Abstract
DNA double-strand breaks (DSBs) can lead to instability of the genome if not repaired correctly. The MRE11/RAD50/NBS1 (MRN) complex binds DSBs and initiates damage-induced signaling cascades via activation of the ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia- and rad3-related (ATR) kinases. Mutations throughout MRE11 cause ataxia-telangiectasia-like disorder (ATLD) featuring cerebellar degeneration, and cancer-predisposition in certain kindreds. Here, we have examined the impact on DNA damage signaling of several disease-associated MRE11A alleles to gain greater understanding of the mechanisms underlying the diverse disease sequelae of ATLD. To this end, we have designed a system whereby endogenous wild-type Mre11a is conditionally deleted and disease-associated MRE11 mutants are stably expressed at physiologic levels. We find that mutations in the highly conserved N-terminal domain impact ATM signaling by perturbing both MRE11 interaction with NBS1 and MRE11 homodimerization. In contrast, an inherited allele in the MRE11 C-terminus maintains MRN interactions and ATM/ATR kinase activation. These findings reveal that ATLD patients have reduced ATM activation resulting from at least two distinct mechanisms: (i) N-terminal mutations destabilize MRN interactions, and (ii) mutation of the extreme C-terminus maintains interactions but leads to low levels of the complex. The N-terminal mutations were found in ATLD patients with childhood cancer; thus, our studies suggest a clinically relevant dichotomy in MRE11A alleles. More broadly, these studies underscore the importance of understanding specific effects of hypomorphic disease-associated mutations to achieve accurate prognosis and appropriate long-term medical surveillance.
INTRODUCTION
DNA damage is commonplace and results from a variety of endogenous and exogenous sources (1). DNA double-strand breaks (DSBs) are a highly toxic form of damage and arise from replication fork collapse, ionizing radiation (IR) or as intermediates in programmed rearrangements during meiosis and lymphocyte development. If not properly repaired, these lesions may result in deletion, duplication or translocation of genomic material leading to cellular dysfunction and potentially neoplastic transformation. Hence, maintenance of genomic integrity is essential for cellular and organismal viability. To deal with the constant threat of DNA damage, organisms have evolved elaborate DNA damage response (DDR) machinery to attempt DNA repair, and if unsuccessful, remove cells from the proliferative pool.
The MRE11/RAD50/NBS1 (MRN) complex is central to the detection and repair of DSBs, as well as the restart of stalled replication forks (2). This complex serves as a hub for regulation of kinases involved in damage responses. When DSBs arise, MRN directly binds DNA ends and facilitates activation of ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia- and rad3-related (ATR). These are the apical kinases of the DDR that are responsible for cell cycle arrest, initiation of DNA repair and, if necessary, apoptosis. DNA ends are bound directly and stabilized in proximity by a heterotetramer composed of two MRE11 and two RAD50 protomers. When MRN is engaged at DSBs, it recruits and activates ATM through direct interaction with the NBS1 subunit as well as other possible contacts (3–5). MRE11 possesses endo- and exo-nuclease activities that initiate resection of DNA ends (6,7). This end processing facilitates generation of single-stranded DNA upon which ATR can be subsequently loaded and activated (3,8). MRN-dependent resection is promoted by ATM; thus, DSB-induced ATR activation is also downstream of ATM activity (9,10).
Inherited deficiencies in DNA damage signaling are responsible for a spectrum of disorders with diverse sequelae involving the central nervous system, the immune system and musculoskeletal development (1). Biallelic mutations in ATM or NBN (NBS1) result in ataxia-telangiectasia (AT; MIM 208900) or Nijmegen breakage syndrome (NBS; MIM 251260), respectively (11–13). These syndromes are marked by neurologic phenotypes (cerebellar ataxia and microcephaly, respectively), immunodeficiency, cellular genomic instability and IR hypersensitivity (14,15). As anticipated from the important role of DNA repair in maintaining genomic stability, AT and NBS patients display strong predisposition to malignancy. In fact, a single hypomorphic ATM or NBS1 allele can be sufficient to increase cancer risk (16–24). This cancer predisposition is also apparent in mouse models. Though Nbs1-null mice are not viable (25), mice with Nbs1 heterozygosity or hypomorphism are predisposed to oncogenesis (26,27). Furthermore, Atm-null mice are predisposed to lymphoma (28), and mice carrying one hypomorphic Atm allele are cancer predisposed (29). Other components of the DSB repair machinery are also tumor suppressors—for example, BRCA1 and BRCA2 (30,31).
Given that MRE11 functions in a complex with NBS1, it would be anticipated that inherited MRE11 deficiencies would phenocopy NBS. However, this is not commonly observed. Germline biallelic MRE11A (MRE11) mutation results in a syndrome more similar to AT than NBS and has thus been termed ataxia-telangiectasia-like disorder (ATLD; MIM 604391) (32–37). Like AT, ATLD is characterized by cerebellar ataxia which develops after birth through neurodegeneration rather than the microcephaly at or shortly after birth associated with NBS. In addition, the earliest identified ATLD patients appear to differ from both AT and NBS in their cancer predisposition. ATLD patients homozygous for the first identified ATLD allele, MRE11AATLD1, have not been reported to develop early cancer, nor are Mre11aATLD1/ATLD1 mice predisposed to the development of lymphoma—the malignancy to which AT and NBS patients are most markedly predisposed (32,38). However, MRE11AATLD1 has been associated with a familial cancer syndrome inherited in an autosomal dominant manner (39). Also, more recently identified ATLD patients harboring novel MRE11A mutations do seem cancer prone. Two ATLD-afflicted brothers both died of lung cancer (pulmonary adenocarcinoma) at the remarkably young ages of 9 and 16 (35). These two individuals had cerebellar degeneration but additionally had developmental defects suggestive of NBS.
Comparison of these DDR disorders makes clear that while there are some common features, there are also critical differences among specific patient groups. Complete absence of any MRN component is early embryonic lethal in mice; thus, the disease alleles are partial loss-of-function mutations that must preserve essential functions of the complex (25,40,41). It is likely that the differing impact of each of the mutations is largely responsible for the varied spectrum of sequelae in the resulting disorders. Understanding and predicting these differences has important implications for prognosis and treatment for these disorders. Given that mutations in AT and MRN components can affect the wider population through haplo-insufficiency and somatic mutation, greater understanding of these factors has important health implications.
