Abstract
Most patients with advanced breast cancer develop osteolytic bone metastases, which have numerous complications. Because current therapies are not curative, new treatments are needed. Conditionally replicating adenoviruses (CRAds) are anticancer agents designed to infect and lyse tumor cells. However, in spite of their promise as selective cancer therapeutics, replicating adenoviruses have shown limited efficacy in the clinical setting. We hypothesized that a CRAd armed with osteoprotegerin (OPG) would eradicate bone metastases of breast cancer both directly, by oncolysis, and indirectly, by inhibiting osteoclastic bone resorption and thus reducing the tumor burden. We constructed an armed CRAd (Ad5-Δ24-sOPG-Fc-RGD) by replacing viral E3B genes with a fusion of the ligand-binding domains of OPG and the Fc portion of human IgG1. Conditional replication was conferred by a 24-base pair deletion within E1A (Δ24), which prevents the binding of E1A to the retinoblastoma tumor suppressor/cell cycle regulator protein and limits replication in normal cells. Enhanced infection of cells expressing low levels of the primary Ad5 receptor was conferred by incorporating an RGD peptide sequence into the fiber knob to mediate binding to αv integrins. After characterization of the armed CRAd, we demonstrated that infection of breast cancer cells by Ad-Δ24-sOPG-Fc-RGD both killed the infected cells by oncolysis and inhibited the formation of osteoclasts in an in vitro co-culture model. In a murine model of osteolytic bone metastases of breast cancer, the CRAd armed with sOPG-Fc reduced tumor burden in the bone and inhibited osteoclast formation more effectively than an unarmed CRAd.
Keywords: adenovirus, bone metastases, breast cancer, osteoprotegerin, virotherapy
Introduction
The majority of patients with advanced breast cancer develop bone metastases, which can be associated with complications such as severe bone pain, pathological fractures, leukoerythroblastic anemia, bone deformity, nerve compression and hypercalcemia.1 These debilitating complications lead to a decrease in the quality of life and are a significant cause of morbidity for patients with late-stage disease. Current treatments for breast cancer bone metastasis, such as radiotherapy and bisphosphonates, may alleviate symptoms and delay progression of the disease but are not curative.2,3 Moreover, these treatments can have undesirable side effects.2,3 Therefore, new therapies for breast cancer bone metastases are needed.
Conditionally replicating adenoviruses (CRAds) of human serotype 5 are a class of anticancer agents with great potential.4 The selective replication of the viruses in cancer cells amplifies the initial inoculum and destroys the cells by lysis. The viral progeny are then released, enabling them to infect neighboring cells, which results in multiple self-perpetuating rounds of infection, replication, lysis and spread throughout the tumor, all while sparing normal cells. CRAds have already been used in clinical trials in which their safety has been demonstrated. However, they have yet to dramatically alter the course of disease when administered as single agents.5 Thus, the antitumor efficacy of CRAds must be improved.
One means of augmenting efficacy utilizes the CRAd as a platform for the delivery of a therapeutic transgene, thereby creating a so-called armed replicating adenovirus in which the input dose of the transgene is amplified by replication of the virus.6 It is rational that the therapeutic transgene used to arm a CRAd intended for breast cancer bone metastasis should reflect the unique properties of the bone microenvironment. Breast cancer bone metastases are predominantly osteolytic in nature; tumor cells interfere with normal bone remodeling by accelerating bone resorption.7 Breast cancer cells in the bone produce several factors that can affect bone resorption, with parathyroid hormone-related protein (PTHrP) perhaps the most important. PTHrP promotes the upregulation of receptor-activator of NF-κB ligand (RANKL) on the surface of osteoblasts.8 The binding of RANKL to its receptor RANK on the surface of osteoclast precursors stimulates the development and activation of mature osteoclasts. The increased bone resorption mediated by osteoclasts then releases growth factors from the bone matrix that further stimulate tumor cell proliferation and PTHrP production, establishing a so-called “vicious cycle.”9 Osteoprotegerin (OPG) is a soluble inhibitor of bone resorption that is secreted by osteoblasts and acts as a decoy receptor for RANKL.10,11 The administration of recombinant OPG has been shown to decrease skeletal tumor burden both in animal models and clinical trials.12,13 We hypothesize that a CRAd armed with OPG will inhibit the progression of breast cancer bone metastasis by two distinct actions: both directly, due to lysis of tumor cells by the replicating adenoviruses, and indirectly, due to the activity of OPG in the bone microenvironment inhibiting osteoclastic bone resorption and thus reducing the tumor burden.
The armed CRAd used in this study carries a fusion gene consisting of the N-terminal RANKL-binding domains of OPG fused to the Fc portion of human IgG1. This shortened form of OPG (sOPG-Fc) therefore lacks the C-terminus which has been shown to promote survival in some cancer cell types through its ability to bind tumor necrosis factor-related apoptosis-inducing ligand (TRAIL).14,15 The sOPG-Fc transgene was inserted into the CRAd backbone in place of the E3B region that encodes three proteins (RIDα, RIDβ and 14.7k) which protect infected cells from lysis and consequently are redundant in oncolytic adenoviruses. The sOPG-Fc transgene was placed under the control of the endogenous E3 promoter, splicing and polyadenylation signals, a strategy that has been shown to yield high levels of transgene expression, late in the infection cycle of an armed CRAd.16 This approach retained the gene encoding the E3-11.6k protein, the so-called adenovirus death protein (ADP), which mediates efficient lysis and release of progeny virus from infected cells.17
The sOPG-Fc-armed CRAd was based on a previously described platform, Ad5-Δ24RGD.18 This Ad5-based virus is rendered conditionally-replicative in cells with dysregulated cell cycles by means of a 24-base pair deletion in the CR2 region of the viral E1A gene.19 This mutation renders the E1A protein unable to bind and inactivate the retinoblastoma tumor suppressor/cell cycle regulator protein, Rb, and therefore precludes efficient viral replication in cells with an intact G1-S phase checkpoint (i.e., non-neoplastic or “normal” cells). An arginine-glycine-aspartic acid (RGD) peptide is incorporated into the HI loop of the fiber knob20 of the Ad5-Δ24RGD platform to allow enhanced infectivity via a pathway independent of the native primary cellular receptor for Ad5, the coxsackievirus and adenovirus receptor (CAR), which is downregulated in breast cancer cell lines and primary tumors.21,22 This tropism-modification redirects initial binding of the virus to αVβ3 and αVβ5 integrins, which are often elevated on cancer cells, with αVβ3 integrin associated with breast cancer bone metastasis.23
Materials and Methods
Cells
Human A549 lung carcinoma cells, MCF-10A human breast epithelial cells, and the human breast cancer cell lines MDA-MB-435, MDA-MB-361, MCF-7 and ZR-75 were purchased from the American Type Culture Collection (ATCC; Manassas, VA). The 293 human embryonic kidney cell line was purchased from Microbix (Toronto, ON, Canada). The 911 human embryonic retinoblast cell line was provided by Dr. Alex J. Van Der Eb (Leiden University, Leiden, Netherlands). A subline of MDA-MB-231 cells with an enhanced propensity for bone metastasis has been described previously.24 MDA-MB-435 cells stably expressing luciferase (MDA-MB-435-LUC)25 were provided by Dr. Kurt Zinn (University of Alabama at Birmingham [UAB], Birmingham, AL). ST2 murine bone marrow stromal cells were from Riken Cell Bank, Japan. Primary human dermal fibroblasts were provided by Dr. N. Sanjib Banerjee (UAB). MCF-10A cells were propagated in serum-free complete mammary epithelial growth medium (Cambrex, Walkersville, MD), supplemented with cholera toxin (100 ng/ml; Calbiochem, San Diego, CA). ST2 cells were propagated in α-minimum essential medium (α-MEM). All other cells were cultured in a 50:50 mixture of Dulbecco’s modified Eagle medium (DMEM) and Ham’s F-12 medium. Media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), L-glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 μg/ml). Cells were cultured at 37 °C in a humidified atmosphere, with ST2 cells maintained at 8% CO2 and all others at 5% CO2. Media and supplements were from Mediatech (Herndon, VA) unless noted.
Viruses
The wild-type human adenovirus serotype 5, Adwt300, was purchased from ATCC. The tropism-modified control virus Ad5-RGD, with wild-type E1 and E3 regions and an RGD peptide in the HI loop of the fiber knob, was generated by transfecting A549 cells with Pac I-digested pVK503C.20 Ad5-Δ24 and Ad5-Δ24RGD are CRAds with a 24-bp deletion in the CR2 region of E1A and have been described previously.26 Ad5-Δ24RGD also has an RGD peptide in the HI loop of the fiber knob.
