Abstract
Smooth muscle cell (SMC) proliferation is a hallmark of vascular injury and disease. Global hypomethylation occurs during SMC proliferation in culture and in vivo during neointimal formation. Regardless of the programmed or stochastic nature of hypomethylation, identifying these changes is important in understanding vascular disease, as maintenance of a cells' epigenetic profile is essential for maintaining cellular phenotype. Global hypomethylation of proliferating aortic SMCs and concomitant decrease of DNMT1 expression were identified in culture during passage. An epigenome screen identified regions of the genome that were hypomethylated during proliferation and a region containing Collagen, type XV, alpha 1 (COL15A1) was selected by ‘genomic convergence’ for characterization. COL15A1 transcript and protein levels increased with passage-dependent decreases in DNA methylation and the transcript was sensitive to treatment with 5-Aza-2′-deoxycytidine, suggesting DNA methylation-mediated gene expression. Phenotypically, knockdown of COL15A1 increased SMC migration and decreased proliferation and Col15a1 expression was induced in an atherosclerotic lesion and localized to the atherosclerotic cap. A sequence variant in COL15A1 that is significantly associated with atherosclerosis (rs4142986, P = 0.017, OR = 1.434) was methylated and methylation of the risk allele correlated with decreased gene expression and increased atherosclerosis in human aorta. In summary, hypomethylation of COL15A1 occurs during SMC proliferation and the consequent increased gene expression may impact SMC phenotype and atherosclerosis formation. Hypomethylated genes, such as COL15A1, provide evidence for concomitant epigenetic regulation and genetic susceptibility, and define a class of causal targets that sit at the intersection of genetic and epigenetic predisposition in the etiology of complex disease.
INTRODUCTION
Vascular smooth muscle cells (SMCs) are an essential structural component of the artery and provide the necessary contractile function to maintain blood flow (1). The cells themselves lay quiescent until activated by a number of events at the onset of atherosclerosis (2). Activation leads to SMC proliferation and migration into the intima and through a complex cascade of events involving multiple cell types ultimately the production of an atherosclerotic plaque (2). SMCs have the ability to change between different phenotypic states (3), and thus, play a number of roles in vessel stability and disease. Therefore, these cells make for an exquisite model for the study of epigenetic changes associated with their phenotypic plasticity and disease.
One of the key features of a stable atherosclerotic plaque is the presence of a SMC-rich fibrous cap (4,5). SMCs isolated from the atherosclerotic plaque and fibrous cap are pre-senescent (6) and globally hypomethylated (7). As such, they exhibit reduced proliferative capacity and lifespan (8). The pre-senescent state can be achieved through multiple rounds of SMC replication in vitro (6) and comparison of early and late replicated cells has served as a model of SMC replicative aging (6,9,10). In addition, like all cells that lack telomerase activity, SMCs have a lifespan that is limited by the number of cell divisions, resulting in replicative senescence (11). Cell division requires the faithful replication of epigenetic marks such as histone modifications and DNA methylation, a process that cells undergoing replicative senescence do inefficiently (12–15).
Proliferation of SMCs has been linked to changes in DNA methylation, and these changes have been implicated in early atherosclerosis formation and coronary artery disease (CAD) (9,16,17). This process has also been likened to the proliferative state of cancer cells within a solid tumor (18). We hypothesized that DNA methylation changes in the proliferating SMC could regulate SMC phenotype, and that genetic variability within regions of DNA methylation change could account for some of the phenotypic variability and complexity of atherosclerosis. To test this hypothesis and initially characterize genomewide SMC DNA methylation, we sought to identify differential patterns of DNA methylation in replicating, pre-senescent SMCs.
We report herein regions of the genome that change DNA methylation state with SMC passage. Second, we have characterized a novel atherosclerosis gene, COL15A1, involved in SMC phenotype that is regulated by epigenetic state in passaged cells and present in atherosclerotic tissue. Finally, we have identified a common polymorphism associated with atherosclerosis in aged individuals that resides in an epigenetically regulated region of COL15A1 and show that its epigenetic modification correlates with the disease state.
RESULTS
Replicative senescence as a model of SMC aging
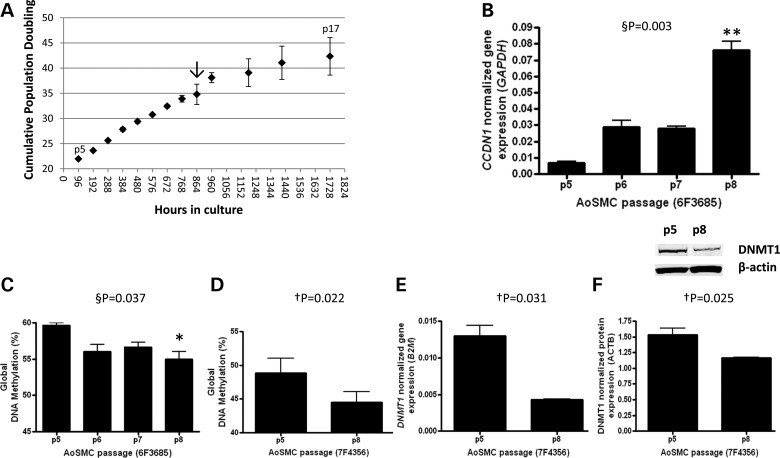
Aortic SMCs (AoSMCs) isolated from a 22-year-old male Caucasian (6F3685) were obtained frozen at Passage 3 from Lonza after undergoing eighteen population doublings (PDs). Cells were plated, counted and allowed to grow for 3 days to recover. These proliferating cells were then plated at 3500 cells per cm2 in triplicate and grown to 80% confluence, which equated to 96 h in culture during exponential growth. This process was repeated until cells no longer reached 80% confluency. The first time point measured in triplicate was at Passage 5 (p5) and the last was at p17 (Fig. 1). In order to identify the molecular changes that lead to senescence, we sought to obtain cells that were at early, pre-senescent states of aging. To do this, senescence was monitored by evaluating cell growth and cyclin D1 (CCDN1) gene expression, a known marker of SMC senescence and the pre-senescent state (19). As previously reported (19), changes in cell proliferation were apparent at ∼35–40 PDs (Fig. 1A, arrow), whereas CCDN1 levels displayed a significant 8-fold increase between p5 and p8 consistent with a critical molecular change to the proliferating cells between 20 and 30 PDs (Fig. 1B). We hypothesized that global change in DNA methylation may be an important molecular event that takes place before cells senesce, and that this change may be apparent by p8. We assayed global methylation levels in passaged SMCs using the LUMA assay (20) and detected a significant loss of methylation (∼5% of 2.4 million CpG sites assayed (21)) between p5 and p8 (Fig. 1C). We replicated this change in a genetically unique AoSMC line (7F4356) derived from a 21-year-old male Caucasian (Fig. 1D). Concomitantly, we identified a decrease in expression of the maintenance methyltransferase DNMT1 at both the transcript (Fig. 1E) and protein levels (Fig. 1F). We have thus categorized p5 and p8 as young and aged, respectively, and refer to the assay as replicative aging.
Figure 1.
Proliferating aortic SMCs exhibit early molecular signs of senescence. (A) Aortic SMCs were grown in culture in triplicate under proliferating conditions and passaged when cells reached 80% confluence. PD with time was calculated. Arrow indicates first change in cell growth between 35 and 40 PDs. (B) Cyclin D1 (CCDN1) transcript levels were assayed by passage to assess changes in the senescent state. (C) Global DNA methylation level was measured by a LUMA assay by passage and (D) in a second AoSMC line for validation. (E) DNMT1 transcript level and (F) protein level was measured in the AoSMC validation line. A representative western blot is shown for DNMT1 and loading control β-actin. §P, repeated measures ANOVA, Bonferroni's multiple comparison test *P < 0.05, **P < 0.01; †P, paired t-test, two-tailed.
A screen for epigenetic changes with SMC replicative aging
We sought to identify regions of the genome that change the DNA methylation state with SMC replicative aging. We hypothesized that DNA methylation changes associated with contextural aging in tissue are pivotal to the development of a complex disease. In order to identify these changes, we conducted an epigenome-wide screen for changes in DNA methylation with SMC replicative aging using a restriction enzyme-based methodology (22). Briefly, DNA isolated from the previously characterized young and aged AoSMCs was subject to sequential digestion with SmaI and XmaI. XmaI adaptors were ligated on to XmaI ends, which represent methylated CG sites in the context of CCCGGG sequence across the genome. Young and aged samples were amplified, labeled and cohybridized to a human BAC microarray (23). Three biological replicates of the same genetic background were assayed and hybridization of the young samples to themselves served as the baseline control. As this experiment was exploratory in nature, an a priori threshold of P ≤ 0.01 (two-tailed t-test) was used to detect significantly differentially methylated regions between young and aged cells without correction for multiple comparisons. The genes within these regions were mapped to the human genome assembly hg18/build36. A total of 344 regions were detected to change with AoSMC replicative age (P < 0.01, two-tailed t-test). Of that group, 176 were hypermethylated and 168 were hypomethylated in aged cells (Supplementary Material, Table S1).
