at 2.9 mW/cm2) on cell viability (Figure 3k–m). We observed that neurons were viable after 30 seconds of light exposure applied during microfabrication. The viabilities were observed to be 85.54 ± 7.15%, 76.10 ± 12.22%, and 22.33 ± 3.64% at 20, 30, and 60 s, respectively (n = 3–8 samples of each kind). Sixty seconds of exposure resulted in significantly less (p < 0.05) cell viability compared to 20 and 30 seconds (Figure 3k). Therefore, results indicated that less than 30 seconds had minimal effect (p < 0.05) on neurons with a higher viability compared to 60 s of light exposure. Next, we assessed the effect of UV light intensity (2.9–6.9 mW/cm2 for 20 s) on cell viability in neuron encapsulating elements (Figure 3l). We observed no statistically significant effect (p > 0.05, n = 3) on cell viability as a result of increasing intensity. The neuron cell viability was 83.35 ± 2.22% at an intensity of 6.9 mW/cm2. To achieve high cell viability and minimize potential effects of UV light, we used 2.9 mW/cm2 intensity and 20 seconds crosslinking duration, which resulted in a viability of 85.54 ± 7.15%, not significantly different compared to controls that were not exposed to light (89.24 ± 3.84%; p > 0.05; n = 3 photocrosslinked cubes, n = 3 control samples). Cell viability assays were conducted at different time points (Figure 3m): day 0 (immediately after photocrosslinking at 2.9 mW/cm2 intensity and 20 seconds crosslinking duration), day 3 (after 3 days of culture) and day 10. We observed similar (p > 0.05, n = 3–8) cell viabilities at day 3 (82.82 ± 8.23%) and day 10 (89.80 ± 5.26%) compared to day 0 (85.54 ± 7.15%). These results demonstrated that the photopolymerization parameters of photocrosslinkable hydrogels have minimal effect on neural cell viability both in short and long term.
To demonstrate the ability to digitally sculpt complex tissues with multiple cell types, we next created engineered 3D tissue constructs (Figure 3n) composed of three cell types (Figure 3o), encapsulating ESCs, HUVECs and NIH 3T3 fibroblasts mimicking the complexity in studies aiming to regenerate myocardium. [42] We observed that each cell type was spatially confined in individual elements, as indicated by cell tracker staining (Figure 3o). We have shown that cellular composition as well as cellular concentration in each building block can be controlled spatially (Figure 3o,p). We also observed that cells were viable in tissue prototypes after microfabrication (Figure 3p). The quantitative viability analysis showed that all cell types remained viable at the end of three day in vitro culture: 92% (± 5%), 74% (±8%), and 95% (± 3%) for ESCs, and 3T3s, and HUVECs, respectively (Figure 3q).
To demonstrate the 3D tissue sculpting capability of the method, we created multiple layer 3D tissue constructs (Figure 4). In this multilayer fabrication process, after the first layer fabrication, the lithography process is repeated to create a second and then a third layer (Figure 4a). In this process, compositions of each element were alternated in different layers to increase the degree of complexity achievable. With this process, large number of complex multilayer structures can be fabricated simultaneously (Figure 4b), in various geometries and architecture (Figure 4c-i), indicating the high throughput potential of this manufacturing platform. The same system and approach would be simply scaled up in size to achieve millions of tissue prototypes. For instance, with a standard 12 inch mask used in the semiconductor industry, 145 thousand tissue prototypes with an element size of 400 μm and separation of 400 μm can be fabricated. This number increases to 2.3 million tissue prototypes with an element size of 100 μm and a separation of 100 μm using the presented process with a 12 inch mask. Thus, our technology advances the scales of fabrication in engineering complex tissue architectures.
Figure 4.