The two armed CRAds were constructed as follows. Human OPG cDNA was amplified by RT-PCR from mRNA isolated from MG-63 cells. The sOPG-Fc fusion gene, encoding the extracellular domain of OPG (amino acids 1-201 [ref 10]) linked to glutamic acid residue 226 of the human IgG1 hinge-Fc region27 was generated by overlap extension PCR. Amino acid residue 230 of the Fc region was mutated from cysteine to serine to prevent dimerization of the Fc domains. The sOPG-Fc gene was subcloned in place of the E3B region into pZErO-2 E3 6.9, a vector containing a 6.9 kbp fragment of the Ad5 genome including the E3 region (Dr. Nik Korokhov, VectorLogics, Inc., Birmingham, AL). The resultant plasmid was linearized with BamH I and cotransformed into E. coli BJ5183 (Stratagene, La Jolla, CA) with Swa I-linearized pVK500CΔE3, a plasmid containing the Ad5 genome deleted for E3 and the fiber gene.20 The resulting plasmid was cotransformed into E. coli BJ5183 with Pme I-linearized pShuttle-Δ24, a plasmid containing a Δ24-modified E1 region of the Ad5 genome.26 The resultant plasmid, pVK500C-Δ24-sOPG-Fc, was was subjected to a final round of recombination by linearization with Swa I and cotransformation into E. coli BJ5183 with pNEB.PK.FHIRGD, a plasmid which contains the Ad5 fiber gene with RGD in the HI loop.20 This final plasmid, pVK500C-Δ24-sOPG-Fc-RGD, was linearized with Pac I and used to transfect A549 cells to generate the tropism-modified armed CRAd, Ad5-Δ24-sOPG-Fc-RGD. The armed CRAd with native tropism, Ad5-Δ24-sOPG-Fc, was generated in a similar fashion by the recombination of Swa I-linearized pVK500C-Δ24-sOPG-Fc with a shuttle plasmid containing the wild-type Ad5 fiber gene.
The E1-deleted vector Ad-CMV-sOPG-Fc-RGD was generated by subcloning the sOPG-Fc fusion gene into pAdenoVator-CMV5-IRES-GFP (Qbiogene, MP Biomedicals, Irvine, CA), under control of the CMV promoter. This plasmid was recombined with pVK503C20 and the recombinant was linearized with Pac I and used to transfect 911 cells. Ad-CMV-OPG-Fc-RGD, with full-length OPG fused to human IgG1 Fc, was constructed similarly.
Viruses were amplified in the respective mammalian cell lines, purified by two rounds of cesium chloride density centrifugation and titered.28,29
Quantitative reverse transcriptase polymerase chain reaction (QRT-PCR) for expression of sOPG-Fc and viral genes
MDA-MB-231 and -435 cells were infected with Ad5-Δ24-sOPG-Fc, Ad5-Δ24-sOPG-Fc-RGD or Adwt300 at a multiplicity of infection (MOI) of 0.1 IU per cell. At 4, 8, 12, 24 and 36 h postinfection, total cellular RNA was isolated from cell lysates (RNeasy Mini Kit, Qiagen, Valencia, CA) and subjected to real-time QRT-PCR (LightCycler 480 system, Roche Diagnostics, Indianapolis, IN). RNA from cells infected with armed CRAds was assayed for expression of sOPG-Fc (forward primer: 5′-GGAGGTGCATAATGCCAAG-3′; reverse primer: 5′-CTGACCACACGGTACGTGCT-3′; probe: 5′-6FAM-TACTGCTCCTCCCGCGGCTTTG-TAMRA-3′). Samples from cells infected with Adwt300 were assayed for expression of E3B genes 14.7k (5′-CAGAGCAGCGCCTGCTAGA-3′; 5′-TGGAGCTCTTGATTCATGCG-3′; 5′-6FAM-TGCTCGGCCGCTGCCCTG-TAMRA-3′) and RIDβ (5′-GCTGGAAACGAATAGATGCCA-3′; 5′-GTTGCAGTGGAAGCATAGCG-3′; 5′-6FAM-ACCACCCAACTTTCCCCGCGC-TAMRA-3′). All samples were analyzed for E3 gp 19k (5′-CCTAGGTTTACTCACCCTTGCG-3′; 5′-CAGGCTGGCTCCTTAAAATCC-3′; 5′-6FAM-CAGCCCACGGTACCACCCAAAAGG-TAMRA-3′), ADP (5′-TCTGCTGCCTAAAGCGCAA-3′; 5′-TTTGGGTGTAGCACAATGATGG-3′; 5′-6FAM-CGCGCCCGACCACCCATCTATAGT-TAMRA-3′) and fiber (5′-TGATGTTTGACGCTACAGCCATA-3′; 5′-GATTTGTGTTTGGTGCATTAGGTG-3′; 5′-6FAM-ACCAAATTCAAGCCCATCTCCTGCATTAATG-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-GGTTTACATGTTCCAATATGATTCCA-3′; 5′-ATGGGATTTCCATTGATGACAAG-3′; 5′-6FAM-CGTTCTCAGCCTTGACGGTGCCAT-3′) was the control. Results are expressed as copy number/ng total RNA.
Secretion and activity of sOPG-Fc
Monolayers of MDA-MB-231 cells in 24-well plates were infected with Ad5-Δ24, Ad5-Δ24-RGD, Ad5-Δ24-sOPG-Fc or Ad5-Δ24-sOPG-Fc-RGD at an MOI of 0.1 IU per cell in DMEM/F12 with 2% (v/v) FBS. Cells were incubated for 1 h at 37 °C before the infection mixtures were removed and replaced with serum-free growth medium with supplements. At various intervals postinfection (24, 36, 48 and 60 h), medium samples were collected and stored at −80 °C. After the final time point, samples were thawed and diluted 1:300. Samples were then assayed for the presence of osteoprotegerin with a Human Osteoprotegerin ELISA kit (RayBiotech, Inc., Norcross, GA). Additionally, samples of conditioned medium collected on day 6 from both the murine and human osteoclast formation experiments were diluted 1:500 and assayed as above.
The ability of sOPG-Fc to bind RANKL was verified. Forty-eight hours postinfection of MDA-MB-231 cells at an MOI of 0.1 IU per cell with Ad5-Δ24-sOPG-Fc or Ad5-Δ24-sOPG-Fc-RGD, medium was harvested and 500 μl aliquots were added to two Eppendorf tubes. One tube of each pair received 1 μg recombinant soluble human RANKL (Research Diagnostics Inc., Concord, MA). Tubes were incubated overnight at 4 °C before adding 30 μl of a protein G-coated agarose bead slurry (EZview Red Protein G Affinity Gel; Sigma-Aldrich, St. Louis, MO). Samples were incubated for 2 h at 4 °C with rotation. The beads were washed twice in PBS, resuspended in Laemmli sample buffer with β-mercaptoethanol (Sigma) and boiled for 5 min. RANKL was detected by SDS-PAGE followed by immunoblotting using a goat anti-human RANKL primary antibody (PeproTech, Rocky Hill, NJ; diluted 1:1000) and rabbit anti-goat alkaline phosphatase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA; diluted 1:4000). Blots were developed with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium chloride.
Viral DNA replication
Monolayers of MDA-MB-231, -435 and MCF-10A cells in 24-well plates were infected with Adwt300, Ad5-Δ24, Ad5-Δ24RGD, Ad5-Δ24-sOPG-Fc or Ad5-Δ24-sOPG-Fc-RGD at an MOI of 0.1 IU per cell and overlaid with 1 ml medium. Liver samples were obtained from a human cadaver in accordance with federal and institutional guidelines and slices measuring 8 mm in diameter and 250 μm in thickness were prepared by a Krumdieck tissue slicer (Alabama Research and Development, Munford, AL) using techniques described previously.30 Liver slices were infected with 1 × 106 IU of Adwt300 or each of the CRAds. At 2 and 4 days postinfection, DNA was extracted from 200 μl samples of medium (QIAamp DNA Blood Mini Kit, Qiagen) and analyzed by real-time Q-PCR for the Ad5 E4 gene as an indicator of viral replication, using previously described primers and probe.31 Results are expressed as copy number/ng total DNA.