Gene expression changes with aortic SMC replicative age and chemical demethylation
We hypothesized that candidates that show changes in genome level DNA methylation would also show concomitant changes in gene expression. As such, RNA isolated from young and aged SMCs was subjected to gene expression analysis by Illumina beadchip hybridization (48 687 probes assayed). Three biological replicates of the same genetic background were assayed for both young and aged SMC groups. In addition, each set of young and aged cells was treated with 5-Aza-2′-deoxycytidine, a DNA demethylating agent, and RNA was isolated and assayed on the same bead chip as the other samples. Chemical demethylation allows for the identification of loci that are potentially under the control of DNA methylation (direct or indirect), thus adding an additional level of candidate validation. The number of DNA methylation changes in the genome for this assay far exceeds the number of gene expression changes possible, therefore our a priori P-value cutoff was set higher at P ≤ 0.05 for these analyses. A comparison of gene expression changes between young and aged aortic SMCs identified 64 significant changes (P < 0.05, two-tailed t-test) in gene expression representing 60 genes (Supplementary Material, Table S2); none of which met Bonferroni correction (α = 0.05, α′ = 1.03 × 10−6). A comparison of gene expression changes between 5-Aza-2′-deoxycytidine-treated (p5Aza) and -untreated cells (p5) identified 3301 probes that increased significantly with treatment in the young cells (P < 0.05, two-tailed t-test) (Supplementary Material, Table S3). Nine hundred and nine probes met Bonferroni correction (α = 0.05, α′ = 1.03 × 10−6). A comparison of gene expression changes between 5-Aza-2′-deoxycytidine-treated (p8Aza) and -untreated cells (p8) identified 348 probes that increased significantly with treatment in the aged cells (P < 0.05, two-tailed t-test) (Supplementary Material, Table S3). Thirty-nine probes met Bonferroni correction (α = 0.05, α′ = 1.03 × 10−6). Three hundred and forty-five probes overlapped between the two groups, indicating that the majority of epigenetic changes detected with passage were hypomethylation events.
Genomic convergence identifies COL15A1 as an epigenetically regulated target of aging
We combined gene lists sorted by uncorrected P-values from each experiment described (DNA methylation, gene expression changes with passage and gene expression changes with hypomethylation in young or aged cells) with a list of genes that change with disease in human aorta (24). A single gene, COL15A1, exhibited significant loss of DNA methylation and increased gene expression with atherosclerosis and age, as well as a response to 5-Aza-2′-deoxycytidine in young cells (summarized in Table 1). This was the only gene of the group that met all of the criteria. To assess significance across experiments, a meta-analysis was performed combining P-values from the differential methylation and passage expression experiments. Because the measures of expression and methylation are different in each dataset and the anticipated effects and effect sizes are very different in the two datasets, we created a summary measure of evidence across the two datasets using Fisher's formula to combine P-values for heterogeneous datasets (25). Five genes met Bonferroni significance (Table 1, bold line) including COL15A1. This suggests that epigenetic and gene expression changes in COL15A1 occur with replicative age and atherosclerosis, and that we have uncovered a novel molecule that may be involved in SMC phenotype and atherosclerosis.
Table 1.
Meta-analysis of candidate genes that change methylation with SMC age and atherosclerosis
| Gene name | Clone name | Chromosome position (hg19) | Clone methylation P-value | Aorta expression P-value | SMC p5 v p8 expression P-value | Decitabine expression P-value | Meta-analysis P-value |
|---|---|---|---|---|---|---|---|
| FDXR | Chr17tp-12D6 | chr17:72752767-72973410 | 0.003 | < 0.001 | 0.713 | 3.678E−38 | 7.900E−101 |
| FDXR | Chr17tp-3E10 | chr17:72755352-72928265 | 0.009 | < 0.001 | 0.713 | 3.678E−38 | 3.551E−99 |
| ILF3 | Chr19tp-2B6 | chr19:10659649-10837601 | 0.009 | < 0.01 | 0.387 | 4.171E−08 | 4.330E−13 |
| COL15A1 | Chr9tp-4G1 | chr9:101666190-101848490 | 0.009 | < 0.05 | 0.031 | 8.517E−07 | 1.128E−12 |
| SCP2 | Chr1tp-27H8 | chr1:53305823-53420058 | 0.001 | < 0.05 | 0.219 | 5.431E−06 | 3.442E−11 |
| SRC | Chr20tp-7D9 | chr20:35834692-36001722 | 0.009 | < 0.05 | 0.832 | 1.933E−06 | 3.522E−09 |
| CUX1 | Chr7tp-4A2 | chr7:101389746-101602926 | 0.001 | < 0.001 | 0.073 | 2.550E−02 | 1.020E−05 |
| MXRA7 | Chr17tp-3A10 | chr17:74597354-74758571 | 0.001 | <0.001 | 0.504 | 1.026E−02 | 3.282E−05 |
| OAT | Chr10tp-12A6 | chr10:126018450-126199404 | 0.009 | <0.001 | 0.565 | 1.860E−03 | 1.238E−04 |
| IGFBP4 | Chr17tp-10D10 | chr17:38465526-38637706 | 0.001 | <0.05 | 0.057 | 4.580E−01 | 8.327E−04 |
| SKP1 | Chr5tp-22E12 | chr5:133317466-133505837 | 0.003 | <0.05 | 0.961 | 1.600E−02 | 9.354E−04 |
| PSMB7 | Chr9tp-3E12 | chr9:126966658-127144068 | 0.002 | <0.05 | 0.628 | 3.254E−02 | 9.627E−04 |
| TNRC6B | Chr22tp-8E6 | chr22:40465006-40493898 | 0.004 | <0.05 | 0.576 | 2.765E−02 | 1.855E−03 |
| AGAP1 | Chr2tp-24E1 | chr2:236878894-237041900 | 0.001 | <0.05 | 0.849 | 5.907E−02 | 1.913E−03 |
| FGL2 | Chr7tp-5B12 | chr7:76653720-76848379 | 0.003 | <0.01 | 0.251 | 9.569E−02 | 2.307E−03 |
| MFN2 | Chr1tp-25F3 | chr1:11983047-12107571 | 0.008 | <0.05 | 0.160 | 2.007E−01 | 9.114E−03 |
| PMM1 | Chr22tp-8E7 | chr22:41894136-41999953 | 0.002 | <0.01 | 0.356 | 4.807E−01 | 1.283E−02 |
| PEX3 | Chr6tp-16H5 | chr6:143702511-143834084 | 0.002 | <0.05 | 0.607 | 4.090E−01 | 1.999E−02 |
| GPR161 | Chr1tp-31E9 | chr1:167986178-168084802 | 0.005 | <0.05 | 0.175 | 7.304E−01 | 2.349E−02 |
| LILRB3 | Chr19tp-8A11 | chr19:54527514-54723456 | 0.001 | <0.01 | 0.896 | 7.791E−01 | 3.335E−02 |
| SYK | Chr9tp-4A4 | chr9:93520347-93708269 | 0.006 | <0.05 | 0.376 | 5.270E−01 | 4.598E−02 |
| MRPL33 | Chr2tp-4F10 | chr2:27799947-28002099 | 0.002 | <0.001 | 0.940 | 7.105E−01 | 4.600E−02 |
| RHOQ | Chr2tp-10A8 | chr2:46693747-46873151 | 0.005 | <0.05 | 0.594 | 4.951E−01 | 4.785E−02 |
| HTR2B | Chr2tp-31A3 | chr2:231840654-231999694 | 0.006 | <0.05 | 0.951 | 2.826E−01 | 5.565E−02 |
| CD3E | Chr11tp-17H10 | chr11:118065102-118257538 | 0.005 | <0.01 | 0.371 | 8.433E−01 | 5.888E−02 |
| LILRB3 | Chr19tp-2B12 | chr19:54668579-54771282 | 0.003 | <0.01 | 0.896 | 7.791E−01 | 7.671E−02 |
| HTR2B | Chr2tp-17C10 | chr2:231817067-231997557 | 0.009 | <0.05 | 0.951 | 2.826E−01 | 7.980E−02 |
| UBE2E3 | Chr2tp-17B3 | chr2:181866626-182017989 | 0.008 | <0.05 | 0.915 | 3.430E−01 | 8.028E−02 |
| ATP10A | Chr15tp-11D10 | chr15:25878086-26048915 | 0.009 | <0.05 | 0.33 | 7.80E−01 | 8.08E−02 |
Bold indicates gene chosen for follow up studies; Bold line indicates regions that survive Bonferroni correction.