We endeavored to define the specific defects in DNA damage signaling caused by several MRE11A alleles associated with diverse clinical outcomes. To this end, we designed a structure–function system to express mutant alleles at physiologic levels in murine cells harboring an endogenous Mre11a conditional Cre/LoxP allele (Mre11acond) engineered in our laboratory (41). We find that the MRE11 mutants have distinct consequences on stability of the MRN complex and DNA damage signaling, which appear to correlate with differences in cancer predisposition. Furthermore, these studies provide evidence that MRE11 dimer formation and MRE11–NBS1 interaction are mechanistically linked in mammals thus providing insight into how certain MRE11A mutations abrogate DNA damage signaling.
RESULTS
Selection of four distinct disease-associated MRE11A mutant alleles
We sought to determine if disease-associated MRE11A mutations impact DNA damage signaling in differing ways and if unique functional defects relate to variability in disease sequelae (35,39,42). Unfortunately, most ATLD-associated MRE11A mutations cause low levels of the entire MRN complex, preventing structure–function analyses through study of patient cell lines (32,33,35–37). To circumvent this limitation, we engineered cell lines to express MRE11 mutants of interest at approximately physiologic levels and in which endogenous wild-type Mre11a could be inactivated through Cre/LoxP-mediated deletion (41).
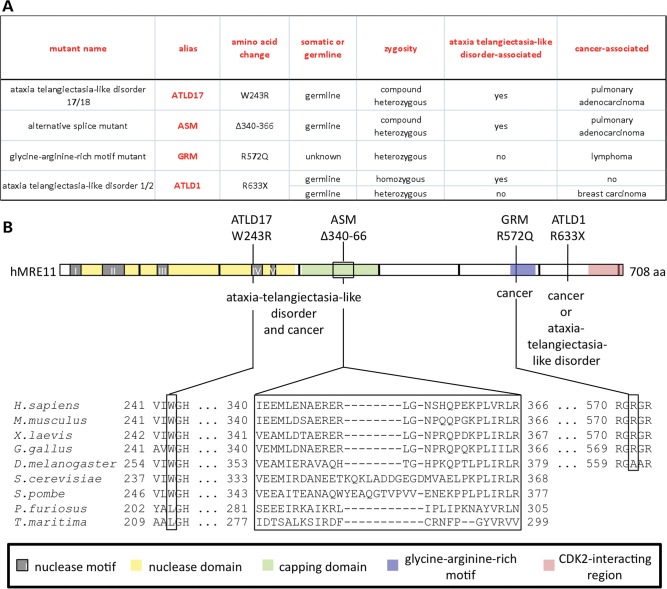
Inherited alleles of interest were chosen based on maximum differences in patient outcome (Fig. 1A). To this end, we compared the impact of the first reported ATLD allele, MRE11AATLD1, to that of recently identified alleles in compound heterozygotes, MRE11AATLD17 and MRE11AASM (Fig. 1B). MRE11AATLD1 contains a nonsense mutation that truncates 76 amino acid residues from the MRE11 C-terminus (32). Although MRE11AATLD1/ATLD1 patients and Mre11aATLD1/ATLD1 mice exhibited cellular hallmarks of cancer, they do not appear to be cancer predisposed (32,38). In contrast, two brothers harboring both MRE11AATLD17 and MRE11AASM died of pulmonary adenocarcinoma at the ages of 9 and 16 years (35). While this patient cohort is small, the early age of lung cancer is striking given that it is normally a disease of old age (43). The brothers' wild-type MRE11A heterozygous parents and sibling did not have ATLD or lung cancer at last report (44).
Figure 1.
Disease-associated MRE11 mutants in this study. (A) Summary of the name, nickname (alias), amino acid change and clinical context for each mutant. (B) A HsMRE11 stick diagram with pertinent domains and motifs. Note that the phosphodiesterase domain includes both the nuclease (catalytic) domain and the capping domain. Amino acid changes are labeled along with mutant aliases and associated human diseases (top). Alignments of the residues surrounding the mutant sites are shown (bottom).
The MRE11AATLD17 mutation (c.727T>C) results in substitution at a highly conserved residue (p.W243R) near motif IV of the N-terminal phosphodiesterase domain (Fig. 1B). MRE11AASM possesses an intronic mutation, c.1098+5G>A, near a splice donor site associated with skipping of the 81-nucleotide exon 10. The resulting mRNA maintains the open reading frame, thus encoding a predicted protein lacking 27 amino acid residues (p.Δ340–366). The deleted residues reside in the capping domain, a structural feature unique to MRE11 compared with other known phosphoesterases (45,46).
In addition to inherited mutations causing ATLD, MRE11A has been found mutated in the context of familial and sporadic cancers. Mutational analysis of MRE11A in unselected primary tumors revealed a heterozygous mutation (c.1715G>A) encoding MRE11 R572Q in a lymphoma (42). While it is not known whether this allele was inherited or if it played a causal role in this lymphoma, the mutation nonetheless resides in an interesting region of mammalian MRE11. R572 is in a glycine-arginine-rich (GAR) motif, portions of which are conserved among metazoans. Methylation of GAR motif arginines has been shown to be important for MRE11 exonuclease activity and activation of the ATR kinase (47–50). On the organismal level, alteration of the GAR motif arginines caused radiation hypersensitivity, a trait common among cancer-prone genomic instability syndromes. However, similar to most studied ATLD alleles, the murine MRE11 protein lacking GAR motif arginines was present at low levels relative to wild-type MRE11 (50). Thus, we engineered the lymphoma-associated MRE11 GAR motif mutant (MRE11GRM) into our expression system to facilitate study of this mutant with the protein at physiologic levels.