Cytopathic effect
MDA-MB-231, -435, -361, MCF-7, ZR-75, primary fibroblasts and MCF-10A (both confluent, non-proliferating and subconfluent, proliferating) cells were infected with Ad5-Δ24-sOPG-Fc-RGD and each of the replicating control viruses at MOIs of 1, 0.1 and 0.01 IU per cell. Eight days postinfection, cells were fixed and stained with crystal violet.
Osteoclast formation
In vitro osteoclast formation assays were performed using murine32 and human precursors. Macrophages from the bone marrow of 4- to 8-week-old female athymic nude Foxn1nu mice (Harlan, Indianapolis, IN) were cultured overnight in α-MEM containing 10% FBS and 10 ng/ml recombinant murine macrophage colony stimulating factor (M-CSF; R & D Systems Inc., Minneapolis, MN). The following day, non-adherent cells were purified on a lymphocyte separation medium (density 1.077 g/ml; Mediatech) gradient. Macrophages were collected from the interface and 2 × 105 macrophages cocultured with 2 × 104 ST2 murine bone marrow stromal cells per well of a 12-well plate, in α-MEM containing 10% FBS, 1 × 10−8 M 1,25-dihydroxyvitamin D3 (Biomol Research Laboratories Inc., Plymouth Meeting, PA) and 1 × 10−6 M dexamethasone (Sigma). Cocultures were given 24 h to allow attachment of cells before porous (0.4 μm pore size) Transwell® inserts 12 mm in diameter (Corning; Corning, NY) were added to the wells. The inserts carried monolayers of MDA-MB-231 or -435 cells infected immediately prior to transfer at an MOI of 0.1 IU per cell with CRAds or Ad-CMV-OPG-Fc-RGD.
Macrophages were prepared from fresh human bone marrow (Lonza, Walkersville, MD). Cells were purified as described above and cultured overnight in α-MEM with 50 ng/ml recombinant human M-CSF (R & D Systems, Inc.). The following day, non-adherent macrophages were harvested, seeded at 5 × 104 cells per well of a 24-well plate, and cultured in α-MEM containing 10 % FBS, 10 ng/ml human M-CSF and 25 ng/ml recombinant human RANKL (R & D Systems, Inc.) for 48 h to allow attachment. Monolayers of MDA-MB-231 or -435 cells, on porous (0.02 μm pore size) 10 mm diameter Anopore® inserts (Nalge Nunc International; Rochester, NY) were infected as described above and transferred to the 24-well plates.
Cultures were maintained in the respective osteoclastogenic media. Every 3 days, 1 ml of conditioned medium in each well was replaced with 1 ml of fresh medium. After 9 days, the inserts carrying cancer cells were stained with crystal violet. Medium from day 9 was assayed for the osteoclast-specific protein tartrate-resistant acid phosphatase 5b33 (TRAP5b) using a murine MouseTRAP or human BoneTRAP ELISA kit (Immuno-diagnostic Systems Inc., Fountain Hills, AZ).
Murine model of osteolytic bone metastasis
Animal experiments were performed in accordance with federal and institutional guidelines for animal care. Osteolytic lesions were established in thirty 4- to 5-week-old female athymic nude Foxn1nu mice (Harlan) by injecting 1 × 105 MDA-MB-435-LUC cells into the left tibia.34 After 10 days, MDA-MB-435-LUC tumors were visualized by noninvasive bioluminescence imaging.35,36 Mice with luciferase-expressing tumors were divided randomly into three groups of 10 animals and given intratibial injections of Ad5-Δ24-sOPG-Fc-RGD, Ad5-Δ24RGD (1 × 106 IU in 20 μl PBS) or PBS only. Luciferase expression was monitored by bioluminescent imaging. Mice were sacrificed on the tenth day post-treatment and the left tibia dissected and preserved in 10% (w/v) neutral buffered formalin (Fisher Scientific, Pittsburgh, PA).
Histology
Tibiae were gently decalcified in EDTA for 14 days and then processed through graded alcohols and xylenes using a VIP 1000 Tissue Processor (Sakura Finetek, Torrance, CA), and embedded in paraffin. Longitudinal, frontal sections were cut at 5 microns, at approximately the meta-diaphyseal level of the tibia. Consecutive sections were stained with hematoxylin and eosin (H & E) and for TRAP with 1.1 mg/ml fast red TR salt in 0.2 M acetate buffer. Another consecutive section was immunostained for Ad5 capsid proteins using rabbit Ad5 antiserum (Cocalico Biologicals, Inc., Reamstown, PA), biotinylated goat anti-rabbit secondary antibodies, and streptavidin peroxidase (Biogenex Laboratories, Inc., San Ramon, CA) with diaminobenzidine (Lab Vision Corporation, Fremont, CA) as substrate.
Histomorphometry
Histomorphometry was performed in the UAB Center for Metabolic Bone Disease Histomorphometry and Molecular Analysis Core Laboratory by individuals blinded to the identity of the experimental groups. All animals for which the inoculation of tumor cells within the bone was confirmed by H & E staining were considered for analysis. This excluded 5 animals from the untreated group, 4 animals from the Ad5-Δ24RGD group and 3 animals from the Ad5-Δ24-sOPG-Fc-RGD group. A region of interest (ROI) was selected that was exactly 0.25 mm distal to the proximal growth plate and extended 4 mm distally (thereby avoiding the primary spongiosa) through the entire medullary portion of the metaphysis and diaphysis of the tibia, but excluding the cortical bone. Standard bone histomorphometry was performed by the methods of Parfitt et al.37 using Bioquant Image Analysis software (R & M Biometrics, Nashville, TN). A two-dimensional histological section displayed profiles of three-dimensional structures. Four primary measurements were made: total tissue area (entire ROI), total tumor area, trabecular bone perimeter, and number of osteoclasts (taken from the TRAP-stained section). Only bone spicules enveloped or adjacent to tumor were considered for analysis. Likewise, only osteoclasts that were within or juxtaposed to the tumor area and were in contact with a visible trabecular bone surface were considered. The primary measurements were used to calculate the percentage of total tissue (including all marrow space and trabecular bone within the ROI) encompassed by tumor and the number of osteoclasts per mm of bone.
Statistical analysis
Data from in vitro osteoclast formation assays were analyzed using a Student’s-Fisher t test. Histomorphometry data were subjected to Wilcoxon two-sample analysis. Data points that were outside of two standard deviations from the mean were considered outliers and were therefore removed from further statistical analysis. Differences were considered significant when P ≤ 0.05.
Results
Construction and characterization of tropism-modified, sOPG-Fc-armed CRAd
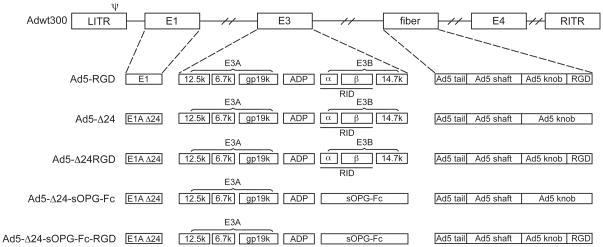
The genomes of the tropism-modified, sOPG-Fc-armed CRAd and the control viruses are shown schematically in Fig. 1. The tropism-modified, armed CRAd, Ad5-Δ24-sOPG-Fc-RGD, was constructed by replacing the E3B region of the Ad5 genome with an sOPG-Fc fusion gene, while retaining the native gene expression control elements. This CRAd was based on the previously described Ad5-Δ24RGD platform and thus has a 24-base pair deletion in the CR2 region of the viral E1A gene and an RGD peptide inserted into the HI loop of the fiber knob. An armed CRAd with native tropism, Ad5-Δ24-sOPG-Fc, was constructed as a control. Unarmed CRAds with native and modified tropism, designated Ad5-Δ24 and Ad5-Δ24RGD, respectively, had been generated previously.18 Ad5-RGD is a tropism-modified control virus, with wild-type E1 and E3 regions.
Figure 1.
Schematic of viral genomes.