Validation of COL15A1 epigenetic and expression changes
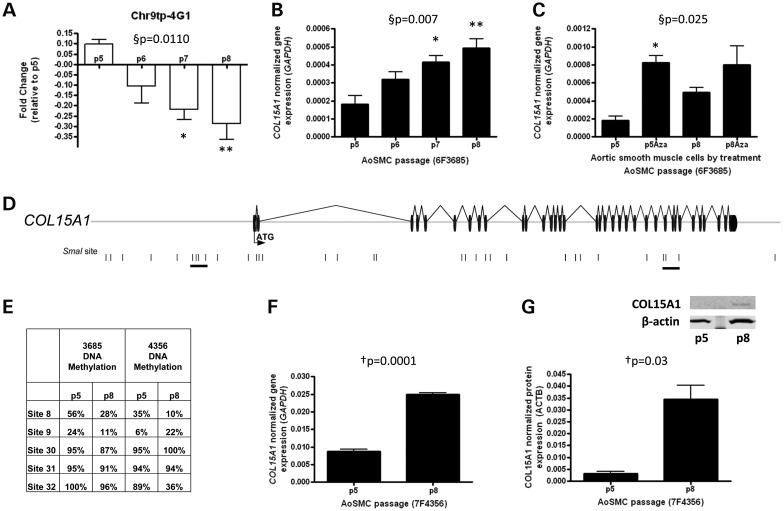
To verify our DNA methylation results, bisulfite-cloned sequencing was performed on young and aged aortic SMCs at the 33 SmaI (CCCGGG) sites contained within the clone containing COL15A1 (Chr9tp-4G1) that was identified in the DNA methylation screen (Fig. 2A). No fewer than 20 clones were sequenced for each sample and site (Supplementary Material, Table S4). This analysis identified 13 sites that lost methylation with age; however, two regions contained groups of sites: 6, 8, 9; and 30, 31, 32 (underlined in Fig. 2D), where at least two SmaI sites that changed were within 3000 bp of one another, allowing for the amplification of a double stranded molecule in the assay used to screen the genome. These sites cluster in two regions of the gene: upstream of the transcription start site (sites 6, 8 and 9) and at the 3′ end of the gene within the introns that surround exon 31 (sites 30 and 31); site 32 falls within exon 35. We also performed quantitative real-time PCR (RT-PCR) to detect COL15A1 gene expression changes with age (Fig. 2B) and with 5-Aza-2′-deoxycytidine treatment (Fig. 2C). We validated these changes in a second SMC line previously described and show hypomethylation with passage at sites 8 and 32 (Fig. 2E) with concomitant changes in transcription (Fig. 2F) and protein levels of COL15A1 (Fig. 2G). The directionality of the gene expression and DNA methylation changes suggests that hypomethylation of COL15A1 can lead to increases in gene expression with replicative age in SMCs.
Figure 2.
Identification of COL15A1 as a gene that is epigenetically regulated in smooth muscle replicative aging. (A) The clone containing COL15A1 exhibits decreased methylation with passage. This was assessed by microarray hybridization of p5, p6, p7 and p8 methylated DNA and displayed as fold change (log2[p8/p5]). (B) COL15A1 gene expression was measured by RT-PCR with age of SMC (n = 3) and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (C) Normalized COL15A1 gene expression was measured by RT-PCR in both young and aged cells in the absence and presence of a demethylating chemical, 5-Aza-2′-deoxycytidine (Aza). Data were normalized to GAPDH. (D) A schematic of the clone containing COL15A1 identified in a screen for genes that change DNA methylation with SMC age. The start codon (ATG) is contained within the first exon; exons are denoted as black discs and introns as lines. SmaI sites contained within this region are denoted by a black vertical line. Groups of SmaI sites that are hypomethylated with age are underlined. (E) Bisulfite cloned sequencing results from two passaged AoSMCs lines. Indicated sites are part of the two underlined clusters in the gene schematic. (F) Validation of COL15A1 expression changes with passage in second AoSMC line (7F4356). (G) Total cellular COL15A1 protein levels were assayed in young (p5) and aged (p8) SMCs by western blot. Quantitation (n = 3) was performed on a LI-COR Odyssey Infrared Imaging System. Data were normalized to β-actin (ACTB). A representative western blot is shown for COL15A1 and loading control β-actin. §P, repeated measures ANOVA, *Bonferroni's Multiple Comparison test P < 0.05; **P < 0.01. †P, paired t-test (n = 3), two-tailed.
COL15A1 levels affect aortic SMC phenotype
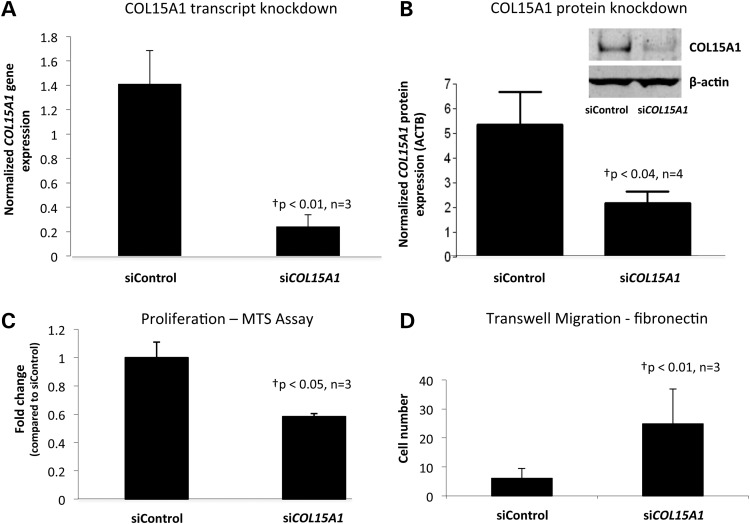
In our model of replicative aging, aging of SMCs requires proliferation; therefore, we hypothesized that COL15A1 levels may regulate the proliferation of SMCs, thus changing the migratory state of the cell. To test this hypothesis we reduced the levels of COL15A1 in the cell using siRNA (Fig. 3A and B) and compared the proliferative and migratory abilities of these cells to a knockdown control that expresses COL15A1 at normal levels. Cells with decreased levels of COL15A1 displayed a reduction in proliferation (Fig. 3C) and an increase in migration to fibronectin (Fig. 3D). These data indicate that levels of COL15A1 are important for key smooth muscle phenotypes.
Figure 3.
COL15A1 levels modify SMC phenotype. (A) Human AoSMCs (7F4356) were transfected with siCOL15A1 or siControl and the level of COL15A1 (A) gene expression normalized to Beta-2 Microglobulin (B2M) and (B) protein expression normalized to β-actin (ACTB) were measured. A representative western blot is shown for COL15A1 and loading control β-actin. (C) COL15A1 control or knockdown cells were assayed for proliferation using the MTS assay from Promega. Data were normalized to the siControl. (D) COL15A1 control and knockdown cells were assayed for their ability to migrate through a permeable membrane in response to fibronectin. †P, paired t-test (n = 3), two-tailed.
COL15A1 expression increases in atherosclerotic lesions
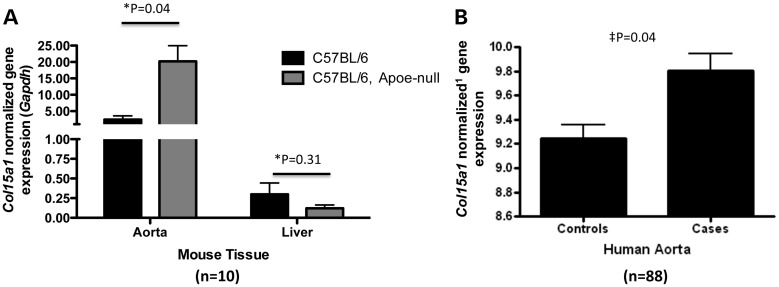
Male Apoe null mice were fed a Western diet for 13 weeks to advance atherosclerosis. Similarly, C57BL/6 mice were fed a chow diet for the same period. Aortas were harvested and quantitation of Col15a1 transcript levels in aorta and liver (n = 5) of wild-type or Apoe null mice confirmed that vessels with disease have higher levels of Col15a1 compared with a tissue not involved with vessel atherosclerosis (liver) (Fig. 4A). This was also true in human atherosclerotic vessels (n = 88) (Fig. 4B). These data provide in vivo evidence for the increase of COL15A1 transcript in response to atherosclerotic disease formation.
Figure 4.
COL15A1 expression increases with atherosclerosis. (A) Col15a1 mRNA levels were measured by RT-PCR in thoracic aorta or liver taken from wild type (C57/BL6) chow-fed mice (n = 5) or Apoe null mice (n = 5) after 13 weeks of a high fat Western diet feeding. Expression of Col15a1 in the thoracic aorta but not liver is significantly increased with accelerated atherosclerosis (high fat diet feeding in an Apoe null background). (B) Microarray gene expression data for COL15A1 from aorta tissue with disease (Cases, n = 26) or without disease (Controls, n = 62). Disease was quantitated using Sudan IV staining for lipids and by counting raised lesions. Controls showed neither lipid staining nor raised lesions. *P, unpaired t-test, two-tailed. ‡P, mixed model (see methods). 1microarray data normalization provided in methods.