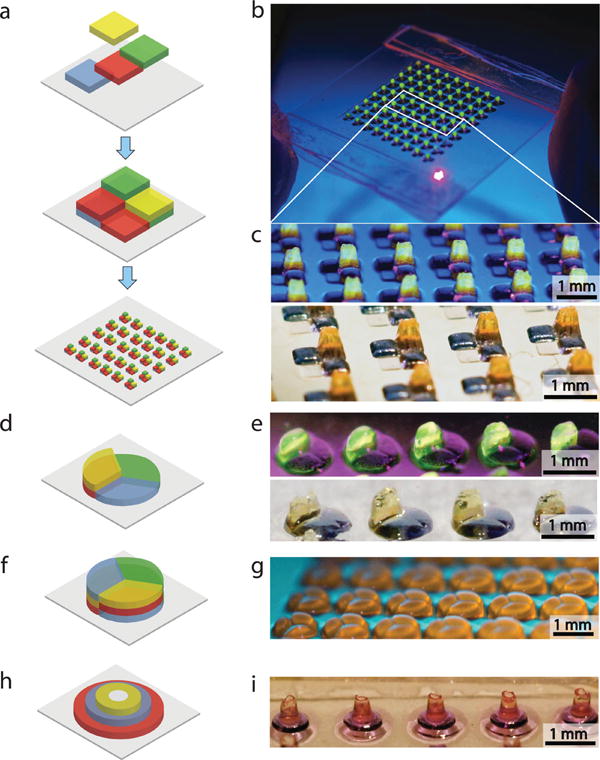
Multilayer 3D digitally specified spatially heterogeneous tissue sculpting. (a) Fabrication of 3Dsquare geometry tissue prototypes using multilayer photolithography. (b,c) Micro-fabricated array of multilayer digitally specified 3D tissue prototypes. (d–g) Side view of 3D digitally sculpted spatially heterogeneous radial circular tissue construct at different stages of microfabrication. (h,i) Side view of three layered concentric circular microfabricated tissue construct.
The multilayer photolithography digital sculpting method presented here utilizes multiple masks to create individual elements in a tissue construct (Figures S1 and S2). Since the masks have designated openings to create individual elements in a layer, the cells in an individual element are exposed to light only once during the fabrication of a layer. On the other hand, photocrosslinked hydrogels have been commonly used in the literature to encapsulate various cells, including neural cells,[43] for applications in tissue engineering.[44,45] Potential effects of UV on cells during the fabrication process of photocrosslinked materials can be eliminated by replacing the UV light source with a visible light source and utilizing visible light photoinitiators, such as eosin-Y, triethanolamine, or camphorquinone as reported earlier.[46–49]
In this work, we have demonstrated the compatibility of our digital tissue sculpting technology with neural cells. To achieve this, we developed digitally specifiable 3D hydrogel scaffolds that can be used as neural cell carriers, which supported neural cell survival and development with a naturalistic ratio of multiple cell-types. The engineered neural tissue presented here offers new opportunities as a platform technology for 3D in vitro studies of neural networks. Further, the in situ engineered neural tissue may enable broad clinical applications for engineering neural implants, providing an important step forward in transplantation and cell therapy for neural degenerative diseases, as well as injuries of brain and spinal cord. In combination with optogenetic methods for activating defined cell types,[50] as well as robotic methods for analyzing single neurons in intact tissues in an integrative fashion,[51] the described methodologies may open up neural tissue engineering to the regime of highly accurate 3D architectures capable of exhibiting naturalistic cell type distributions. With this system, long-term culture studies of neurons may be performed, which enable gaining insight to neuronal and neurite development in precisely controlled geometries. Importantly, these new insights and the digitally specifiable neural tissue could act as testbeds that can lead to disruptive advances such as bioengineered brain with a natural diversity of neuron types that would enable restoration of brain function after neurodegeneration, stroke, traumatic brain injury, spinal cord injury, or amyotrophic lateral sclerosis.