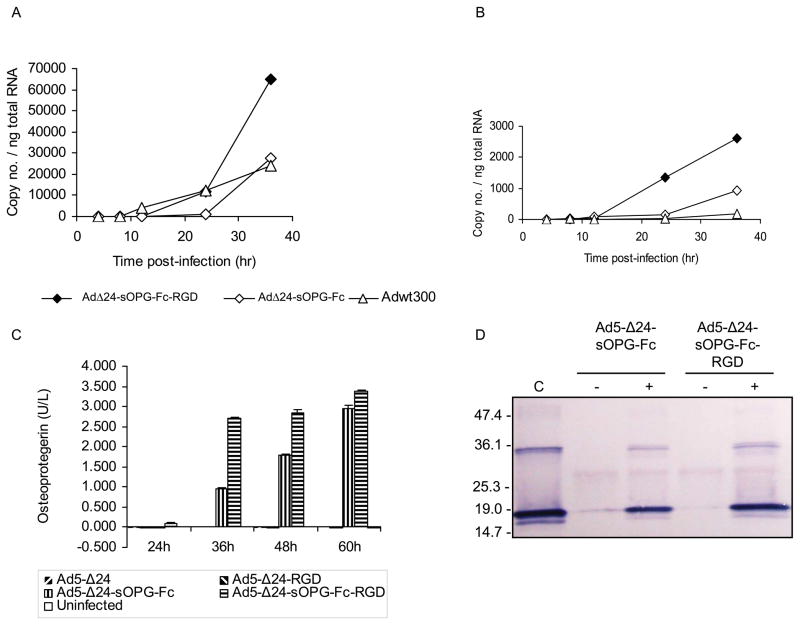
Quantitative RT-PCR confirmed that expression of sOPG-Fc from the E3B region of Ad5-Δ24-sOPG-Fc-RGD and Ad5-Δ24-sOPG-Fc resembled expression of the native E3B genes 14.7k and RIDβ from wild-type Ad5 (Adwt300) in both timing and amount (results shown Fig. 2A for 14.7k). Expression of sOPG-Fc by the armed CRAds did not inhibit expression of the viral ADP gene (Fig. 2B), the E3 gp19k gene or the fiber gene, which is immediately downstream of the transgene (not shown).
Figure 2.
Characterization of armed CRAds. A, B, MDA-MB-231 cells were infected with Ad5-Δ24-sOPG-Fc, Ad5-Δ24-sOPG-Fc-RGD or Adwt300. At the indicated times, cellular RNA was subjected to QRT-PCR to detect expression of: A, the sOPG-Fc gene (cells infected with Ad5-Δ24-sOPG-Fc or Ad5-Δ24-sOPG-Fc-RGD) or the 14.7k gene (cells infected with Adwt300); and B, the ADP gene. The copy number was normalized to total cellular RNA. C, Secretion of sOPG-Fc. MDA-MB-231 cells were infected with Ad5-Δ24, Ad5-Δ24RGD, Ad5-Δ24-sOPG-Fc or Ad5-Δ24-sOPG-Fc-RGD. At the indicated times, medium was harvested, diluted 1:300 and expression of sOPG-Fc was detected in an ELISA. D, sOPG-Fc binds RANKL. Medium from MDA-MB-231 cells 48 h postinfection was incubated in the presence (+) or absence (−) of sRANKL and pulled down with protein G-agarose. Proteins were released and subjected to immunoblot using anti-RANKL primary antibody. C: sRANKL.
MDA-MB-231 cells infected with the armed CRAds secreted sOPG-Fc, which was detectable in the medium from 24 hours postinfection (Fig. 2C). Moreover, cells infected with the tropism-modified armed CRAd yielded higher amounts of sOPG-Fc than the isogenic CRAd with wild-type fibers. The ability of sOPG-Fc expressed by infected cells to bind its cognate ligand, RANKL, was verified. Forty-eight hours postinfection of MDA-MB-231 cells with Ad5-Δ24-sOPG-Fc or Ad5-Δ24-sOPG-Fc-RGD, medium was harvested and incubated in the presence or absence of recombinant soluble human RANKL. Protein G-coated agarose beads, which recognize the Fc domain of sOPG-Fc and RANKL:sOPG-Fc complexes but fail to bind free RANKL, were then employed in a pull-down assay. Proteins were then released from the protein G beads, and SDS-PAGE followed by immunoblotting was performed to detect RANKL (Fig. 2D). This assay confirmed that the sOPG-Fc secreted by cells infected with the armed CRAds was able to bind its native ligand, RANKL. In all cases, similar results were obtained when MDA-MB-435 cells were infected (not shown).
Expression of sOPG-Fc does not enhance the oncolytic potency of a CRAd
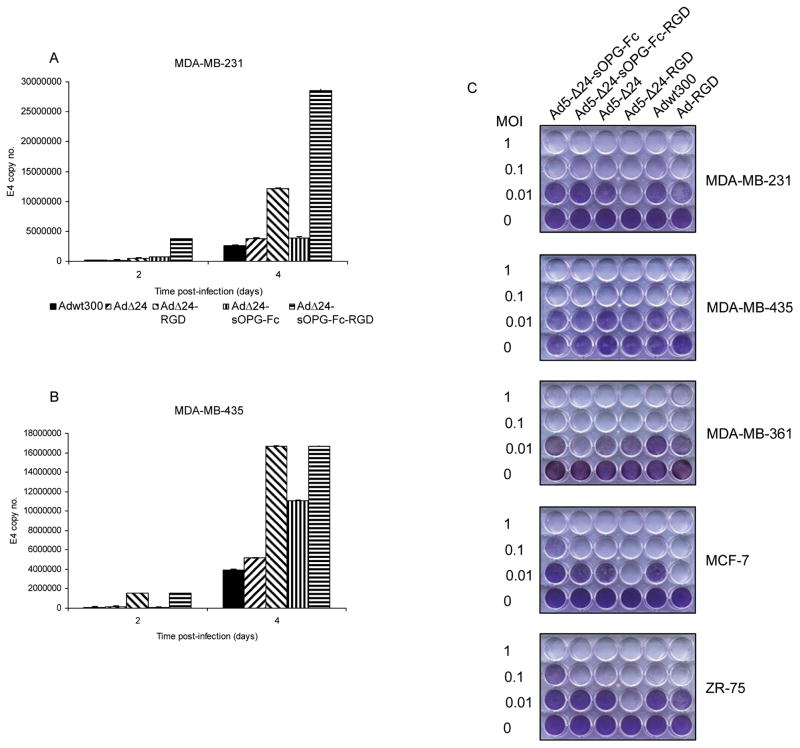
MDA-MB-231 and -435 breast cancer cells were infected with tropism-modified and -unmodified versions of the armed CRAds and the unarmed CRAds, or with a wild-type adenovirus, Adwt300. DNA was isolated from conditioned medium two and four days postinfection and subjected to Q-PCR using primers specific for the Ad5 E4 gene, as an indicator of viral replication. The sOPG-Fc-armed CRAds with either tropism-modified or wild-type fibers replicated at levels similar to their respective unarmed control platforms in both MDA-MB-231 (Fig. 3A) and -435 (Fig. 3B) cells. In addition, both Ad5-Δ24-sOPG-Fc-RGD and Ad5-Δ24RGD exhibited superior replication to their respective controls with wild-type fibers, Ad5-Δ24-sOPG-Fc or Ad5-Δ24, reflecting the enhanced infectivity of the tropism-modified CRAds in these CAR-deficient breast cancer cell lines.
Figure 3.
Oncolytic potency of armed CRAds. MDA-MB-231 (A) and MDA-MB-435 (B) cells were infected. DNA was extracted from medium 2 and 4 days postinfection and subjected to Q-PCR to detect the E4 gene. Results are means ± SD of duplicate determinations. Representative results of 3 separate experiments are shown. C, a panel of breast cancer cell lines was infected at the indicated MOIs. Eight days postinfection, cells were stained with crystal violet. Representative results of 3 separate experiments are shown.
These observations were confirmed in a qualitative determination of oncolytic potency in which a panel of breast cancer cell lines (MDA-MB-231, MDA-MB-435, MDA-MB-361, MCF-7 and ZR-75) was infected. Eight days postinfection, the cytopathic effect was monitored by staining viable cells with crystal violet (Fig. 3C). In total, this set of experiments demonstrated that the oncolytic potency of the CRAds was not enhanced by the expression of sOPG-Fc.