Generation of cell lines expressing Mre11a mutant alleles
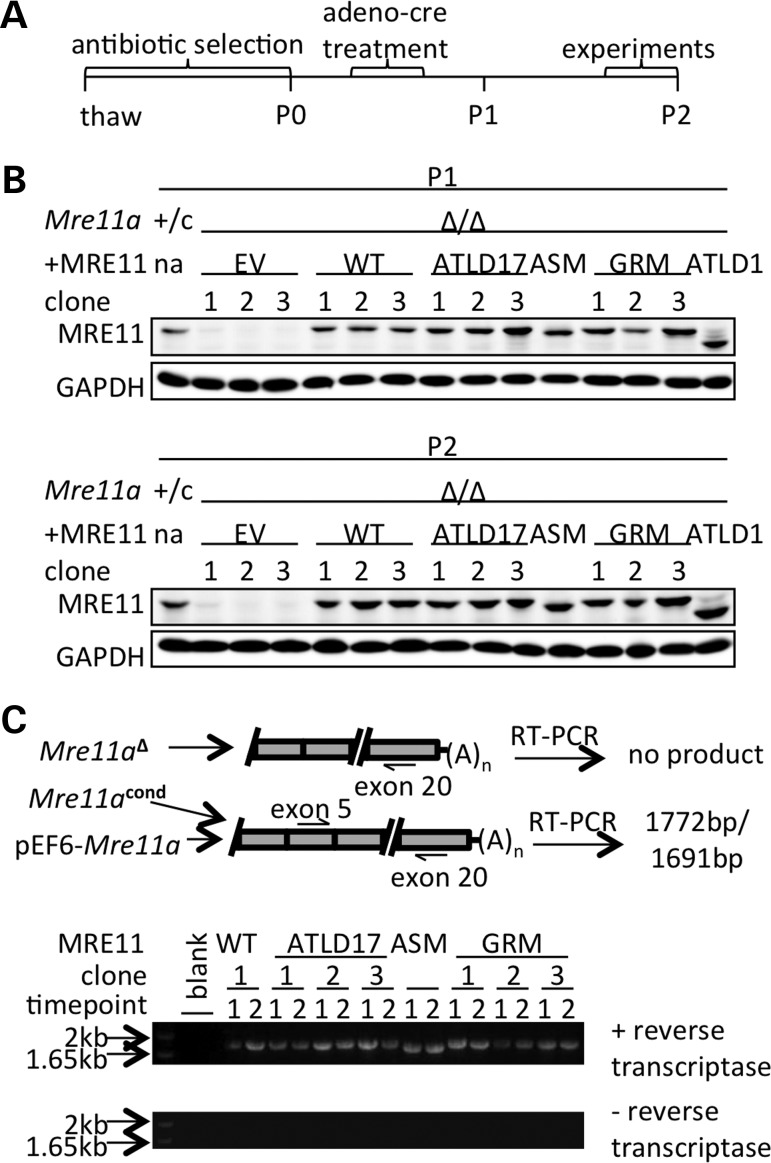
Immortalized murine embryonic fibroblasts (MEFs) harboring one Mre11a allele rendered null in the germline (Mre11a–) and one floxed conditional allele (Mre11acond) (41) were stably transfected with pEF6-Mre11a constructs expressing the mutants of interest from cDNA. Clones were isolated and exposed to adenovirus expressing Cre recombinase to convert the endogenous Mre11acond allele to null (Mre11aΔ). Clones expressing at least physiologic levels of mutant MRE11 protein were used for experimentation, and protein levels were confirmed before and after each experimental time period (Fig. 2A). Untagged proteins were expressed to avoid disruption of MRE11 interactions as previously reported (51). For MRE11ATLD1 and MRE11ASM, gel mobility distinguished these from wild-type MRE11 (Fig. 2B). For the missense mutants MRE11ATLD17 and MRE11GRM, expression was assessed by an RT-PCR-sequencing strategy (Fig. 2C and Supplementary Material, Fig. S1) combined with confirmation of endogenous allele deletion by genomic PCR (data not shown) and by comparison with Mre11acond/− cells treated in parallel during each experiment.
Figure 2.
Generation of MRE11 mutant-expressing cell lines. (A) The peri-experimental timeline. Cells were passaged (P) as indicated. (B) Western blots demonstrating approximately physiologic levels of MRE11 protein expressed from stably integrated plasmid constructs. Endogenous Mre11a was inactivated in these lines through cre-loxP mediated deletion (Mre11aΔ/Δ). Physiologic MRE11 levels encoded by endogenous alleles are shown at the far left (Mre11a+/c harbors one wild-type and one floxed conditional allele). EV denotes empty vector controls. (C) The RT-PCR strategy to type Mre11a mRNA of MRE11-expressing clones is shown (above) along with RT-PCR results (below). Bands of the expected sizes were detected for each clone (upper panel). No bands were detected in the absence of reverse transcriptase (lower panel).
Activation of ATM and ATR kinases
MRN recruits ATM to DSBs and facilitates kinase activation through direct interaction of ATM with NBS1 (3–5). ATM activation and activity can be monitored by measuring ATM autophosphorylation at S1987 (52) and ATM phosphorylation of KRAB associated protein (KAP1, also called TIF1β, TRIM28 and KRIP-1) (53). Also, ATM kinase and MRE11 nucleolytic activities together mediate DNA resection (9,10,41). The resulting replication protein A-coated ssDNA can be loaded with ATRIP-ATR at which time ATR phosphorylates substrates such as the CHK1 kinase (3,8,10). To determine the ability of each MRE11 mutant to activate ATM and ATR in the presence of DSBs, pATM S1987, pKAP1 S824 and pCHK1 S345 induction was measured.
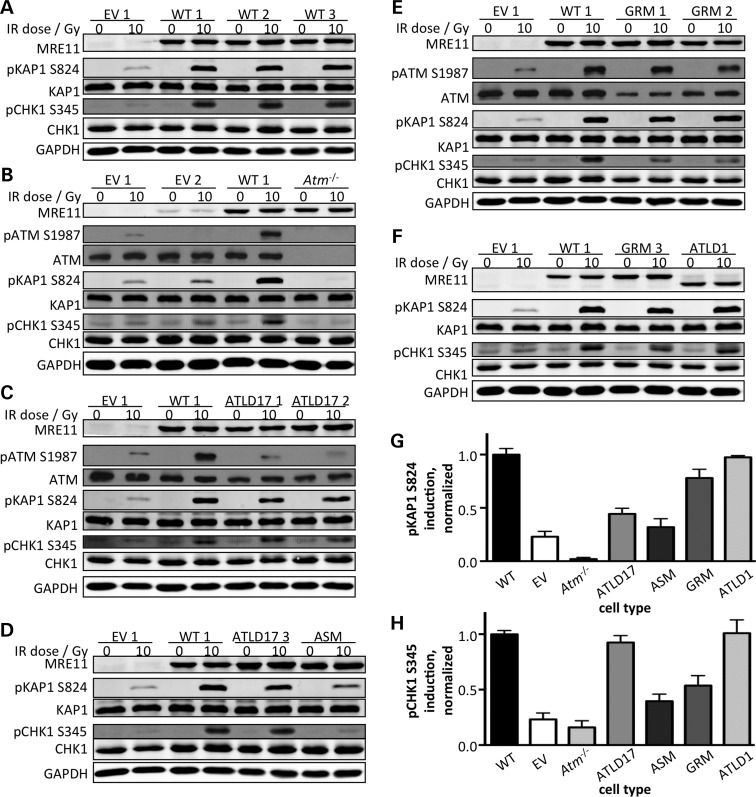
We used IR to experimentally induce DSBs, and observed robust KAP1 and CHK1 phosphorylation in the presence of wild-type MRE11, but only weak induction in the absence of MRE11 or ATM (Fig. 3A and B). We next assessed the impact of MRE11A alleles found in the ATLD siblings who succumbed to pulmonary adenocarcinoma. Cells expressing MRE11ATLD17 showed deficiencies in pATM and pKAP1 induction but not pCHK1 induction (Fig. 3C and D). Decreased pKAP1 induction was observed following IR doses ranging from 0.5 to 10 Gy and following recovery times from 6 min to 8 h (Supplementary Material, Fig. S2A–C). Induction of pKAP1 was also substantially abrogated in cells expressing MRE11ASM (Fig. 3D and Supplementary Material, Fig. S2D). In addition, MRE11ASM-expressing cells showed reduced levels of IR-induced pCHK1, suggesting this mutant was defective in facilitating ATR activity. Hence, whereas both alleles in the ATLD patients with pulmonary adenocar-cinoma had a negative impact on ATM activity, only MRE11ASM-expressing cells had deficient ATR activity.