COL15A1 is expressed in SMC rich regions of the aorta and fibrous cap
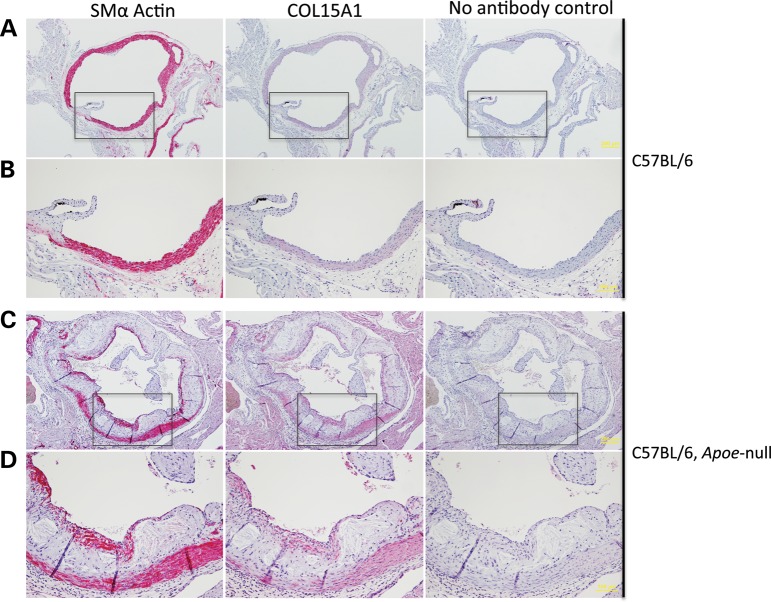
In order to determine the expression pattern of Col15a1 in an intact vessel and in the context of atherosclerosis, we isolated the aortic root from a mouse model of diet-induced atherosclerosis and control mice. Male Apoe null mice were fed a Western diet for 18 weeks to advance atherosclerosis. Similarly, C57BL/6 mice were fed a chow diet for the same period. Aortas were harvested, and the aortic root sectioned and stained for an SMC marker (Acta2) or Col15a1 (Fig. 5). Staining showed that both Acta2 and Col15a1 were expressed in similar regions of the SMC rich media in control mice (Fig. 5A and B). Western diet fed Apoe null mice displayed atherosclerotic plaque formation, medial Acta2- and Col15a1-positive cells as seen in controls, and a cap predominantly constituted of Acta2- and Col15a1-positive cells (Fig. 5C and D). Localized expression of the protein in the lesion cap may suggest an important protective role for COL15A1 in lesion stability.
Figure 5.
Col15a1 is expressed in SMCs in the mouse aorta. Transverse sections of the aortic root were taken from a wild-type C57BL/6 chow-fed mouse (A and B) or a mouse model of atherosclerosis (C57BL/6 Apoe null) fed a high fat Western diet (C and D). Eighteen weeks of Western diet feeding in the Apoe null background induced atherosclerosis (A versus C). Regions boxed in A and C are shown enlarged in B and D, respectively. Scale is indicated in yellow. Four micron sections were stained with an antibody to smooth muscle alpha actin (Acta2) or Col15a1.
Polymorphisms in COL15A1 are associated with atherosclerosis
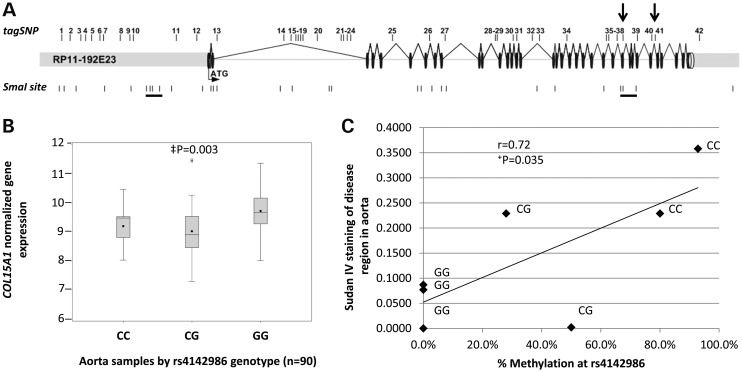
Given the effects of COL15A1 levels on smooth muscle phenotype, we hypothesized that sequence variants within COL15A1 that affect transcription of the molecule may also be associated with atherosclerosis. We identified 42 variants with low linkage disequilibrium (LD) that represent the known common genetic variation in the COL15A1 gene and assayed them in a well-characterized sample of aged atherosclerosis (CATHGEN) (26). Approximately 500 Caucasian individuals aged >55 years were genotyped for genetic association analysis using an additive model to identify variants that are over-represented in individuals with atherosclerosis (Table 2). Cases and controls were chosen on the basis of extent of CAD as measured by CAD index (CADi, see Methods). We identified one variant (rs4142986, P = 0.017) that was significantly associated with atherosclerosis and one variant (rs1413298, P = 0.052) with borderline significance by logistic regression after adjusting for sex and known CAD risk factors (history of hypertension, history of diabetes mellitus, body mass index, history of dyslipidemia and smoking history). The rs4142986 variant is not in strong LD with any of the other variants tested. The maximum r2 observed for rs4142986 is with rs1413298 (r2 = 0.2), raising the possibility that the association seen for rs1413298 may be due to this very low level of LD with rs4142986. Interestingly, both of these variants fall within a CpG site and, therefore, have the potential to be methylated. However, only one of these variants, rs4142986, also reside within one of differentially methylated regions we defined in our epigenetic screen (between SmaI sites 30 and 31, see Figs 2 and 6A). We hypothesized that the methylation state of the risk allele (C allele) of rs4142986 could play a role in expression of COL15A1, and that this could explain its association with disease. That is, if increased COL15A1 expression is atheroprotective, then methylation of rs4142986 may lead to decreased expression and make an individual more prone to disease.
Table 2.
Association analysis of SNPs in COL15A1
| Controls |
Cases |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| tagSNP | PROBE | P-value | Odds ratio | Individuals assayed | Minor allele | Minor allele frequency (%) | Individuals assayed | Minor allele frequency | Minor allele frequency (%) |
| 1 | RS932670 | 0.103 | 0.714 | 275 | C | 14.7 | 214 | C | 11.9 |
| 2 | RS7874187 | 0.606 | 1.077 | 281 | G | 37.2 | 215 | G | 40.0 |
| 3 | RS12379516 | 0.681 | 1.100 | 281 | C | 10.1 | 223 | C | 11.7 |
| 4 | RS4743294 | 0.359 | 1.153 | 272 | A | 29.0 | 213 | A | 31.9 |
| 5 | RS10819454 | 0.165 | 1.480 | 282 | C | 6.4 | 220 | C | 8.0 |
| 6 | RS10819456 | 0.117 | 1.573 | 279 | T | 5.9 | 223 | T | 8.1 |
| 7 | RS7470221 | 0.131 | 0.810 | 281 | C | 47.9 | 221 | C | 46.2 |
| 8 | RS13288865 | 0.755 | 1.083 | 282 | G | 7.4 | 221 | G | 7.7 |
| 9 | RS17710605 | 0.876 | 0.961 | 276 | G | 9.2 | 216 | G | 7.9 |
| 10 | RS10819460 | 0.246 | 1.268 | 282 | C | 14.5 | 221 | C | 15.8 |
| 11 | RS17783501 | 0.247 | 0.842 | 274 | C | 37.6 | 212 | C | 36.1 |
| 12 | RS7045933 | 0.362 | 1.155 | 277 | T | 30.9 | 221 | T | 31.7 |
| 13 | RS6478963 | 0.487 | 1.112 | 280 | C | 29.8 | 221 | C | 31.7 |
| 14 | RS1413294 | 0.612 | 1.092 | 281 | G | 20.3 | 217 | G | 21.4 |
| 15 | RS2050257 | 0.394 | 0.879 | 277 | A | 38.3 | 217 | A | 34.8 |
| 16 | RS4743299 | 0.167 | 1.239 | 282 | T | 25.5 | 222 | T | 28.4 |
| 17 | RS4743300 | 0.759 | 1.045 | 279 | T | 35.1 | 222 | T | 35.4 |
| 18 | RS882727 | 0.445 | 0.851 | 279 | A | 13.6 | 220 | A | 13.2 |
| 19 | RS882728 | 0.992 | 1.002 | 281 | G | 26.2 | 221 | G | 25.1 |
| 20 | RS10819537 | 0.305 | 1.176 | 282 | T | 27.5 | 220 | T | 29.8 |
| 21 | RS1572136 | 0.507 | 1.108 | 276 | C | 28.3 | 220 | C | 29.3 |
| 22 | RS7027650 | 0.840 | 0.971 | 280 | T | 39.8 | 221 | T | 38.5 |
| 23 | RS4743301 | 0.736 | 1.090 | 280 | T | 8.4 | 220 | T | 8.6 |
| 24 | RS989392 | 0.495 | 1.134 | 280 | A | 16.4 | 222 | A | 17.3 |
| 25 | RS2416664 | 0.943 | 0.985 | 278 | T | 12.4 | 221 | T | 12.2 |
| 26 | RS3780622 | 0.374 | 0.860 | 283 | C | 22.3 | 221 | C | 19.9 |
| 27 | RS1889268 | 0.629 | 0.928 | 280 | C | 32.7 | 219 | C | 30.1 |
| 28 | RS10819587 | 0.327 | 0.787 | 277 | A | 10.8 | 220 | A | 9.1 |
| 29 | RS1041631 | 0.430 | 0.856 | 282 | A | 16.5 | 220 | A | 14.5 |
| 30 | RS12352939 | 0.674 | 1.096 | 279 | T | 11.1 | 222 | T | 11.9 |
| 31 | RS7028686 | 0.652 | 1.088 | 280 | G | 14.6 | 221 | G | 15.4 |
| 32 | RS4742755 | 0.563 | 0.883 | 281 | G | 13.3 | 221 | G | 12.9 |
| 33 | RS12684946 | 0.085 | 0.637 | 280 | A | 9.1 | 221 | A | 7.0 |
| 34 | RS17711860 | 0.514 | 1.137 | 283 | G | 13.4 | 220 | G | 14.1 |
| 35 | RS869776 | 0.496 | 0.889 | 278 | G | 23.9 | 219 | G | 22.6 |
| 36 | RS12352174 | 0.528 | 1.134 | 282 | G | 14.4 | 220 | G | 15.0 |
| 37 | RS12216916 | 0.323 | 0.800 | 280 | T | 12.0 | 221 | T | 10.0 |
| 38 | RS4142986 | 0.017 | 1.434 | 280 | C | 30.5 | 219 | C | 37.0 |
| 39 | RS11789689 | 0.108 | 0.689 | 282 | A | 11.3 | 222 | A | 9.7 |
| 40 | RS7047475 | 0.305 | 0.827 | 280 | G | 21.1 | 221 | G | 19.5 |
| 41 | RS1413298 | 0.052 | 0.738 | 281 | C | 32.7 | 221 | C | 27.4 |
| 42 | RS7040869 | 0.827 | 0.954 | 281 | A | 13.9 | 219 | A | 13.2 |
Bold indicates significantly associated SNP.