Expression of sOPG-Fc does not alter the selectivity of Δ24-mutant CRAds
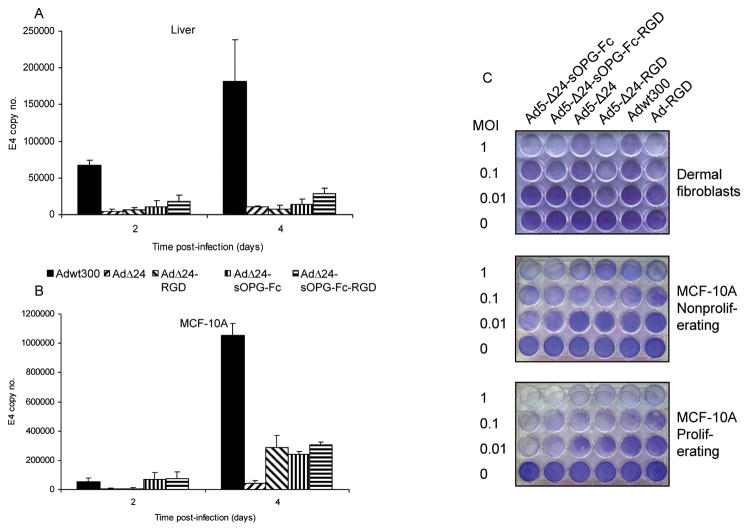
We wished to verify that expression of sOPG-Fc by the armed CRAds did not affect the selectivity of viral replication conferred by the 24-base pair deletion in the E1A gene. Human liver slices and MCF-10A normal human breast epithelial cells were infected with tropism-modified and -unmodified versions of the armed CRAd and the unarmed CRAd, or with a wild-type adenovirus, Adwt300. The conditioned culture medium was harvested at 2 and 4 days post-infection. DNA was extracted and subjected to PCR using primers specific for the E4 region of the adenoviral genome, as a measure of viral DNA replication. The wild-type adenovirus, Adwt300, replicated efficiently in the human liver slices (Fig. 4A), in accordance with previous studies.38 The unarmed CRAds, Ad5-Δ24 and Ad5-Δ24-RGD, exhibited reduced replication in human liver slices with respect to Adwt300. The armed CRAds, Ad5-Δ24-sOPG-Fc and Ad5-Δ24-sOPG-Fc-RGD, retained the selectivity of the CRAd platform. Similar results were obtained in MCF-10A normal breast epithelial cell cells (Fig. 4B).
Figure 4.
Selectivity of the armed CRAds. Viral replication. Human liver slices (A) and MCF-10A normal breast epithelial cells (B) were infected with the indicated viruses. The conditioned culture medium was harvested at 2 and 4 days postinfection. DNA was extracted and subjected to Q-PCR to detect the E4 gene as a measure of viral DNA replication. Results are the means ± SD of duplicate determinations. Representative results of 3 separate experiments are shown. C, a panel of non-neoplastic cells was infected at the indicated MOIs. Eight days postinfection, viable cells were fixed and stained with crystal violet. Representative results of 3 separate experiments are shown.
A qualitative assay demonstrated that non-neoplastic cells, fibroblasts and confluent, non-proliferating and subconfluent, proliferating MCF-10A cells, were more resistant than breast cancer cell lines to replication of Δ24-mutant CRAds and the unarmed control viruses (Fig. 4C). As previously reported, replication of the Δ24 mutant CRAds was greater in proliferating normal cells than in matched, non-proliferating normal cells.39
These studies therefore confirmed that expression of sOPG-Fc did not alter the selectivity of replication of the Δ24-mutant CRAds.
CRAds armed with sOPG-Fc inhibit osteoclast formation in vitro
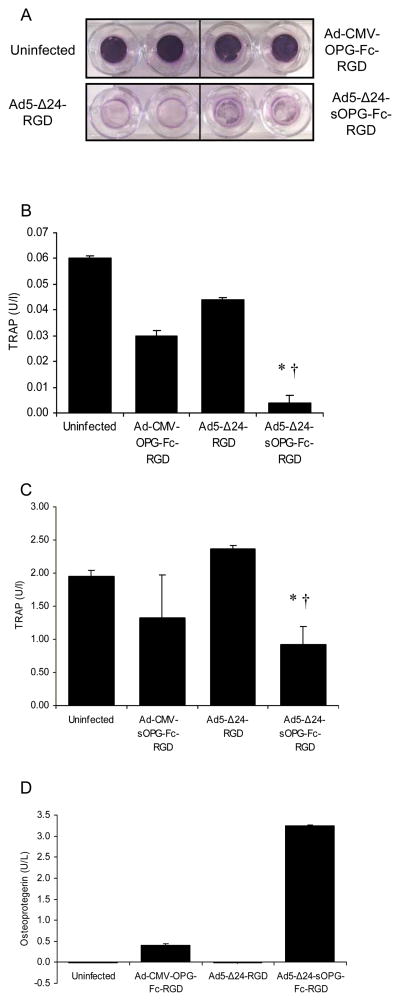
Our hypothesis is that the CRAd armed with sOPG-Fc will eradicate bone metastases of breast cancer both directly, by oncolysis, and indirectly, by inhibiting osteoclastic bone resorption and thus reducing the tumor burden. We next sought to demonstrate in vitro that the sOPG-Fc-armed CRAd could inhibit osteoclast formation while simultaneously replicating in and lysing breast cancer cells. To this end, we employed two in vitro model systems, murine and human. MDA-MB-231 or -435 breast cancer cells growing on permeable cell-culture inserts were first infected with the tropism-modified armed or unarmed CRAds, Ad5-Δ24-sOPG-Fc-RGD and Ad5-Δ24RGD respectively, which possessed increased oncolytic potency relative to the isogenic viruses with native tropism. A tropism-modified, E1-deleted replication-deficient vector expressing OPG-Fc (murine experiment) or sOPG-Fc (human experiment) from the CMV promoter was used as a control. The inserts were then placed in wells containing osteoclast precursors. The murine model system consisted of cocultures of murine bone marrow macrophages and ST2 murine bone marrow stromal cells. The osteoclast precursors in the human model system were human bone marrow macrophages plus recombinant soluble RANKL. Cultures were maintained in osteoclastogenic medium. Under these conditions, osteoclasts are expected to form within 7 to 10 days. Expression of sOPG-Fc was measured in medium samples from day 6. After 9 days, osteoclast formation was assayed by an ELISA for the osteoclast-specific marker protein TRAP5b33 in the medium.
In the model systems, the breast cancer cells were lysed by the unarmed and sOPG-armed CRAds, whereas the replication-defective vectors expressing OPG failed to lyse the breast cancer cells (MDA-MB-435 cells shown in Fig 5A). The level of TRAP5b in the medium was significantly reduced in both the murine (Fig. 5B) and human (Fig. 5C) coculture systems overlaid with MDA-MB-435 breast cancer cells infected by the armed CRAds compared with the unarmed CRAd platform, indicating a reduction in the number of osteoclasts (P < 0.05 for all pair-wise comparisons within an experiment, by Student’s-Fisher t test). This reduction in osteoclast formation correlated with the expression of sOPG-Fc by the armed CRAds (shown for murine system in Fig. 5D). Similar results were obtained when MDA-MB-231 cells were infected. Hence, the sOPG-Fc-armed CRAds could inhibit osteoclast formation while simultaneously lysing breast cancer cells.
Figure 5.
CRAd armed with sOPG-Fc lyses cancer cells and inhibits osteoclast formation in vitro. MDA-MB-435 cells were infected with Ad5-Δ24-sOPG-Fc-RGD, Ad5-Δ24RGD or replication-defective vectors and grown for 9 days on inserts overlaying cocultures of murine osteoclast precursors and ST2 bone marrow stromal cells or human osteoclast precursors and RANKL. A, Cancer cells on the inserts were fixed and stained with crystal violet. The osteoclast marker protein TRAP5b was assayed by ELISA in the murine (B) or human (C) systems. Results are means ± SD of duplicate determinations. Representative results of 2 separate experiments are shown. Significant differences for sOPG-Fc-armed CRAd vs. uninfected (*) and unarmed CRAd (†) are indicated; P ≤ 0.05, determined by Student’s-Fisher t test. D, expression of sOPG-Fc in the medium of the murine system was quantified in an ELISA.
Tropism-modified CRAd armed with sOPG-Fc inhibits breast cancer bone metastasis in vivo
We next sought to determine whether the tropism-modified, armed CRAd, Ad5-Δ24-sOPG-Fc-RGD, would more effectively inhibit the growth of osteolytic breast cancer bone metastases in vivo than its unarmed control, Ad5-Δ24RGD.