Figure 3.
Assessment of DNA damage-induced ATM/ATR kinase activation. (A–F) Western blot analyses for representative substrates of the ATM and ATR kinases. The indicated cell lines (top) were either untreated or treated with 10 Gy IR and allowed to recover for 30 min. Whole cell lysates were immunoblotted for the proteins indicated (left). GAPDH is a loading control. Samples were loaded in various combinations to allow accurate comparison of phospho-protein levels across multiple gels. Immunoblots are representative of at least three independent experiments. EV, empty vector controls. (G, H) Quantitation of pKAP S824 (G) and pCHK S345 (H) induction from immunoblots. After accounting for protein loading, induction was normalized to the weighted mean induction of the wild-type lines. Each bar represents at least three independent experiments. Error bars represent standard error of the mean.
The MRE11GRM mutant identified in a lymphoma supported approximately wild-type levels of ATM activation (Fig. 3E and F). A modest 10% reduction in KAP1 phosphorylation was noted (Fig. 3G) but is likely not biologically significant. In contrast, pCHK1 induction was reduced to 50% of wild-type (Fig. 3E and F, and Supplementary Material, Fig. S2E), indicating reduced ATR activation. This implies that even a subtle disruption of the MRE11 GAR motif is sufficient to compromise ATR activity.
Finally, we analyzed cells expressing MRE11ATLD1, a mutant from ATLD patients who do not appear to be cancer prone. In stark contrast to the mutants described above, MRE11ATLD1 expression was able to complement pATM, pKAP1 and pCHK1 induction (Fig. 3F and (51)). Therefore, this disease-associated mutant retains the ability to activate both ATM and ATR kinases. Quantitation of pKAP1 and pCHK1 induction is shown in Figure 3G and H. ATM/ATR activation results are summarized in Table 1.
Table 1.
Comparison of ATM/ATR activation among the four disease associated Mre11 alleles in this study
| Assay | ATLD17 | ASM | GRM | ATLD1 | ||
| ATM activity | pKAP1 induction | + | <+ | +++ | ++++ | |
| pATM induction | + | ND | ++++ | ND | ||
| G2/M checkpoint | ++++ | ++ | ++++ | ++++ | ||
| ATR activity | pCHK1 induction | ++++ | + | + | ++++ | |
| Symbol | + | ++ | +++ | ++++ | ND | |
| % of wild-type | 12.5, 37.5% | 37.5, 62.5% | 62.5, 87.5% | 87.5, 112.5% | Not determined |
Assessment of the G2/M cell cycle checkpoint
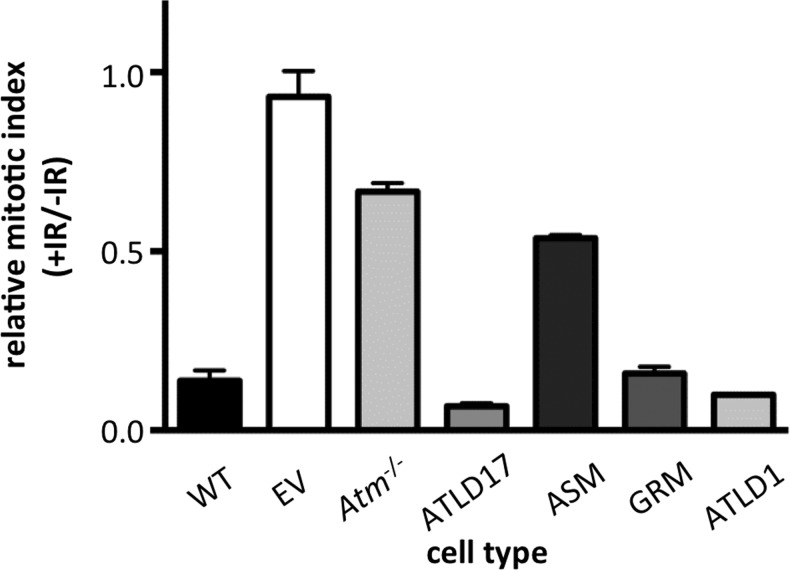
Upon DNA damage, ATM triggers cell cycle arrest at the G2/M transition (54,55). We therefore assessed the competency of the early G2/M checkpoint in our mutant MRE11-expressing lines. Cells were mock-treated or treated with 10 Gy IR, and the mitotic index was calculated based on the percentage of cells positive for the mitosis-specific phospho-histone H3 S10 marker (56). Activation of the G2/M checkpoint is reflected by a significant drop in mitotic index 60 min post-IR. A defective checkpoint is identified by a significantly higher mitotic index post-IR compared with wild-type control. Consistent with previous reports, we found that cells lacking MRN or ATM had a higher mitotic index post-IR compared with wild-type, reflecting the defective G2/M checkpoint (Fig. 4 and Table 1) (38,54).
Figure 4.