We digitally sculpted multiple cell types in elements within a single tissue prototype in a high throughput and repeatable manner. This method is based on multilayer photolithography used in the semiconductor industry and presents great potential to become a broadly applicable method for high-throughput applications in tissue engineering, regenerative medicine, and pharmacological studies. This photolithographic approach enables digital modulation of biological and chemical materials being instantaneously incorporated into the engineered architecture as a function of time and space allowing microscale precision without requiring specialized optics or robotics. This advancement allows us to envision building millions of digitally specified tissue prototypes with predetermined biomaterials, encapsulated molecules and cell types at complexity and throughput levels not attained before for in vitro cultures. The developed method would become a broadly enabling platform for studying cell–cell and cell-matrix inter actions in a variety of physiologic (e.g., stem cell differentiation) and pathological (e.g., cancer) settings, in vitro drug screening applications in pharmaceutical industry, and in advanced biomanufacturing.
Supplementary Material
Acknowledgments
This work was performed at Demirci Bio-Acoustic MEMS in Medicine (BAMM) Laboratories at the Division of Biomedical Engineering at Brigham and Women's Hospital, Harvard Medical School. Dr. Demirci acknowledges that this material is based in part upon work supported by the NSF CAREER Award Number 1150733, NIH R21-HL112114 and R21-AI087107. ESB was supported by the Paul Allen Family Foundation, New York Stem Cell Foundation, NIH, NSF, the IET AF Harvey Prize, and MIT Lincoln Laboratory.
Footnotes
Supporting Information: Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Umut Atakan Gurkan, Harvard Medical School, Division of Biomedical Engineering at Brigham and Women's Hospital, Bio-Acoustic-MEMS in Medicine (BAMM) Laboratory, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
Yantao Fan, Harvard Medical School, Division of Biomedical Engineering at Brigham and Women's Hospital, Bio-Acoustic-MEMS in Medicine (BAMM) Laboratory, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
Feng Xu, Harvard Medical School, Division of Biomedical Engineering at Brigham and Women's Hospital, Bio-Acoustic-MEMS in Medicine (BAMM) Laboratory, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
Burcu Erkmen, Harvard Medical School, Division of Biomedical Engineering at Brigham and Women's Hospital, Bio-Acoustic-MEMS in Medicine (BAMM) Laboratory, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
Emel Sokullu Urkac, Harvard Medical School, Division of Biomedical Engineering at Brigham and Women's Hospital, Bio-Acoustic-MEMS in Medicine (BAMM) Laboratory, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
Gunes Parlakgul, Harvard Medical School, Division of Biomedical Engineering at Brigham and Women's Hospital, Bio-Acoustic-MEMS in Medicine (BAMM) Laboratory, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
Jacob Bernstein, Media Lab and McGovern Institute, Departments of Brain and Cognitive Sciences and Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Wangli Xing, Email: wlxing@tsinghua.edu.cn, Medical Systems Biology Research Center, School of Medicine, Tsinghua University, Beijing 100084, PR China, National Engineering Research Center for Beijing Biochip Technology, 18 Life Science Parkway, Beijing, 102206, P. R. China.
Edward S. Boyden, Email: edboyden@mit.edu, Media Lab and McGovern Institute, Departments of Brain and Cognitive Sciences and Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Utkan Demirci, Email: udemirci@rics.bwh.harvard.edu, Harvard Medical School, Brigham and Women's Hospital, Harvard-MIT Health Sciences & Technology, 65 Landsdowne St. PRB 252, Cambridge, MA 02139, USA.