Osteolytic tumors were established in female athymic mice by injecting MDA-MB-435-LUC cells into the left tibia. After 10 days, xenografts were confirmed by bioluminescent imaging and mice injected intratibially with Ad5-Δ24RGD, Ad5-Δ24-sOPG-Fc-RGD or PBS only.
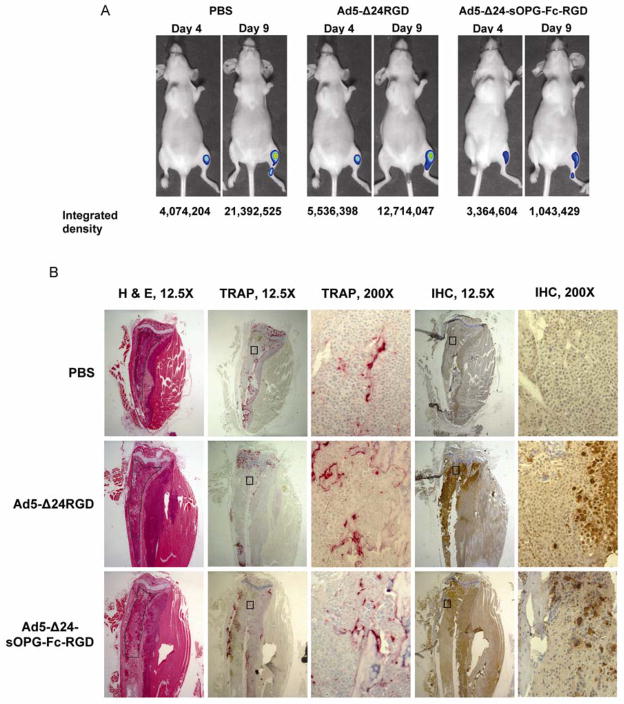
Representative bioluminescence images of mice from each group at day 4 and 9 post-treatment are shown in Fig. 6A. Examination of luciferase images gave some indication that tumors treated with Ad5-Δ24-sOPG-Fc-RGD grew more slowly than did those treated with Ad5-Δ24RGD. Overall, signal intensities increased over time, indicating that tumors continued to grow in all treatment groups.
Figure 6.
Intratibial tumors of MDA-MB-435-LUC cells in nude mice were treated with Ad5-Δ24-sOPG-Fc-RGD, Ad5-Δ24RGD or PBS. A, bioluminescent imaging on days 4 and 9. B, histology of consecutive sections, shown at 12.5X and 200X. H & E: hematoxylin and eosin; TRAP: detection of TRAP osteoclast marker protein with 1.1 mg/ml fast red TR salt in 0.2 M acetate buffer; IHC; immunohistochemistry for Ad5 capsid proteins using rabbit Ad5 antiserum, biotinylated goat anti-rabbit secondary antibodies, and streptavidin peroxidase. Tissue samples were harvested 10 days after treatment.
At the completion of the experiment, mice were sacrificed, and tissue sections of the tibiae prepared. Consecutive sections were stained for tumor with H & E or for osteoclasts by TRAP. An additional section was stained for Ad5 capsid proteins, confirming viral replication in the tumors. Representative sections from mice of each group are shown in Fig. 6B, which correspond to mice shown in Fig. 6A. Mice from each group exhibited extensive bone destruction, with tumor frequently invading the cortex and surrounding muscle. Within each treatment group, a range of tumor sizes was observed histologically.
Histomorphometric determination of tumor area as a percentage of total tissue (Fig. 7A) showed that mice treated with Ad5-Δ24-sOPG-Fc-RGD had significantly less tumor burden in the bone than the untreated group (27.5 ± 16.3 vs. 57.1 ± 30.4, mean ± SD; P = 0.038) or the Ad5-Δ24RGD-treated group (53.2 ± 25.8, P = 0.038). These represent 51% (relative to untreated) and 48% (relative to Ad5-Δ24RGD treatment) decreases in tumor burden for the group treated with Ad5-Δ24-sOPG-Fc-RGD. Mice treated with Ad5-Δ24RGD exhibited a slight reduction in tumor burden relative to the untreated group, but this difference was not significant (P = 0.341).
Figure 7.
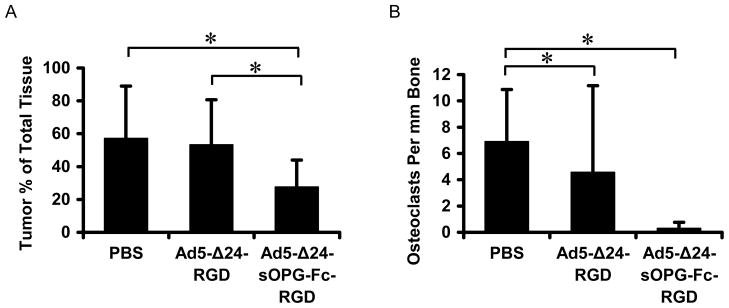
Histomorphometric analysis of stained tissue sections to measure tumor burden (A) and osteoclast formation (B). Shown are group means ± SD, n = 4 (PBS), 5 (Ad-Δ24RGD) and 6 (Ad-Δ24-sOPG-Fc-RGD), *Significant difference, P < 0.05, by Wilcoxon two-sample analysis. Tissue samples were harvested 10 days after treatment.
All tumor-bearing bones showed a decreased amount of trabecular bone and associated osteoclasts in comparison with sections of normal bone (not shown), indicating substantial bone loss. Mice treated with either virus showed a further reduction in osteoclasts per mm bone in comparison to untreated mice (Fig. 7B). Furthermore, treatment with Ad5-Δ24-sOPG-Fc-RGD reduced osteoclast formation as compared to the untreated group (0.28 ± 0.47 vs. 6.89 ± 3.98, P = 0.025) to a greater extent than did treatment with Ad5-Δ24RGD (4.56 ± 6.60, P = 0.049). A reduction in osteoclasts per mm bone was also seen in mice treated with Ad5-Δ24-sOPG-Fc-RGD in comparison to mice treated with Ad5-Δ24RGD (0.28 ± 0.47 for the armed CRAd vs. 4.56 ± 6.60 for the unarmed CRAd), although this effect was not significant (P = 0.07).
Thus, treatment of breast cancer bone metastases with an sOPG-Fc-armed CRAd reduced tumor burden in the bone more effectively than did an unarmed CRAd. Moreover, the armed CRAd reduced osteoclast formation on tumor-associated bone.
Discussion
A number of studies in animal models have shown that arming a CRAd with a rationally chosen therapeutic transgene can improve its antitumor efficacy over that of an unarmed CRAd.40 In terms of Paget’s hypothesis,41 the CRAd armed with sOPG-Fc employed in this study was designed to target both the “seed”, the metastatic breast cancer cell, and the “soil”, the bone microenvironment. We hypothesized that Ad5-Δ24-sOPG-Fc-RGD, a CRAd armed with sOPG-Fc, would inhibit breast cancer bone metastasis both directly, by lysing tumor cells, and indirectly, due to sOPG-Fc in the bone microenvironment inhibiting bone resorption. Using murine and human models of osteoclastogenesis in vitro, we showed that Ad5-Δ24-sOPG-Fc-RGD inhibits the formation of osteoclasts while lysing tumor cells. Our in vivo results showed that expression of sOPG-Fc by a CRAd enhances its ability to control bone metastases, by reducing tumor burden in the bone and inhibiting osteoclast formation. These findings supported our hypothesis that a CRAd armed with sOPG-Fc would have enhanced efficacy against bone metastases, due to both direct and indirect action. This conclusion is further supported by the in vitro study which indicated that the armed CRAd had slightly reduced oncolytic potency relative to the unarmed control: the enhanced efficacy due to inhibition of osteoclast formation could only be observed in the treatment of bone metastases in vivo.
We sought to treat established bone metastases with an sOPG-Fc-armed CRAd. Because these tumors had already progressed from micrometastases at the time of treatment, this more closely reflects the clinical situation than other studies involving OPG and bone metastasis. In most studies, recombinant OPG has been administered in a preventative protocol, with treatment initiated either before or on the same day as the injection of the tumor cells. In these situations, OPG is highly effective at reducing tumor burden, and has been shown to prevent establishment of bone metastases in some cases.12,42–45 Models employing OPG as a treatment for established bone metastases have also been described, and while OPG has been shown to reduce skeletal tumor burden in this setting, complete elimination of bone metastases from treatment with OPG alone has not been reported.44–47 This reflects the difficulty of treating such well-established tumors and is not surprising, given that the ability of OPG to reduce tumor burden in the bone is the product of its antiresorptive activity rather than direct cytotoxicity.46 We suggest that the delivery of sOPG-Fc by a CRAd has the potential to augment the therapeutic efficacy due to the directly oncolytic effect of viral replication.