Assessment of the DNA damage-induced G2/M checkpoint. Cells were either mock treated or treated with 10 Gy IR, allowed to recover for 60 min and then stained for the mitosis-specific p-histone H3S10 modification. p-H3S10 was detected by flow cytometry, and mitotic index was determined by the percentage of p-H3S10 positive cells. Relative mitotic index (Y-axis) reflects the ratio of the mitotic index of IR-treated cells to untreated cells for a given line. MRE11 wild-type cells (WT) show a significant reduction in mitotic index after IR, reflecting activation of the G2/M checkpoint. Absence of MRE11 (EV, empty vector) or ATM (Atm−/−) shows significantly higher relative mitotic indices compared with WT controls, reflecting a defective G2/M checkpoint. A minimum of three independent experiments were performed for each cell line. Error bars represent standard error of the mean.
MRE11ASM, which conferred defects in both ATM and ATR kinase activation, caused a defective G2/M checkpoint (Fig. 4 and Table 1). In fact, the impact was similar to that in ATM-deficient cells. MRE11ATLD17 did not have a measurable impact on the G2/M checkpoint, despite the reduced activation of ATM. This mutant did permit greater ATM activity than MRE11ASM; thus it is likely that the observed difference in checkpoint proficiencies is due to differences in ATM activation. MRE11ATLD1, which supported ATM and ATR activation, and MRE11GRM, which supported normal ATM activation, both appeared capable of maintaining the G2/M checkpoint (summarized in Table 1).
MRE11/RAD50/NBS1 complex stability
The findings above support the notion that unique MRE11A mutations can impact ATM and ATR functions in differing ways. Kinase activation by MRN involves conformational changes to all three complex components upon recognition of a DSB (57–62). To gain understanding of the mechanism by which the mutants impact signaling, we compared the stabilities of MRN complexes containing each mutant.
Previous studies have demonstrated that disrupted MRN complex formation can cause abnormal ratios of the complex components; patients with MRE11A mutations that compromise MRE11–NBS1 interaction have reduced NBS1:MRE11 molar ratios (34,35,37). We assessed cellular RAD50:MRE11 and NBS1:MRE11 ratios to determine if they suggested defects in MRN complex stability.
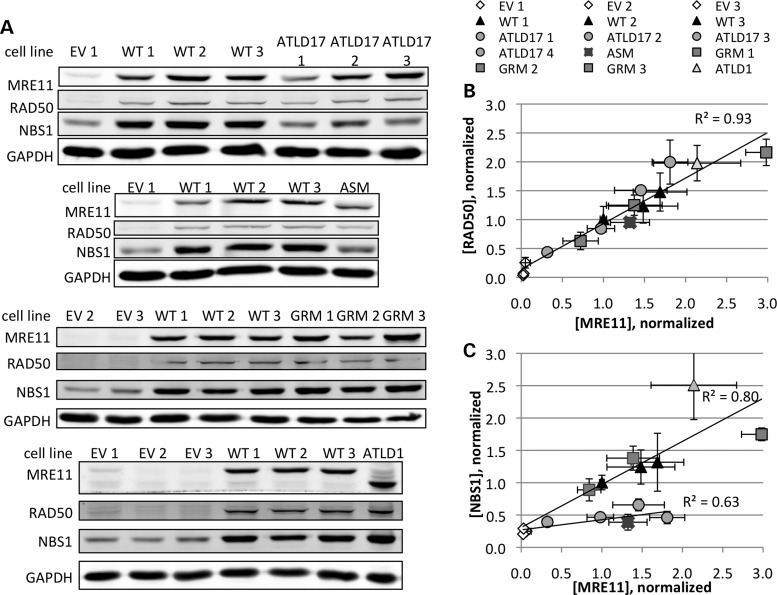
Mre11a deletion resulted in proportionally decreased RAD50 levels (Fig. 5A and B). Expression of wild-type MRE11, or each of the four MRE11 mutants, resulted in a proportional increase in RAD50 levels. A least-squares best fit for all Mre11a alleles yielded a line with an R2 = 0.93 (Fig. 5B). RAD50:MRE11ATLD17 and RAD50:MRE11ATLD1 ratios were consistent with those found in ATLD17 and ATLD1 patient cells (32,35). Thus, each mutant can maintain RAD50 levels to a similar extent as wild-type MRE11.
Figure 5.
Comparison of MRE11/RAD50/NBS1 molar ratios. (A) Western blots comparing protein levels of the MRN components (left). Cell lines (top) are: EV (empty vector controls), WT (exogenously-expressed wild-type MRE11) and the four Mre11a mutant alleles as described in the text. Endogenous wild-type Mre11a has undergone cre/loxP-mediated deletion in EV and the four mutant lines. (B, C) Quantitation of cellular MRN complex component levels from immunoblots as shown in (A). MRE11, RAD50 (B), and NBS1 (C) levels were normalized to those in wild-type-expressing clone 1 (WT 1) whole cell lysate after accounting for protein loading. Each data point represents a minimum of three measurements (range: 3–15, median: 5), and error bars represent standard error of the mean. The trend lines shown with R2 values represent the least-squares best fits. Note the reduced NBS1:MRE11 ratios in MRE11ATLD17- and MRE11ASM-expressing lines (C).
Depletion of MRE11 also resulted in decreased NBS1 levels (Fig. 5A and C). In contrast to what was seen for RAD50:MRE11, cells expressing either MRE11ATLD17 or MRE11ASM had reduced NBS1:MRE11 ratios suggestive of compromised interaction with NBS1. The least squares fit for wild-type cell lines had a slope appreciably different from the best fit for MRE11ATLD17- or MRE11ASM-expressing lines (Fig. 5C). Cells expressing MRE11ATLD1 or MRE11GRM had wild-type-like NBS1:MRE11 ratios, implying normal interactions. In support of our observations, ATLD17/18 patient cells were found to have reduced NBS1:MRE11 ratios (35), whereas ATLD1/2 patient cells had normal NBS1:MRE11 ratios (32) despite both lines having reduced overall levels of MRN. These findings are summarized in Table 2.
Table 2.