References
- 1.Pampaloni F, Reynaud EG, Stelzer EHK. Nat Rev Mol Cell Biol. 2007;8:839. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 2.Yamada KM, Cukierman E. Cell. 2007;130:601. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Geckil H, Xu F, Zhang X, Moon S, Demirci U. Nanomedicine (Lond) 2010;5:469. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurkan UA, Tasoglu S, Kavaz D, Demirel MC, Demirci U. Adv Healthcare Mater. 2012;1:149. doi: 10.1002/adhm.201200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon S, Hasan SK, Song YS, Xu F, Keles HO, Manzur F, Mikkilineni S, Hong JW, Nagatomi J, Haeggstrom E, Khademhosseini A, Demirci U. Tissue Eng Part C Methods. 2010;16:157. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song YS, Lin RL, Montesano G, Durmus NG, Lee G, Yoo SS, Kayaalp E, Haeggstrom E, Khademhosseini A, Demirci U. Anal Bioanal Chem. 2009;395:185. doi: 10.1007/s00216-009-2935-1. [DOI] [PubMed] [Google Scholar]
- 7.Anghelina M, Krishnan P, Moldovan L, Moldovan NI. Am J Pathol. 2006;168:529. doi: 10.2353/ajpath.2006.050255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuong C, Wu P, Plikus M, Jiang T, Bruce Widelitz R, Gerald PS. Current Topics in Developmental Biology. Vol. 72. Academic Press; 2005. p. 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu F, Finley TD, Turkaydin M, Sung Y, Gurkan UA, Yavuz AS, Guldiken RO, Demirci U. Biomaterials. 2011;32:7847. doi: 10.1016/j.biomaterials.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu F, Wu CA, Rengarajan V, Finley TD, Keles HO, Sung Y, Li B, Gurkan UA, Demirci U. Adv Mater. 2011;23:4254. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceyhan E, Xu F, Gurkan UA, Emre AE, Turali ES, El Assal R, Acikgenc A, Wu CaM, Demirci U. Lab Chip. 2012 doi: 10.1039/c2lc40523g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon S, Ceyhan E, Gurkan UA, Demirci U. PLoS ONE. 2011;6:e21580. doi: 10.1371/journal.pone.0021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tasoglu S, Kavaz D, Gurkan UA, Guven S, Chen P, Zheng R, Demirci U. Adv Mater. 2012 doi: 10.1002/adma201200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hui EE, Bhatia SN. Proc Natl Acad Sci USA. 2007;104:5722. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith LG, Swartz MA. Nat Rev Mol Cell Biol. 2006;7:211. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 16.Birgersdotter A, Sandberg R, Ernberg I. Sem Cancer Biol. 2005;15:405. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Gurkan U, Kishore V, Condon K, Bellido T, Akkus O. Calcified Tissue Int. 2011;88:388. doi: 10.1007/s00223-011-9467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavik E. Neural Tissue Engineering. Springer; New York: 2011. [Google Scholar]
- 19.Musick K, Khatami D, Wheeler BC. Lab Chip. 2009;9:2036. doi: 10.1039/b820596e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silver J, Miller JH. Nat Rev Neurosci. 2004;5:146. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 21.Kim DH, Viventi J, Amsden JJ, Xiao JL, Vigeland L, Kim YS, Blanco JA, Panilaitis B, Frechette ES, Contreras D, Kaplan DL, Omenetto FG, Huang YG, Hwang KC, Zakin MR, Litt B, Rogers JA. Nat Mater. 2010;9:511. doi: 10.1038/nmat2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. Nat Mater. 2011;10:799. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 23.Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R. Lab Chip. 2010;10:2062. doi: 10.1039/c004285d. [DOI] [PubMed] [Google Scholar]