Figure 6.
A genetic variant in COL15A1 is associated with atherosclerosis, leads to reduced COL15A1 gene expression in human aorta and its methylation state is correlated with disease. (A) Forty-two tag SNPs were selected to cover all of the known genetic variation in COL15A1. These SNPs were genotyped in an atherosclerosis sample (CATHGEN) and allelic association with atherosclerosis was assessed using multivariable logistic regression modeling adjusting for sex and known CAD risk factors. Significant (SNP38) and trending (SNP 41) SNPs are indicated with arrows. Underlined SmaI sites indicate sites that change DNA methylation with passage. (B) COL15A1 gene expression was assessed in aorta tissue regardless of disease and the data were grouped by genotype. C allele carriers have a significant reduction in COL15A1 gene expression. (C) The C allele of rs4142986 is the minor allele and creates a putative DNA methylation site in a region of the gene that changes methylation with age. DNA methylation at rs4142986 was measured in aorta samples scored for disease (Sudan IV). DNA Methylation (%) is plotted versus disease for individuals of each genotype. There is a significant correlation with disease and increased methylation. ‡P, mixed model (see methods); †P, Pearson's correlation, two-tailed.
The risk allele of rs4142986 is associated with a reduction in COL15A1 gene expression in human aorta and its methylation level is correlated with early atherosclerotic disease
In order to test the hypothesis that rs4142986 could play a causative role in the development of atherosclerosis by changing COL15A1 gene expression, we genotyped rs4142986 in 88 human aorta samples and assessed COL15A1 expression levels in relation to genotype (Fig. 6B). Carriers of the rs4142986_C risk allele exhibit significantly decreased (P = 0.003) levels of COL15A1, indicating that this polymorphism, or an SNP in LD with it, may control COL15A1 gene expression. The risk variant is a C/G SNP that resides in a CG site in the 30th intron of COL15A1. This SNP is located 33 bp downstream of the CpG site in SmaI site 30, which is a part of the cluster of sites in the 3′ end of the gene identified to change methylation status with age. We therefore hypothesized that the C allele of rs4142986 could be methylated, and that its methylation level would correlate with disease severity. DNA methylation was assessed at rs4142986 in aorta samples that were scored for disease using Sudan IV staining as a measure of lipid in the vessel (24). We identified samples with each rs4142986 genotype and performed bisulfite cloned sequencing to assess the methylation levels in C/G and C/C individuals. DNA methylation was highest in C/C individuals, intermediate in C/G individuals, and not found in G/G individuals. The level of methylation correlated with the extent of disease; C/C individuals are the most methylated and most diseased (Fig. 6C).
DISCUSSION
We report the results of a convergent analysis of genome-wide gene epigenetic and expression screens in aortic SMCs that identified a novel atherosclerosis susceptibility gene, COL15A1. We show that regions within and flanking COL15A1 change DNA methylation with replicative aging of SMCs, and that a polymorphism associated with atherosclerosis in aged individuals falls within one of these epigenetically regulated regions. This SNP, rs4142986, resides within a CpG site at which the methylation status is associated with atherosclerotic disease. Finally, we provide evidence that COL15A1is implicated in SMC phenotype by inversely modulating the cells ability to proliferate and migrate. These experiments demonstrate the feasibility of in vitro epigenetic and expression screening for a cellular phenotype can lead to the identification of novel genes and causative alleles for complex human disease research.
Known role of collagen type XV alpha 1
COL15A1 is a non-fibrillar member of the collagen superfamily that self-trimerizes via a central triple helix domain (27,28). This molecule is found in the basement membrane of many tissues (29,30) and forms bridges between large collagenous bundles in vivo (31). Although the exact cellular role of COL15A1 is unclear, its location and bridging ability suggest a role for the full-length protein in the stability of cellular matrices, allowing for resilience to mechanical forces (31). A conventional knockout of Col15a1 exhibits subtle, complex phenotypes in both the cardiovascular and nervous systems (32–35). The mice are viable; however, under exercise-induced stress, they develop heart failure, muscle atrophy, and capillary rupture (32). Under normal conditions, these mice exhibit a lowered cardiac ejection fraction at 1 month, which is compensated for by 5 months, in addition to poor capillary perfusion, which persists throughout the lifetime of the mouse (34). In addition, the capillaries in cardiac muscle are tortuous and prone to rupture, which is most likely a function of the impaired basement membrane formation (32,34). Purified recombinant COL15A1 also has demonstrated effects on cell (fibrosarcoma HT1080 cells) adhesion and migration (36). The addition of COL15A1 to a BSA (bovine serum albumin )/fibronectin coating on cell culture plates or a transwell plate significantly decreased the adhesion and migration of human fibrosarcoma cells, respectively (36). Intriguingly, COL15A1 also produces a cleavage product called restin that shares 72% identity with COL18A1's endostatin molecule (37). Injections of recombinant mouse endostatin significantly reduce angiogenesis and tumor growth in a mouse orthotopic injection model (38), and overexpression of human COL15A1 in cervical carcinoma cells completely ablates their tumorigenicity (39). Although both restin and endostatin have interesting antitumor effects, it is not yet known whether the previously described adhesive and migratory effects of COL15A1 on human fibrosarcoma cells are due to the full-length collagen, or to this cleavage product. The current literature is mixed on whether recombinant mouse restin has anti-proliferative or anti-migratory effects on endothelial cells (40,41). This is clearly a rich avenue for further research, and COL15A1 or more likely, restin, may be a novel target for drug therapy for atherosclerotic diseases.
SMC-specific expression of COL15A1
The expression of Col15a1/COL15A1 in SMCs is of particular interest in the context of atherosclerosis. The process by which SMCs migrate from the media into the intima, and subsequently through the forming plaque to form a stabilizing fibrous cap (3,42–44), is dependent on extracellular matrix components (45). COL15A1 is an extracellular matrix protein that affects both the proliferative and migratory phenotypes of proliferating SMCs, indicating that it may play an important role in the initiation of atherosclerosis. Its presence in the fibrous cap further suggests a role in stabilization and potential control of SMC phenotype. Given the weight of these data, we propose that epigenetic changes associated with replicative senescence of SMCs can control the phenotypic state and may play an essential role in SMC activation in the development of atherosclerosis.
The intersection of genetic and epigenetic susceptibility in atherosclerosis
It is clear that genetic predisposition plays a significant role in the etiology of complex disease. It is also increasingly apparent that epigenetic changes contribute to the pathophysiology of disease development (46). However, chronic diseases such as obesity, atherosclerosis and diabetes, rather than being caused by these individual mechanisms, may also be partly driven by the intersection between the altered chromatin structure and their heritable genetic components (46). Our approach began with the integration of genome-wide epigenetic and expression data to identify the molecular and cellular mechanisms underlying complex disease in an etiologically relevant tissue using a cellular model of age-related atherosclerosis. A single gene, COL15A1, satisfied numerous a priori molecular criteria at nominal significance levels. Further, we carried out genetic analysis and identified significant association of a nucleotide polymorphism that falls within a methylated CpG dinucleotide that is differentially methylated and contributes to the regulation of the gene in an allele specific manner. Our study highlights the importance of integrated analyses using independent datasets, particularly from genetic and epigenetic sources. The individual analysis of these datasets of complex disease may identify nominal significance of a given platform, however recognizing the intersection of genetic predisposition, aberrant epigenetic regulation and the functional surrogacy of differential expression, will facilitate the identification of novel genes and functionally important pathways, even in a model with the complexity of aging-dependent cardiovascular disease.
MATERIALS AND METHODS
Aortic SMC culture
Cryopreserved human aortic SMCs lot #6F3685 (derived from a 22-year old, Caucasian male) were obtained from Lonza (Walkersville, MD) at Passage 3 and cultured at Duke University. Cells were grown in T-75 SoLo Flasks Nunclon™Δ (NUNC, Rochester, NY, USA) according to manufacturer's instructions (www.lonzabioscience.com). The initial proliferating culture was grown to 80% confluency in one T75 flask. These cells were counted one day after plating to determine the starting number of viable plated cells (∼300 000 cells), which was used to determine starting PD at Passage 5 (∼22 PD). From this initial culture, 3500 cells /cm2 were plated in three separate flasks and were incubated at 37°C for 96 h (∼80% confluency). Media was replaced every 48 h. Cells were grown in this fashion and reached 80% confluency in 4 days until Passage 14. At this point, cells were given additional time to reach 80% confluency as indicated. Each flask represents an individual biological replicate of the same genetic background, and these three replicates were used for the genome screen. Cryopreserved human aortic SMCs lot #7F4356 (derived from a 22-year old, Caucasian male) were obtained from Lonza (Walkersville, MD) at Passage 3 and cultured at the University of Virginia until Passage 8 under the same conditions described.