Comparison of MRN complex stability among the four disease associated Mre11 alleles in this study
| Method | ATLD17 | ASM | GRM | ATLD1 | |
| Homodimerization | Y2H | ++ | <+ | ++++ | ++++ |
| RAD50 interaction | Molar ratios | ++++ | ++++ | ++++ | ++++ |
| Co-IP | +++ | ++ | ++++ | ++++ | |
| NBS1 interaction | Molar ratios | ++ | ++ | ++++ | ++++ |
| Y2H | ++ | <+ | ++++ | ++++ | |
| Co-IP | ≥++ | ≥++ | ++++ | ++++ | |
| Symbol | + | ++ | +++ | ++++ | ND |
| % of wild-type | 12.5%, 37.5% | 37.5%, 62.5% | 62.5%, 87.5% | 87.5%, 112.5% | Not determined |
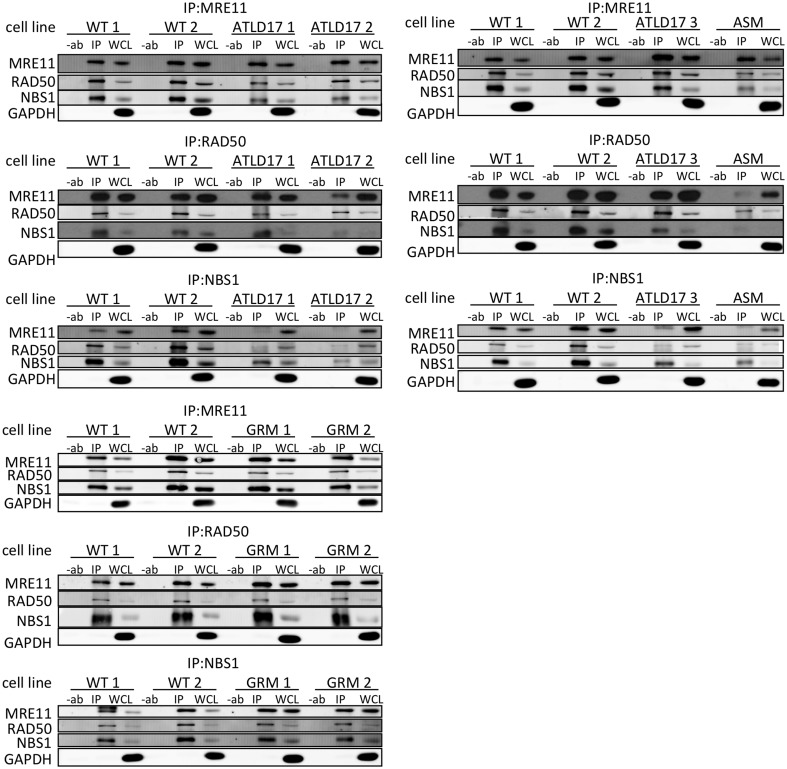
We further probed the stability of MRN complexes by co-immunoprecipitation (co-IP) of MRN components. Co-IP of NBS1 with MRE11ATLD17 or MRE11ASM was substantially reduced compared with wild-type MRE11 (Fig. 6). In the case of MRE11ATLD17, we also performed co-IPs with NBS1 12 and 30 min after a dose of 10 Gy IR, and we found a defect similar to that seen in unirradiated cells (Supplementary Material, Fig. S3). MRE11ATLD17 co-IPed with RAD50 to a slightly lesser degree than wild-type MRE11. However, MRE11ASM displayed dramatically reduced co-IP with RAD50. In contrast, MRE11GRM appeared just as capable as wild-type MRE11 in pulling down and being pulled down by RAD50 and NBS1. Previous work has shown that MRE11ATLD1 does not impact the MRN complex as assessed by co-IP (38,51). See Table 2 for summary.
Figure 6.
MRE11/RAD50/NBS1 co-IP. MRE11, RAD50 or NBS1 were immunoprecipitated from whole cell lysates as indicated above each blot. Immunoblots were performed for each MRN complex component as indicated (left). GAPDH was used as a whole cell lysate loading control. Results shown are representative of a minimum of three co-IPs. Reduced MRE11-NBS1 co-IP is observed in lysates from MRE11ATLD17- and MRE11ASM-expressing lines. Reduced MRE11-RAD50 co-IP is also observed in MRE11ASM lysates.
These studies imply that MRE11ATLD17 and MRE11ASM confer a defect in MRE11–NBS1 binding and, in the case of MRE11ASM, a further impact on RAD50 binding. In contrast, MRE11ATLD1 and MRE11GRM appear not to impact MRN complex stability, at least in a manner which can be revealed by co-IP.
MRE11–NBS1 interaction and MRE11 homodimerization
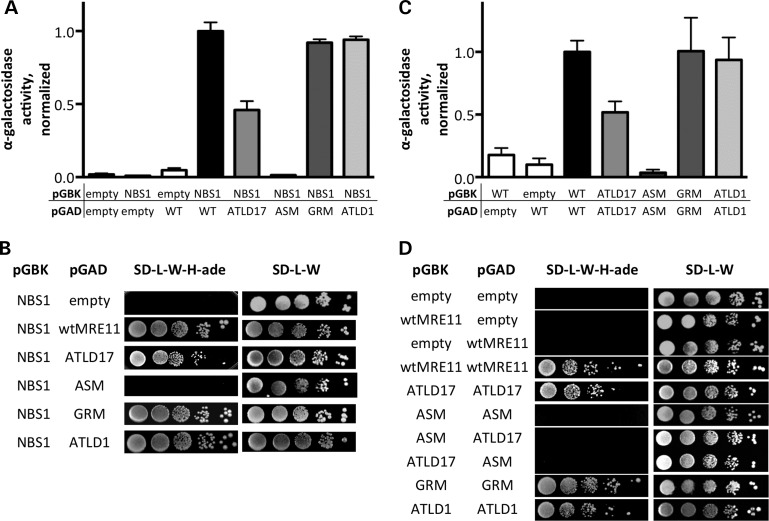
To directly determine whether the four MRE11 mutants in our study impact NBS1 binding, we utilized the yeast two-hybrid system. Wild-type and mutant genes were cloned into pGBK (bait) and pGAD (prey) plasmids, and binding was quantitatively assessed using α-galactosidase expression and qualitatively using colony growth in double selection (-His, -adenine). Both MRE11ATLD17 and MRE11ASM displayed substantial reductions in NBS1 binding (Fig. 7A, B, and Supplementary Material, Fig. S4A). However, MRE11ATLD1 and MRE11GRM interacted with NBS1 to similar extents as wild-type MRE11.
Figure 7.
Direct MRE11–NBS1 interaction and MRE11 homodimerization. Yeast two-hybrid analysis was performed using pGBK and pGAD plasmids—encoding the bait and prey, respectively. Empty vectors are negative controls. Plasmids were selected for by culturing in the absence of leucine and tryptophan (SD-L-W). (A) MRE11–NBS1 interaction quantitated by colorimetric detection of α-galactosidase expression. Bait–prey combinations are shown below each bar, which represents at least three clones per combination with at least three measurements per clone. Error bars represent standard error of the mean. (B) MRE11–NBS1 interaction by Y2H colony growth assay. Bait and prey proteins are indicated on the left. Ten-fold serial dilution series are shown on interaction test plates (SD-L-W-H-ade, center) and plating efficiency control plates (SD-L-W, right). Results are representative of those for at least three clones per bait–prey combination. (C) MRE11 homodimerization by yeast two-hybrid colorimetric assay, as described in (A). (D) Homodimerization assessed by colony growth as described in (B).