- 24.Culver JC, Hoffmann JC, Poché RA, Slater JH, West JL, Dickinson ME. Adv Mater. 2012;24:2344. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visconti RP, Kasyanov V, Gentile C, Zhang J, Markwald RR, Mironov V. Expert Opin Biol Ther. 2010;10:409. doi: 10.1517/14712590903563352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci USA. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Yan Y, Zhang R. Tissue Eng Part B Rev. 2010;16:189. doi: 10.1089/ten.TEB.2009.0576. [DOI] [PubMed] [Google Scholar]
- 29.Smith CM, Stone AL, Parkhill RL, Stewart RL, Simpkins MW, Kachurin AM, Warren WL, Williams SK. Tissue Eng. 2004;10:1566. doi: 10.1089/ten.2004.10.1566. [DOI] [PubMed] [Google Scholar]
- 30.Curley JL, Moore MJ. J Biomed Mater Res Part A. 2011;99A:532. doi: 10.1002/jbm.a.33195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. FASEB J. 2007;21:790. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 32.Zorlutuna P, Jeong JH, Kong H, Bashir R. Adv Funct Mater. 2011;21:3642. [Google Scholar]
- 33.Xu F, Moon SJ, Emre AE, Turali ES, Song YS, Hacking SA, Nagatomi J, Demirci U. Biofabrication. 2010;2:014105. doi: 10.1088/1758-5082/2/1/014105. [DOI] [PubMed] [Google Scholar]
- 34.Xu F, Wu J, Wang S, Durmus NG, Gurkan UA, Demirci U. Biofabrication. 2011;3:034101. doi: 10.1088/1758-5082/3/3/034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore GE. Proc IEEE. 1998;86:82. [Google Scholar]
- 36.Davenport RJ. Science. 2005;309:84. doi: 10.1126/science.309.5731.84. [DOI] [PubMed] [Google Scholar]
- 37.Allen NJ, Barres BA. Nature. 2009;457:675. doi: 10.1038/457675a. [DOI] [PubMed] [Google Scholar]
- 38.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Nat Rev Neurosci. 2004;5:793. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 39.Royce Hynes S, McGregor LM, Ford Rauch M, Lavik EB. J Bio-mater Sci Polym Ed. 2007;18:1017. doi: 10.1163/156856207781494368. [DOI] [PubMed] [Google Scholar]
- 40.Hynes SR, Rauch MF, Bertram JP, Lavik EB. J Biomed Mater Res Part A. 2009;89:499. doi: 10.1002/jbm.a.31987. [DOI] [PubMed] [Google Scholar]
- 41.Valmikinathan CM, Mukhatyar VJ, Jain A, Karumbaiah L, Dasari M, Bellamkonda RV. Soft Matter. 2012;8:1964. doi: 10.1039/c1sm06629c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Condorelli G, Borello U, De Angelis L, Latronico M, Sirabella D, Coletta M, Galli R, Balconi G, Follenzi A, Frati G, Cusella De Angelis MG, Gioglio L, Amuchastegui S, Adorini L, Naldini L, Vescovi A, Dejana E, Cossu G. Proc Natl Acad Sci USA. 2001;98:10733. doi: 10.1073/pnas.191217898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aurand ER, Lampe KJ, Bjugstad KB. Neurosci Res. 2012;72:199. doi: 10.1016/j.neures.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seliktar D. Science. 2012;336:1124. doi: 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- 45.Bryant SJ, Nuttelman CR, Anseth KS. J Biomater Sci Polym Ed. 2000;11:439. doi: 10.1163/156856200743805. [DOI] [PubMed] [Google Scholar]
- 46.Cruise GM, Scharp DS, Hubbell JA. Biomaterials. 1998;19:1287. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 47.Cruise GM, Hegre OD, Lamberti FV, Hager SR, Hill R, Scharp DS, Hubbell JA. Cell Transplant. 1999;8:293. doi: 10.1177/096368979900800310. [DOI] [PubMed] [Google Scholar]
- 48.Ahmed TA, Griffith M, Hincke M. Tissue Eng. 2007;13:1469. doi: 10.1089/ten.2006.0354. [DOI] [PubMed] [Google Scholar]
- 49.Nicodemus GD, Bryant SJ. Tissue Eng Part B Rev. 2008;14:149. doi: 10.1089/ten.teb.2007.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyden ES. F1000 Biol Rep. 1000;3:3. doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR. Nat Methods. 2012;9:585. doi: 10.1038/nmeth.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