In the murine model employed here, cells were injected directly into the tibiae of nude mice and allowed to grow for 10 days before treatment was administered. While there is some controversy over the origins of the MDA-MB-435 cell line,48,49 these cells establish osteolytic lesions in vivo, making them suitable for this study.50 Although this model replicates only the final step of the metastatic cascade, the growth of the tumor at a secondary site, it offers advantages that made it an appropriate means of validating our hypothesis. Intracardiac injection of tumor cells, an alternative approach that is also used in studies of bone metastasis, establishes metastases that are widely distributed throughout the body.25,50 Metastases in major organs could lead to the death of the animals before completion of the experiment or otherwise interfere with a study focused on bone metastases. In contrast, the intratibial injection model allowed us to focus our study on tumors in the appropriate tissue and to establish bone metastases at the same location in each animal. The intratibial model has limitations, though. The tibia represents a very small target for injection, making the procedure technically challenging. As we observed, improper injection of cells can establish tumors in the adjacent muscle, rendering these samples unsuitable for further analysis. Even when the initial injection is in the correct location, tumor cells frequently destroy the cortical bone and invade the surrounding muscle, which was observed by us as well as others employing this model.45,51–53 The escape of the tumor from the skeletal compartment has important ramifications for studies of bone metastasis, since treatments intended to reduce tumor burden by inhibiting bone resorption would not be expected to be effective outside the bone. Hence, treatment at an earlier time point should be considered in future studies.
We observed a wide variation in tumor size within each treatment group, despite our efforts to establish consistently sized tumors in all animals. This variability may reflect the differential growth rates of individual tumors in vivo, as well as the differential response of these tumors to treatment. Also contributing to this variability is the fact that relatively small differences in absolute size are made more pronounced by the small size of intratibial tumors. Other investigators have also reported variable tumor sizes in intratibial models of bone metastasis, which prevented some of the trends observed from reaching statistical significance.44 While variation in tumor sizes may, for biological reasons, be unavoidable, the use of more mice per treatment group would allow for the selection of tumors of comparable size for treatment, and would also increase the statistical power of the study.
In this proof-of-principle study, the CRAds were administered by direct injection into the tibiae harboring osteolytic bone metastases. Such an approach would clearly be less practical in a human clinical situation and highlights the need for adenoviruses capable of selective infection of cancer cells after systemic administration. We and others are currently developing such targeted adenoviruses, either by complexing the virus with adapter proteins with specificity for both the vector and a cellular receptor, or by direct modifications to the capsid proteins.54 The discovery that binding of the Ad5 hexon capsid protein to coagulation factor X mediates hepatic transduction will permit the rational engineering of CRAds with reduced liver sequestration.55,56
Acknowledgments
Sources of Support: NIH grants R01 CA108585, T32 CA075930, P30 AR046031, P50 CA089019, DOD grants DAMD17-03-1-0256 and W81XWH-04-1-0800, Susan G. Komen Breast Cancer Foundation grant BCTR0100406, and Haley’s Hope Memorial Support Fund for Osteosarcoma Research at the University of Alabama at Birmingham, USA.
Footnotes
Conflict of Interest
Joanne T. Douglas holds equity in VectorLogics, Inc.
References
- 1.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002;2:584–593. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 2.Coleman RE. Management of bone metastases. Oncologist. 2000;5:463–470. doi: 10.1634/theoncologist.5-6-463. [DOI] [PubMed] [Google Scholar]
- 3.Pavlakis N, Schmidt R, Stockler M. Bisphosphonates for breast cancer. Cochrane Database Syst Rev. 2005:CD003474. doi: 10.1002/14651858.CD003474.pub2. [DOI] [PubMed] [Google Scholar]
- 4.Alemany R, Balague C, Curiel DT. Replicative adenoviruses for cancer therapy. Nat Biotechnol. 2000;18:723–727. doi: 10.1038/77283. [DOI] [PubMed] [Google Scholar]
- 5.Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned? Gene Ther. 2001;8:89–98. doi: 10.1038/sj.gt.3301377. [DOI] [PubMed] [Google Scholar]
- 6.Hermiston T. Gene delivery from replication-selective viruses: arming guided missiles in the war against cancer. J Clin Invest. 2000;105:1169–1172. doi: 10.1172/JCI9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guise TA, Mundy GR. Cancer and bone. Endocr Rev. 1998;19:18–54. doi: 10.1210/edrv.19.1.0323. [DOI] [PubMed] [Google Scholar]
- 8.Thomas RJ, Guise TA, Yin JJ, Elliott J, Horwood NJ, Martin TJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140:4451–4458. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 9.Kakonen SM, Mundy GR. Mechanisms of osteolytic bone metastases in breast carcinoma. Cancer. 2003;97:834–839. doi: 10.1002/cncr.11132. [DOI] [PubMed] [Google Scholar]
- 10.Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Luthy R, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 11.Udagawa N, Takahashi N, Yasuda H, Mizuno A, Itoh K, Ueno Y, et al. Osteoprotegerin produced by osteoblasts is an important regulator in osteoclast development and function. Endocrinology. 2000;141:3478–3484. doi: 10.1210/endo.141.9.7634. [DOI] [PubMed] [Google Scholar]
- 12.Morony S, Capparelli C, Sarosi I, Lacey DL, Dunstan CR, Kostenuik PJ. Osteoprotegerin inhibits osteolysis and decreases skeletal tumor burden in syngeneic and nude mouse models of experimental bone metastasis. Cancer Res. 2001;61:4432–4436. [PubMed] [Google Scholar]
- 13.Body JJ, Greipp P, Coleman RE, Facon T, Geurs F, Fermand JP, et al. A phase I study of AMGN-0007, a recombinant osteoprotegerin construct, in patients with multiple myeloma or breast carcinoma related bone metastases. Cancer. 2003;97:887–892. doi: 10.1002/cncr.11138. [DOI] [PubMed] [Google Scholar]
- 14.Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 15.Shipman CM, Croucher PI. Osteoprotegerin is a soluble decoy receptor for tumor necrosis factor-related apoptosis-inducing ligand/Apo2 ligand and can function as a paracrine survival factor for human myeloma cells. Cancer Res. 2003;63:912–916. [PubMed] [Google Scholar]
- 16.Hawkins LK, Hermiston T. Gene delivery from the E3 region of replicating human adenovirus: evaluation of the E3B region. Gene Ther. 2001;8:1142–1148. doi: 10.1038/sj.gt.3301509. [DOI] [PubMed] [Google Scholar]
- 17.Tollefson AE, Scaria A, Hermiston TW, Ryerse JS, Wold LJ, Wold WS. The adenovirus death protein (E3-11. 6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K, Fueyo J, Krasnykh V, Reynolds PN, Curiel DT, Alemany R. A conditionally replicative adenovirus with enhanced infectivity shows improved oncolytic potency. Clin Cancer Res. 2001;7:120–126. [PubMed] [Google Scholar]
- 19.Fueyo J, Gomez-Manzano C, Alemany R, Lee PS, McDonnell TJ, Mitlianga P, et al. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene. 2000;19:2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- 20.Dmitriev I, Krasnykh V, Miller CR, Wang M, Kashentseva E, Mikheeva G, et al. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shayakhmetov DM, Li ZY, Ni S, Lieber A. Targeting of adenovirus vectors to tumor cells does not enable efficient transduction of breast cancer metastases. Cancer Res. 2002;62:1063–1068. [PubMed] [Google Scholar]