Within the MRN complex, MRE11 forms a dimer which bridges DNA ends in close proximity and positions DNA termini within the nuclease domain (46,62). Using the yeast two-hybrid approach, we determined whether the four mutants in our study impact the ability of MRE11 to homodimerize. Compared with wild-type MRE11, MRE11ATLD17 homodimerization was reduced (Fig. 7C, D, and Supplementary Material, Fig. S4B). Strikingly, homodimerization was dramatically reduced for MRE11ASM and MRE11ASM with MRE11ATLD17, appearing similar to the results obtained using empty pGBK and pGAD vectors. The MRE11ASM fusion proteins were confirmed to be present in the yeast strains through immunoblotting (Supplementary Material, Fig. S4C and E). No defect in MRE11ATLD1 or MRE11GRM homodimerization was apparent. See Table 2 for summary.
A mammalian MRE11 homodimerization mutant
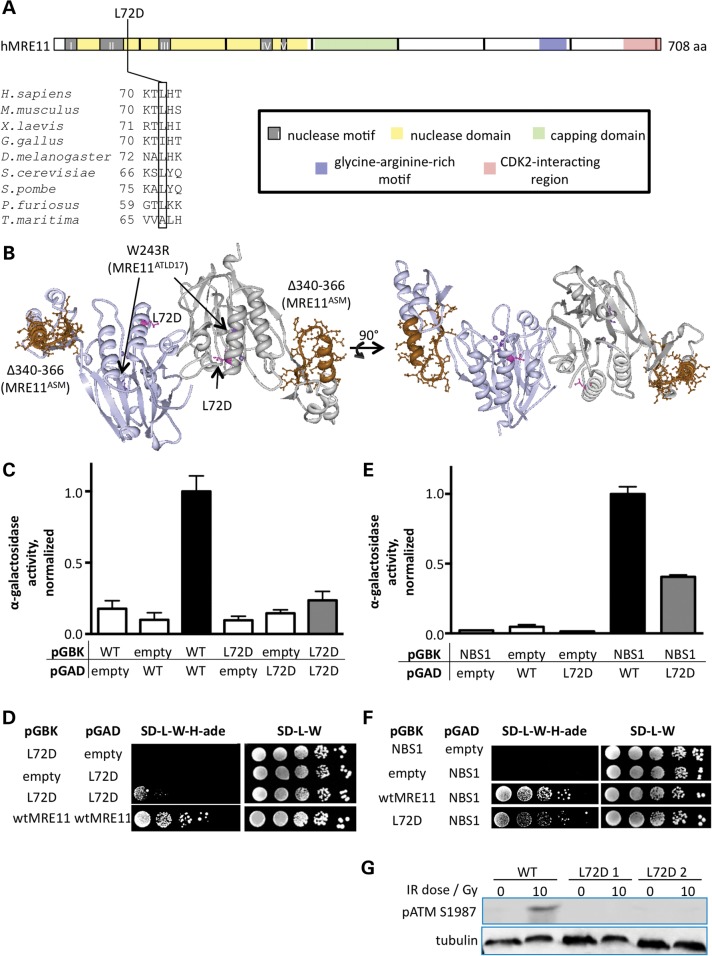
Structural studies of proteins from the single-celled eukaryote Schizosaccharomyces pombe have suggested a mechanistic link between Mre11 dimer stability and binding of Nbs1 to Mre11 (60). The findings from examination of the four mutants in this study suggest that this relationship may hold true in mammals and represent an important cause of variable disease sequelae for syndromes associated with MRN dysfunction. The two mutants that impacted MRE11–NBS1 interaction, MRE11ATLD17 and MRE11ASM, also reduced MRE11 dimer stability, whereas the two remaining mutants, MRE11GRM and MRE11ATLD1, preserved both functions. We therefore wished to determine the extent to which mammalian MRE11 dimer stability and NBS1 interaction are functionally linked. To this end, we sought to design an Mre11a mutation that would specifically disrupt dimer formation, using structural and biochemical studies from the archeal species Pyrococcus furiosus as a guide (62). Archea lack an NBS1 homologue as well as eukaryotic-specific structural motifs of MRE11 involved in NBS1 binding (46,60). Therefore, residues in mammalian MRE11 that are conserved with those required for dimerization in P. furiosus Mre11 likely represent the core features for this function.
We identified murine and human L72 as being homologous to P. furiosus Mre11 L61, the latter of which is necessary for Mre11 dimer stability (Fig. 8A) (62). L72 is an aliphatic hydrophobic residue that participates in formation of a hydrophobic pocket at the homodimer interface (Fig. 8B). We mutated this residue to aspartate and utilized the yeast two-hybrid system to assess MRE11 L72D homodimerization and direct interaction with NBS1. We observed that homodimeration of MRE11 L72D was impaired, using both α-galactosidase levels and colony growth (Fig. 8C and D). Thus, the function of this portion of mammalian MRE11 is conserved despite significant divergence in nearby structures involved in interaction with eukaryote-specific NBS1 (46,60). Importantly, direct interaction with NBS1 was also abrogated by the MRE11 L72D dimerization mutant (Fig. 8E and F).
Figure 8.
Engineered MRE11 homodimerization mutant. (A) MRE11 stick diagram and alignment indicating amino acid residue L72. (B) Crystal structure of the HsMRE11 N-terminal domain homodimer determined by Park et al. (46) with mutation sites indicated as follows: L72D (dimerization mutant, pink), W243R (MRE11ATLD17, violet) and Δ340–366 (MRE11ASM, mocha). (C) Homodimerization assessed by Y2H colorimetric assay as described in legend to Figure 7A. MRE11 L72D homodimerization was similar to background. (D) MRE11 homodimerization assessed by Y2H colony growth assay as described in legend to Figure 7B. (E) MRE11 L72D interaction with NBS1 assessed by Y2H colorimetric assay as described in legend to Figure 7A. MRE11 L72D–NBS1 interaction was reduced by 59% compared with that of wild-type MRE11–NBS1. (F) MRE11 L72D–NBS1 interaction assessed by Y2H colony growth assay as described in legend to Figure 7B. (G) Cells stably expressing either wild-type MRE11 or MRE11 L72D were treated with 10 Gy IR, allowed to recover for 30 min and lysed. MRE11 L72D failed to complement ATM activation as measured by ATM autophosphorylation. Tubulin was used as a loading control.