AoSMC DNA and RNA sample isolation
DNA was isolated using the Gentra Puregene Cell kit from Qiagen (Valencia, CA, USA), and RNA was isolated using RNAlater and a Ribopure kit both from Ambion/Applied Biosystems (Foster City, CA). Complementary DNA (cDNA) was synthesized using an Illumina TotalPrep RNA amplification kit from Ambion/Applied Biosystems. Additionally, all COL15A1 findings were also confirmed using a different AoSMC line, lot 7F4356.
LUMA assay for global DNA methylation
A LUMA assay was performed as described in Karimi et al. (20). Briefly, 200 ng of high-quality genomic DNA was double digested with HpaII and EcoRI or MspI and EcoRI (New England Biolabs, Ipswich, MA, USA) for 4 h at 37°C. Pyrosequencing was performed using a Pyromark Q24 (Qiagen) in an SNP mode assay with the following sequence to be analyzed: ACTCGA. Each reaction was run in triplicate.
5-Aza-2′-deoxycytidine treatment
5-Aza-2′-deoxycytidine was obtained from Sigma-Aldrich (St Louis, MO, USA) and diluted into SMG-2 media to 5 µm. Passage 5 and 8 cells were cultured to 70% confluence, and media with or without (mock control) 5-Aza-2′-deoxycytidine was added to the flask. Cells were cultured for 72 h and harvested for nucleic acid purification.
Epigenetic screen
Methylated DNA from AoSMCs was isolated from cells (lot #6F3685) at Passages 5–8. DNAs from test and reference samples were prepared by the MCA method (47). Briefly, DNA was digested with the methylation-sensitive restriction enzyme, SmaI, generating blunt end cuts followed by digestion with the methylation-insensitive restriction enzyme, XmaI, generating sticky ends. Adapters are ligated to the sticky ends following XmaI digest and the DNA is amplified as described (22) by a PCR using primers that anneal to the adapter sequences. The amplicons from the sample and control are then individually labeled with Cy5 and Cy3 fluorescent dyes. Passage 5 DNA was labeled with Cy5 and Cy3 and co-hybridized to the genomic tilepath microarray, this served as our experimental control of no change. Clones that performed poorly in the self–self experiments were discarded from the dataset (log2 ratio +/− 0.4, variance >0.3). Three biological replicates of the same genetic background at Passages 5 versus 8 were labeled with Cy3 and Cy5, respectively, and co-hybridized to a genomic microarray (23). Data were collected using an Axon scanner and a Bluefuse microarray analysis program was used for data processing. The program incorporates Bayesian statistical models in the analysis and provides: (1) a more uniform analysis of the data by eliminating variability introduced by human examination, (ii) more reproducible results across experiments by normalizing experimental variation (Block Lowess) and (iii) an increased amount of usable data. The software exports the results in excel format. For each clone on the array, a normalized intensity value and user defined log2 ratios (log2[p8 intensity/p5 intensity]) were exported from BlueFuse and the median log2 ratio value for each experimental replicate was calculated. The control experiment, Passage 5 versus Passage 5, the same DNA hybridized to itself provided a baseline of clone variation. The experimental data (Passages 5 versus 8) were compared using a two-tailed t-test to identify significantly differentially methylated clones. We then correlated the sequence contained within these differentially methylated clones to the SmaI sites within the sequence, as well as localized annotated genes and predicted CpG islands, promoters and regulatory regions.
Gene expression using Illumina Beadarray
Global gene expression profiling was conducted for Passages 5 and 8 SMCs using Illumina Human WG-6 V2 BeadArrays as described by the manufacturer (Illumina, Inc., San Diego, CA, USA). Rank invariant normalization was applied to the raw data, and a two-tailed t-test was used to identify significant changes in gene expression between the two time points.
DNA methylation analysis
DNA (200 ng) was bisulfite treated using a MethylCode bisulfite conversion kit from Invitrogen (Carlsbad, CA, USA), and primers were designed to amplify each SmaI site within COL15A1. A PCR was performed on bisulfite converted DNA from Passages 5 and 8 with the following reaction conditions: 95°C for 10 min (min), 35 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 3 min, 72°C for 10 min. PCRs were run on a 2% agarose gel and single bands corresponding to the predicted molecular weight of the product were extracted. DNA was isolated from gel fragments using a Qiagen Gel purification kit. DNA was TOPO-TA cloned using Invitrogen's TOPO-TA cloning kit. Twenty-four clones were picked and sequenced for each site. A minimum of 20 clones were used to determine percent methylation at each site. Primer sequences for each site are listed in Supplementary Material, Table S5.
Quantitative RT-PCR
Applied Biosystems Taqman Gene expression assays were used to perform quantitative RT-PCR (COL15A1- Hs01559630_m1, CCDN1- Hs00765553_m1, DNMT1-Hs00154749, B2M-Hs99999907_m1 and GAPDH - Hs02758991_g1). The following reaction components were used for each probe: 2 µl cDNA, 5 µl Custom TaqMan SNP master mix (Applied Biosystems), 0.5 µl assay and 2.5 µl water. Reactions were performed in a single 384-well plate in triplicate using an ABI PRISM® 7900HT sequence Detection System. PCR conditions were as follows: Step 1: 50°C for 2 min, Step 2: 95°C for 10 min, Step 3: 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Expression relative to GAPDH or B2M was calculated using 2ΔΔCt 42.
Proliferation and migration assays
Cell proliferation assays were performed using Promega's CellTiter 96 Aqueous One Solution Cell Proliferation assay (Madison, WI, USA) according to the manufacturer's directions. Cell migration assays were performed on Millipore MultiScreen-MIC plates containing 8 µm pores. Cells at Passage 5 or 6 were grown to 70% confluence and then switched to serum-free media. A cell suspension (1 × 105 cells/ml, 150 µl) was added to the upper well in serum-free media containing 0.1% BSA (Sigma). Fibronectin (5 µg/ml, Sigma) was added to the bottom chamber in serum-free media with 0.1% BSA (FN-induced chemotaxis). The chambers were incubated at 37°C in a CO2 incubator for 18 h, and fixed in 4% formaldehyde. The non-invaded cells were removed from the upper wells and the invaded cells were stained with 0.2% Crystal Violet solution in 7% ethanol. Cells from 8 to 10 randomly chosen high-power fields (magnification ×20) on the lower surface of the filter were counted (48).
COL15A1 western analysis
Cells were lysed in RIPA with protease inhibitors. Cleared lysates were reduced and electrophoresed on a 3–8% NuPAGE Tris Acetate gel. Proteins were then transferred onto a nitrocellulose membrane. After an initial blocking with 3% dried milk in TBST, membranes were probed with 1 µg/ml Goat anti-COL15A1 (C-20) or anti-DNMT1 (H-300) (both from Santa Cruz Biotechnology, Santa Cruz, CA, USA) diluted in 3% BSA and incubated overnight at 4°C. The membranes were then washed and incubated with a Donkey anti Goat IR-Dye 800CW (LI-COR Biosciences, Lincoln, NE, USA) diluted in blocking buffer. Membranes were then analyzed using the ODYSSEY Infrared Imaging System (LI-COR Biosciences).
Animals
Animal protocols were approved by the Animal Care and Use Committee at the University of Virginia. To accelerate atherosclerosis, mice were fed a Western atherogenic diet (Harlan Teklad, Madison, WI, USA) containing 21.2% fat by weight (0.2% by weight cholesterol, 17.3% by weight protein and 48.5% by weight carbohydrate) for 13 weeks (RNA quantiation) or 18 weeks (immunostaining).
Mouse aorta and liver samples and expression
Apoe null mice (male, 8 weeks of age) were obtained from The Jackson laboratory (Bar Harbor, ME, USA). Mice were fed a Western atherogenic diet for 13 weeks. C57BL/6 mice (male, 21 weeks of age) on a chow diet were used as a control. The Fast Prep FP120 (Q-Biogene) was used to homogenize tissues, and total RNA was prepared from aorta and liver. Col15a1 expression in each sample was normalized to Gapdh levels.