- 22.Anders M, Hansen R, Ding RX, Rauen KA, Bissell MJ, Korn WM. Disruption of 3D tissue integrity facilitates adenovirus infection by deregulating the coxsackievirus and adenovirus receptor. Proc Natl Acad Sci U S A. 2003;100:1943–1948. doi: 10.1073/pnas.0337599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harms JF, Welch DR, Samant RS, Shevde LA, Miele ME, Babu GR, et al. A small molecule antagonist of the alpha(v)beta3 integrin suppresses MDA-MB-435 skeletal metastasis. Clin Exp Metastasis. 2004;21:119–128. doi: 10.1023/b:clin.0000024763.69809.64. [DOI] [PubMed] [Google Scholar]
- 24.Guise TA, Yin JJ, Taylor SD, Kumagai Y, Dallas M, Boyce BF, et al. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J Clin Invest. 1996;98:1544–1549. doi: 10.1172/JCI118947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowey S, Szafran AA, Kappes J, Zinn KR, Siegal GP, Desmond RA, et al. Breast cancer metastasis to bone: evaluation of bioluminescent imaging and microSPECT/CT for detecting bone metastasis in immunodeficient mice. Clin Exp Metastasis. 2007;24:389–401. doi: 10.1007/s10585-007-9076-8. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K, Alemany R, Yamamoto M, Curiel DT. The presence of the adenovirus E3 region improves the oncolytic potency of conditionally replicative adenoviruses. Clin Cancer Res. 2002;8:3348–3359. [PubMed] [Google Scholar]
- 27.Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–14367. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 28.Mittereder N, March KL, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Precious BaR WC. Growth, purification and titration of adenoviruses. In: Mahy BWJ, editor. Virology: A Practical Approach. IRL; Oxford: 1985. pp. 193–205. [Google Scholar]
- 30.Kirby TO, Rivera A, Rein D, Wang M, Ulasov I, Breidenbach M, et al. A novel ex vivo model system for evaluation of conditionally replicative adenoviruses therapeutic efficacy and toxicity. Clin Cancer Res. 2004;10:8697–8703. doi: 10.1158/1078-0432.CCR-04-1166. [DOI] [PubMed] [Google Scholar]
- 31.Rivera AA, Wang M, Suzuki K, Uil TG, Krasnykh V, Curiel DT, et al. Mode of transgene expression after fusion to early or late viral genes of a conditionally replicating adenovirus via an optimized internal ribosome entry site in vitro and in vivo. Virology. 2004;320:121–134. doi: 10.1016/j.virol.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 32.Udagawa N, Takahashi N, Akatsu T, Sasaki T, Yamaguchi A, Kodama H, et al. The bone marrow-derived stromal cell lines MC3T3-G2/PA6 and ST2 support osteoclast-like cell differentiation in cocultures with mouse spleen cells. Endocrinology. 1989;125:1805–1813. doi: 10.1210/endo-125-4-1805. [DOI] [PubMed] [Google Scholar]
- 33.Halleen JM, Alatalo SL, Suominen H, Cheng S, Janckila AJ, Vaananen HK. Tartrate-resistant acid phosphatase 5b: a novel serum marker of bone resorption. J Bone Miner Res. 2000;15:1337–1345. doi: 10.1359/jbmr.2000.15.7.1337. [DOI] [PubMed] [Google Scholar]
- 34.Corey E, Quinn JE, Bladou F, Brown LG, Roudier MP, Brown JM, et al. Establishment and characterization of osseous prostate cancer models: intra-tibial injection of human prostate cancer cells. Prostate. 2002;52:20–33. doi: 10.1002/pros.10091. [DOI] [PubMed] [Google Scholar]
- 35.Le LP, Le HN, Dmitriev IP, Davydova JG, Gavrikova T, Yamamoto S, et al. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006;98:203–214. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- 36.Ono HA, Le LP, Davydova JG, Gavrikova T, Yamamoto M. Noninvasive visualization of adenovirus replication with a fluorescent reporter in the E3 region. Cancer Res. 2005;65:10154–10158. doi: 10.1158/0008-5472.CAN-05-1871. [DOI] [PubMed] [Google Scholar]
- 37.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 38.Rocconi RP, Zhu ZB, Stoff-Khalili M, Rivera AA, Lu B, Wang M, et al. Treatment of ovarian cancer with a novel dual targeted conditionally replicative adenovirus (CRAd) Gynecol Oncol. 2007;105:113–121. doi: 10.1016/j.ygyno.2006.10.057. [DOI] [PubMed] [Google Scholar]
- 39.Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes A, Williams A, et al. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 2000;6:1134–1139. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 40.Cody JJ, Douglas JT. Armed replicating adenoviruses for cancer virotherapy. Cancer Gene Ther. 2009 doi: 10.1038/cgt.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–573. [PubMed] [Google Scholar]
- 42.Zhang J, Dai J, Qi Y, Lin DL, Smith P, Strayhorn C, et al. Osteoprotegerin inhibits prostate cancer-induced osteoclastogenesis and prevents prostate tumor growth in the bone. J Clin Invest. 2001;107:1235–1244. doi: 10.1172/JCI11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanderkerken K, De Leenheer E, Shipman C, Asosingh K, Willems A, Van Camp B, et al. Recombinant osteoprotegerin decreases tumor burden and increases survival in a murine model of multiple myeloma. Cancer Res. 2003;63:287–289. [PubMed] [Google Scholar]
- 44.Tannehill-Gregg SH, Levine AL, Nadella MV, Iguchi H, Rosol TJ. The effect of zoledronic acid and osteoprotegerin on growth of human lung cancer in the tibias of nude mice. Clin Exp Metastasis. 2006;23:19–31. doi: 10.1007/s10585-006-9008-z. [DOI] [PubMed] [Google Scholar]
- 45.Canon JR, Roudier M, Bryant R, Morony S, Stolina M, Kostenuik PJ, et al. Inhibition of RANKL blocks skeletal tumor progression and improves survival in a mouse model of breast cancer bone metastasis. Clin Exp Metastasis. 2008;25:119–129. doi: 10.1007/s10585-007-9127-1. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y, Zhou H, Brennan K, Blair JM, Modzelewski JR, Seibel MJ, et al. Inhibition of bone resorption, rather than direct cytotoxicity, mediates the anti-tumour actions of ibandronate and osteoprotegerin in a murine model of breast cancer bone metastasis. Bone. 2007;40:471–478. doi: 10.1016/j.bone.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Miller RE, Roudier M, Jones J, Armstrong A, Canon J, Dougall WC. RANK ligand inhibition plus docetaxel improves survival and reduces tumor burden in a murine model of prostate cancer bone metastasis. Mol Cancer Ther. 2008;7:2160–2169. doi: 10.1158/1535-7163.MCT-08-0046. [DOI] [PubMed] [Google Scholar]
- 48.Rae JM, Ramus SJ, Waltham M, Armes JE, Campbell IG, Clarke R, et al. Common origins of MDA-MB-435 cells from various sources with those shown to have melanoma properties. Clin Exp Metastasis. 2004;21:543–552. doi: 10.1007/s10585-004-3759-1. [DOI] [PubMed] [Google Scholar]
- 49.Sellappan S, Grijalva R, Zhou X, Yang W, Eli MB, Mills GB, et al. Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64:3479–3485. doi: 10.1158/0008-5472.CAN-3299-2. [DOI] [PubMed] [Google Scholar]
- 50.Harms JF, Welch DR. MDA-MB-435 human breast carcinoma metastasis to bone. Clin Exp Metastasis. 2003;20:327–334. doi: 10.1023/a:1024062911144. [DOI] [PubMed] [Google Scholar]
- 51.van der Pluijm G, Que I, Sijmons B, Buijs JT, Lowik CW, Wetterwald A, et al. Interference with the microenvironmental support impairs the de novo formation of bone metastases in vivo. Cancer Res. 2005;65:7682–7690. doi: 10.1158/0008-5472.CAN-04-4188. [DOI] [PubMed] [Google Scholar]
- 52.Fisher JL, Schmitt JF, Howard ML, Mackie PS, Choong PF, Risbridger GP. An in vivo model of prostate carcinoma growth and invasion in bone. Cell Tissue Res. 2002;307:337–345. doi: 10.1007/s00441-001-0503-x. [DOI] [PubMed] [Google Scholar]
- 53.Fisher JL, Mackie PS, Howard ML, Zhou H, Choong PF. The expression of the urokinase plasminogen activator system in metastatic murine osteosarcoma: an in vivo mouse model. Clin Cancer Res. 2001;7:1654–1660. [PubMed] [Google Scholar]
- 54.Krasnykh VN, Douglas JT, van Beusechem VW. Genetic targeting of adenoviral vectors. Mol Ther. 2000;1:391–405. doi: 10.1006/mthe.2000.0062. [DOI] [PubMed] [Google Scholar]
- 55.Kalyuzhniy O, Di Paolo NC, Silvestry M, Hofherr SE, Barry MA, Stewart PL, et al. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc Natl Acad Sci U S A. 2008;105:5483–5488. doi: 10.1073/pnas.0711757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]