Human donor aorta samples and expression
Human aorta samples were collected from heart transplant donors as previously described (24,26). The study was approved by the Duke institutional review committee and all participants gave informed consent. DNA and RNA were isolated from each sample. Eighty-eight samples in total were analyzed representing 58 unique samples, as 30 individuals had more than one sample. As harvested tissue was obtained from deceased heart donors, the clinical data associated with these aortas are very limited and consists of age, sex and self-reported race (49). All samples were scored for the presence of raised lesions and stained with Sudan IV to detect lipid accumulation as previously reported (49). Cases (n = 26) and controls (n = 62) were determined using the following criteria: any evidence of raised lesions and Sudan IV staining was considered a case, while individuals with no raised lesions (RL = 0) and no Sudan IV staining were considered controls. We were interested in the mean expression level of COL15A1 in the two groups, the standard deviation and then the test of significance of this difference. We performed the test using a mixed model to account for the fact that several individuals have more than one tissue sample and thus more than one expression value. We also adjusted for age, sex and race in the mixed model. Aorta genotyping was performed using Applied Biosystems Taqman allelic discrimination assays, and expression profiling was performed using Affymetrix GeneChip U95Av2 (Affymetrix, Santa Clara, CA, USA). Expression signal intensity values were log2 transformed and normalized using quantile normalization. We analyzed cis effects of COL15A1 variants on COL15A1 expression using the Affymetrix tag 38427_at, representing the 3′ end of COL15A1. Because some subjects had multiple samples while others did not, we treated each individual and sample separately. The expression level of the tag was modeled using multiple linear regression including age, sex, race and additive genotype. To account for repeated measures (i.e. multiple sections per subjects) and as a validation, a mixed model as implemented in the SAS PROC MIXED procedure was utilized (SAS Institute Inc., Cary, NC, USA).
COL15A1 immunostaining
Transverse sections of the aortic root were taken from a wild-type (C57/BL6) chow-fed mouse and an Apoe null mouse fed Western diet for 18 weeks (kindly provided by Dr Coleen McNamara, UVA). Immunostaining was performed on 4 µm paraffin sections that were deparaffinized and incubated in 20 µg/ml proteinase K (Sigma P6556) for 3 min at room temperature, followed by blocking with 2% normal sera and subsequent incubation with 5 µg/ml goat anti-human COL15A1 (C-20, Santa Cruz Biotechnology, Inc.) or biotinylated anti-α SMA (Sigma) overnight at 4°C. After incubation with the AP–ABC complexes, immunoreactivity was visualized by incubation with alkaline phosphatase substrate (Vector Labs). No antibody control experiments were performed in an identical fashion as described for COL15A1 staining, except that COL15A1 antibody was not added.
CAD case–control sample (CATHGEN)
CATHGEN participants (n = 1279) were recruited sequentially through the cardiac catheterization laboratories at Duke University Hospital (Durham, NC, USA) with approval from the Duke Institutional Review Board. All participants undergoing catheterization were offered participation in the study and signed informed consent. Medical history and clinical data were collected and stored in the Duke Information System for Cardiovascular Care database maintained at the Duke Clinical Research Institute. Controls and cases were chosen on the basis of extent of CAD as measured by CADi. CADi is a numerical summary of coronary angiographic data that incorporates the extent and anatomical distribution of coronary disease. Affection status was determined by the presence of significant CAD defined as a CADi ≥ 74, equivalent to three-vessel disease with at least a 95% stenosis in the proximal left anterior descending coronary artery, as we have used for previous studies (26,50). A comparison of clinical characteristics between CATHGEN cases and unaffected controls has previously been presented (50). An aged Caucasian sample was used for the analysis. Medical records were reviewed to determine the age of onset (AOO) of CAD, i.e. the age at first documented surgical or percutaneous coronary revascularization procedure, myocardial infarction (MI) or cardiac catheterization meeting the above-defined CADi thresholds. The CATHGEN cases AOO > 55 years (n = 250) and controls had an age at catheterization ≥60 years (n = 220) and had no CAD as defined by coronary angiography and no documented history of MI, heart transplant, peripheral or cerebrovascular disease, or interventional or surgical coronary revascularization procedures.
SNP genotyping
The Human HapMap project (Release 24/phaseII Nov08) data and the algorithm Tagger (51) were used to generate tagSNPs for RP11-192E23 (clone identified in epigenetic screen, contains COL15A1) using data derived from the CEU population (Utah residents with ancestry from northern and western Europe), given that the CATHGEN sample used was Caucasian (26). A total of 42 SNPs were genotyped at the Center for Human Genetics using Taqman allelic discrimination assays. A total of 15 quality control samples—composed of six reference genotype controls in duplicate, two CEPH pedigree individuals and one no-template sample, were included in each quadrant of the 384-well plate. SNPs that showed mismatches on quality control samples were reviewed by an independent genotyping supervisor for potential genotyping errors. All SNPs examined were successfully genotyped for 95% or more of the individuals in the study. Error rate estimates for SNPs meeting the quality control benchmarks were determined to be <0.2%.
SNP statistical analysis
All SNPs were tested for deviations (P ≤ 0.0001) from Hardy–Weinberg equilibrium (HWE) in the affected and unaffected groups. Two SNPs deviated from HWE, rs7470221 (cases) and rs1413294 (cases). Neither of these SNPs was statistically significant in our association analysis. LD between pairs of SNPs (r2 > 0.7) was assessed using the Graphical Overview of Linkage Disequilibrium package (52). No strong LD (r2 > 0.7) was found, confirming the tagSNP selection. Genetic association in the CATHGEN cases and controls was evaluated using multivariable logistic regression modeling, including the SNP genotype coded for an additive genetic model, i.e. zero, one or two copies of the minor allele. The model was adjusted for sex, and known CAD risk factors (history of hypertension, history of diabetes mellitus, body mass index, history of dyslipidemia and smoking history) as covariates. These adjustments could hypothetically allow us to control for competing genetic pathways that are independent risk factors for CAD, thereby allowing us to detect a separate CAD genetic effect. SAS 9.1 (SAS Institute, Cary, NC, USA) was used for statistical analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health (K99/R00HL089412, HL073389, HL73042 and AG028716).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the individuals who have provided samples to the CATHGEN study. We acknowledge the essential contributions of the following individuals to making this publication possible: Jennifer L. Owens, Brittany Durgin, Elaine Dowdy and the staff at the Center for Human Genetics. We thank Neil Freedman for critical evaluation of the atherosclerotic lesion data. We thank Coleen McNamara for critical reading of the manuscript and for providing slides for immunostaining of Col15a1 in Apoe null and C57/BL6 mice.
REFERENCES
- 1.Owens G.K. Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 2.Watkins H., Farrall M. Genetic susceptibility to coronary artery disease: from promise to progress. Nat. Rev. Genet. 2006;7:163–173. doi: 10.1038/nrg1805. [DOI] [PubMed] [Google Scholar]
- 3.Owens G.K., Kumar M.S., Wamhoff B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 4.Libby P., Geng Y.J., Sukhova G.K., Simon D.I., Lee R.T. Molecular determinants of atherosclerotic plaque vulnerability. Ann. N. Y. Acad. Sci. 1997;811:134–142. doi: 10.1111/j.1749-6632.1997.tb51996.x. discussion 142–5. [DOI] [PubMed] [Google Scholar]
- 5.Davies M.J., Woolf N., Rowles P., Richardson P.D. Lipid and cellular constituents of unstable human aortic plaques. Basic Res. Cardiol. 1994;89(Suppl 1):33–39. doi: 10.1007/978-3-642-85660-0_3. [DOI] [PubMed] [Google Scholar]
- 6.Gorenne I., Kavurma M., Scott S., Bennett M. Vascular smooth muscle cell senescence in atherosclerosis. Cardiovasc. Res. 2006;72:9–17. doi: 10.1016/j.cardiores.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Hiltunen M.O., Turunen M.P., Hakkinen T.P., Rutanen J., Hedman M., Makinen K., Turunen A.M., Aalto-Setala K., Yla-Herttuala S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc. Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 8.Ross R., Wight T.N., Strandness E., Thiele B. Human atherosclerosis. I. Cell constitution and characteristics of advanced lesions of the superficial femoral artery. Am. J. Pathol. 1984;114:79–93. [PMC free article] [PubMed] [Google Scholar]
- 9.Ying A.K., Hassanain H.H., Roos C.M., Smiraglia D.J., Issa J.J., Michler R.E., Caligiuri M., Plass C., Goldschmidt-Clermont P.J. Methylation of the estrogen receptor-alpha gene promoter is selectively increased in proliferating human aortic smooth muscle cells. Cardiovasc. Res. 2000;46:172–179. doi: 10.1016/s0008-6363(00)00004-3. [DOI] [PubMed] [Google Scholar]
- 10.Kim J., Kim J.Y., Song K.S., Lee Y.H., Seo J.S., Jelinek J., Goldschmidt-Clermont P.J., Issa J.P. Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim. Biophys. Acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Matthews C., Gorenne I., Scott S., Figg N., Kirkpatrick P., Ritchie A., Goddard M., Bennett M. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ. Res. 2006;99:156–164. doi: 10.1161/01.RES.0000233315.38086.bc. [DOI] [PubMed] [Google Scholar]
- 12.Calvanese V., Lara E., Kahn A., Fraga M.F. The role of epigenetics in aging and age-related diseases. Ageing Res. Rev. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Fraga M.F., Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Kahn A., Fraga M.F. Epigenetics and aging: status, challenges, and needs for the future. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64:195–198. doi: 10.1093/gerona/gln064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson V.L., Jones P.A. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 16.Turunen M.P., Aavik E., Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim. Biophys. Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Dong C., Yoon W., Goldschmidt-Clermont P.J. DNA methylation and atherosclerosis. J. Nutr. 2002;132:2406S–2409S. doi: 10.1093/jn/132.8.2406S. [DOI] [PubMed] [Google Scholar]
- 18.Hiltunen M.O., Yla-Herttuala S. DNA Methylation, smooth muscle cells, and atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 19.Burton D.G., Sheerin A.N., Ostler E.L., Smith K., Giles P.J., Lowe J., Rhys-Williams W., Kipling D.G., Faragher R.G. Cyclin D1 overexpression permits the reproducible detection of senescent human vascular smooth muscle cells. Ann. N. Y. Acad. Sci. 2007;1119:20–31. doi: 10.1196/annals.1404.026. [DOI] [PubMed] [Google Scholar]
- 20.Karimi M., Luttropp K., Ekstrom T.J. Global DNA methylation analysis using the luminometric methylation assay. Methods Mol. Biol. 2011;791:135–144. doi: 10.1007/978-1-61779-316-5_11. [DOI] [PubMed] [Google Scholar]
- 21.Fazzari M.J., Greally J.M. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 22.Misawa A., Inoue J., Sugino Y., Hosoi H., Sugimoto T., Hosoda F., Ohki M., Imoto I., Inazawa J. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 2005;65:10233–10242. doi: 10.1158/0008-5472.CAN-05-1073. [DOI] [PubMed] [Google Scholar]
- 23.Fiegler H., Redon R., Andrews D., Scott C., Andrews R., Carder C., Clark R., Dovey O., Ellis P., Feuk L., et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo D., Wang T., Dressman H., Herderick E.E., Iversen E.S., Dong C., Vata K., Milano C.A., Rigat F., Pittman J., et al. Gene expression phenotypes of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- 25.Won S., Morris N., Lu Q., Elston R.C. Choosing an optimal method to combine P-values. Stat. Med. 2009;28:1537–1553. doi: 10.1002/sim.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minear M.A., Crosslin D.R., Sutton B.S., Connelly J.J., Nelson S.C., Gadson-Watson S., Wang T., Seo D., Vance J.M., Sketch M.H., Jr, et al. Polymorphic variants in tenascin-C (TNC) are associated with atherosclerosis and coronary artery disease. Hum. Genet. 2011;129:641–654. doi: 10.1007/s00439-011-0959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers J.C., Amenta P.S., Dion A.S., Sciancalepore J.P., Nagaswami C., Weisel J.W., Yurchenco P.D. The molecular structure of human tissue type XV presents a unique conformation among the collagens. Biochem. J. 2007;404:535–544. doi: 10.1042/BJ20070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirz J.A., Boudko S.P., Lerch T.F., Chapman M.S., Bachinger H.P. Crystal structure of the human collagen XV trimerization domain: a potent trimerizing unit common to multiplexin collagens. Matrix Biol. 30:9–15. doi: 10.1016/j.matbio.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D., Clark C.C., Myers J.C. Basement membrane zone type XV collagen is a disulfide-bonded chondroitin sulfate proteoglycan in human tissues and cultured cells. J. Biol. Chem. 2000;275:22339–22347. doi: 10.1074/jbc.M000519200. [DOI] [PubMed] [Google Scholar]
- 30.Kivirikko S., Saarela J., Myers J.C., Autio-Harmainen H., Pihlajaniemi T. Distribution of type XV collagen transcripts in human tissue and their production by muscle cells and fibroblasts. Am. J. Pathol. 1995;147:1500–1509. [PMC free article] [PubMed] [Google Scholar]
- 31.Amenta P.S., Scivoletti N.A., Newman M.D., Sciancalepore J.P., Li D., Myers J.C. Proteoglycan-collagen XV in human tissues is seen linking banded collagen fibers subjacent to the basement membrane. J. Histochem. Cytochem. 2005;53:165–176. doi: 10.1369/jhc.4A6376.2005. [DOI] [PubMed] [Google Scholar]
- 32.Eklund L., Piuhola J., Komulainen J., Sormunen R., Ongvarrasopone C., Fassler R., Muona A., Ilves M., Ruskoaho H., Takala T.E., et al. Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc. Natl Acad. Sci. USA. 2001;98:1194–1199. doi: 10.1073/pnas.031444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ylikarppa R., Eklund L., Sormunen R., Muona A., Fukai N., Olsen B.R., Pihlajaniemi T. Double knockout mice reveal a lack of major functional compensation between collagens XV and XVIII. Matrix Biol. 2003;22:443–448. doi: 10.1016/s0945-053x(03)00074-x. [DOI] [PubMed] [Google Scholar]
- 34.Rasi K., Piuhola J., Czabanka M., Sormunen R., Ilves M., Leskinen H., Rysa J., Kerkela R., Janmey P., Heljasvaara R., et al. Collagen XV is necessary for modeling of the extracellular matrix and its deficiency predisposes to cardiomyopathy. Circ. Res. 2010;107:1241–1252. doi: 10.1161/CIRCRESAHA.110.222133. [DOI] [PubMed] [Google Scholar]
- 35.Rasi K., Hurskainen M., Kallio M., Staven S., Sormunen R., Heape A.M., Avila R.L., Kirschner D., Muona A., Tolonen U., et al. Lack of collagen XV impairs peripheral nerve maturation and, when combined with laminin-411 deficiency, leads to basement membrane abnormalities and sensorimotor dysfunction. J. Neurosci. 2010;30:14490–14501. doi: 10.1523/JNEUROSCI.2644-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurskainen M., Ruggiero F., Hagg P., Pihlajaniemi T., Huhtala P. Recombinant human collagen XV regulates cell adhesion and migration. J. Biol. Chem. 2010;285:5258–5265. doi: 10.1074/jbc.M109.033787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.John H., Radtke K., Standker L., Forssmann W.G. Identification and characterization of novel endogenous proteolytic forms of the human angiogenesis inhibitors restin and endostatin. Biochim. Biophys. Acta. 2005;1747:161–170. doi: 10.1016/j.bbapap.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 38.O'Reilly M.S., Boehm T., Shing Y., Fukai N., Vasios G., Lane W.S., Flynn E., Birkhead J.R., Olsen B.R., Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 39.Harris A., Harris H., Hollingsworth M.A. Complete suppression of tumor formation by high levels of basement membrane collagen. Mol. Cancer Res. 2007;5:1241–1245. doi: 10.1158/1541-7786.MCR-07-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramchandran R., Dhanabal M., Volk R., Waterman M.J., Segal M., Lu H., Knebelmann B., Sukhatme V.P. Antiangiogenic activity of restin, NC10 domain of human collagen XV: comparison to endostatin. Biochem. Biophys. Res. Commun. 1999;255:735–739. doi: 10.1006/bbrc.1999.0248. [DOI] [PubMed] [Google Scholar]
- 41.Xu R., Xin L., Fan Y., Meng H.R., Li Z.P., Gan R.B. Mouse restin inhibits bovine aortic endothelial cell proliferation and causes cell apoptosis. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2002;34:138–142. [PubMed] [Google Scholar]
- 42.Schwartz S.M., Virmani R., Rosenfeld M.E. The good smooth muscle cells in atherosclerosis. Curr. Atheroscler. Rep. 2000;2:422–429. doi: 10.1007/s11883-000-0081-5. [DOI] [PubMed] [Google Scholar]
- 43.Rudijanto A. The role of vascular smooth muscle cells on the pathogenesis of atherosclerosis. Acta Med. Indones. 2007;39:86–93. [PubMed] [Google Scholar]
- 44.Klein L.W. Clinical implications and mechanisms of plaque rupture in the acute coronary syndromes. Am. Heart Hosp. J. 2005;3:249–255. doi: 10.1111/j.1541-9215.2005.03221.x. [DOI] [PubMed] [Google Scholar]
- 45.Adiguzel E., Ahmad P.J., Franco C., Bendeck M.P. Collagens in the progression and complications of atherosclerosis. Vasc. Med. 2009;14:73–89. doi: 10.1177/1358863X08094801. [DOI] [PubMed] [Google Scholar]
- 46.Butcher L.M., Beck S. Future impact of integrated high-throughput methylome analyses on human health and disease. J. Genet. Genomics. 2008;35:391–401. doi: 10.1016/S1673-8527(08)60057-0. [DOI] [PubMed] [Google Scholar]
- 47.Toyota M., Ho C., Ahuja N., Jair K.W., Li Q., Ohe-Toyota M., Baylin S.B., Issa J.P. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 48.Cherepanova O.A., Pidkovka N.A., Sarmento O.F., Yoshida T., Gan Q., Adiguzel E., Bendeck M.P., Berliner J., Leitinger N., Owens G.K. Oxidized phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ. Res. 2009;104:609–618. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connelly J.J., Shah S.H., Doss J.F., Gadson S., Nelson S., Crosslin D.R., Hale A.B., Lou X., Wang T., Haynes C., et al. Genetic and functional association of FAM5C with myocardial infarction. BMC Med. Genet. 2008;9:33. doi: 10.1186/1471-2350-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutton B.S., Crosslin D.R., Shah S.H., Nelson S.C., Bassil A., Hale A.B., Haynes C., Goldschmidt-Clermont P.J., Vance J.M., Seo D., et al. Comprehensive genetic analysis of the platelet activating factor acetylhydrolase (PLA2G7) gene and cardiovascular disease in case–control and family datasets. Hum. Mol. Genet. 2008;17:1318–1328. doi: 10.1093/hmg/ddn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 52.Abecasis G.R., Cookson W.O. GOLD–graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.