An impact on MRE11–NBS1 interaction would be predicted to affect ATM activation. We tested this by stably expressing MRE11 L72D from cDNA in Mre11aΔ/− cells (data not shown). ATM activation was measured by autophosphorylation 30 min after exposure to IR. Indeed, ATM activation was significantly reduced compared with that in cells expressing wild-type MRE11 (Fig. 8G). Thus, our studies have revealed an important functional link between MRE11 homodimerization, MRE11–NBS1 interaction and ATM signaling in mammals.
DISCUSSION
Here, we have shown that unique inherited alleles of MRE11A can have varying impact on control of cellular signaling upon DNA damage. A common theme among the ATLD alleles is reduced activation of the ATM kinase, likely accounting for similar disease sequelae such as cerebellar degeneration. However, the mechanisms differ greatly which may explain clinical variation, such as cancer predisposition. Reduced signaling in MRE11AATLD1/ATLD1 individuals likely results primarily from low levels of the MRN complex because we have shown that expression of MRE11ATLD1 to normal levels supports ATM activity. On the contrary, in MRE11AATLD17/ASM patients, both mutant alleles impact crucial N-terminal structural features required for MRE11–NBS1 interaction and—therefore—ATM activation. The specific defects caused by these two N-terminal mutations suggested a mechanistic link between MRE11 homodimer formation and interaction with NBS1, which we demonstrated through generation of a single-amino acid substitution that abrogates both functions.
The N-terminal half of MRE11 is highly conserved throughout nature. Recent co-crystallization of Mre11 and Nbs1 fragments from the single-celled eukaryote Schizosaccharomyces pombe shows that SpMre11 forms a dimer that interacts with two SpNbs1 fragments (60). Nbs1 binds in a highly asymmetric manner. Although each Nbs1 fragment binds one Mre11 protomer on a peripheral surface, only one Nbs1 fragment binds across the Mre11 dimer interface and makes contact with both Mre11 protomers. This contact occurs at a eukaryote-specific portion of Mre11 recently dubbed ‘latching loops’ (60). The mutant residue in MRE11ATLD17 (HsMRE11 W243, SpMre11 W248) does not lie within the latching loops. Instead, it forms the hydrophobic core of the structural region encompassing the peripheral binding sites. Hence, its substitution could disrupt folding within the N-terminal domain, thereby effecting binding by Nbs1 at the peripheral sites as well as stability of the Mre11 dimer.
Our studies on the MRE11 L72D mutant have illuminated the mechanistic connection between the homodimer interface helices and NBS1 binding. These helices show high conservation, and studies of Mre11 from prokaryotes and S. pombe implicate them in dimer stability independent of the eukaryote-specific latching loops (45,60,63,64). MRE11 L72 lies within these helices, and the impact of MRE11 L72D on homodimerization provides strong evidence that they function in dimer stability in mammals. The influence of this mutant on NBS1 binding indicates that during evolution eukaryotes have built upon the core dimer structure to provide scaffolding for NBS1-dependent functions such as initiation of DNA damage signaling.
The severe impact of the MRE11AATLD17 mutation could in part explain unique aspects of the disease features of the affected patients. MRE11AATLD17 was an allele in the two ATLD17/18 children who succumbed to lung cancer, an outcome not reported in other ATLD patients (32,34–37,65). However, the second allele in these individuals must also be considered. MRE11AASM represents a loss of 27-amino acid residues from a feature unique to MRE11, the capping domain (45). In unicellular organism MRE11 orthologues, the capping domain has DNA- and RAD50-binding sites (58,62,64,66). Therefore, the capping domain is likely to be crucial for folding of the greater N-terminal phosphodiesterase domain and thus required for stability of the MRE11 protein as a whole. In support of this notion, although MRE11AASM mRNA was found in ATLD17/18 patient cells, no evidence of ASM protein was apparent (35,44). Collectively, the available evidence suggests that MRE11AASM may functionally mimic a null allele in the patient cells, leaving the MRE11ATLD17 protein as the disease-causing entity.
In contrast to the well-conserved N-terminus, the C-terminus of MRE11 is divergent in several respects. Prokaryotic Mre11 has little sequence beyond the capping domain, whereas eukaryotes have ∼300 additional residues. The primary sequence of these residues is poorly conserved when comparing different evolutionary groups of eukaryotes, and no structural information is yet available. The Mre11aATLD1 nonsense mutation lies near the 3-prime end of the Mre11a coding sequence and encodes a protein lacking the final 76-amino acid residues (32,38). This truncated protein maintains interactions among MRN components and supports ATM activation. We conclude from this that ATLD1/2 cells have reduced ATM activity and patients have cerebellar degeneration due to low levels of the MRN complex rather than a specific functional defect in the MRE11ATLD1 protein per se. This could provide a rational explanation for the differences in disease course of ATLD1/2 patients compared with syndromes resulting from mutations that cause significant structural alterations in the MRN complex. Examples of the latter would include ATLD17/18—as demonstrated in this study—and NBS resulting from early termination and alterative translation initiation of mutant NBS1 (67). Some ATLD mutations are likely to have intermediate effects compared with those in this study. For example, ATLD3/4 patients harbor a mutation in the N-terminus of MRE11 (N117S), but have not been reported to develop cancer. Interestingly, the ratio of NBS1 to Mre11 proteins in ATLD3/4 is impacted far less than in ATLD17/18, suggesting a lesser impact on MRN structural integrity in ATLD3/4 (32,35). Therefore, we postulate that cancer predisposition may arise in the context of major structural abnormalities in the MRN complex and be less probable when mutations have subtle effects on complex stability or primarily effect MRN protein levels.
How the MRE11AATLD1 nonsense mutation causes low MRE11 levels is not understood. The mammalian MRE11 C-terminus has recently been shown to interact with cyclin-dependent kinase 2 (CDK2) and to be required for CDK2-dependent phosphorylation of the DNA repair factor CTIP (RBBP8) (51). In principle, loss of this interaction could potentially destabilize MRE11 in a manner similar to disrupted interactions with NBS1. However, CDK2 levels are not reduced in MRE11 deficiency, and MRE11 is present at normal levels in CDK2 knockout mouse fibroblasts (data not shown and (51)). It is possible that additional protein interactions occur at the eukaryote-specific extended MRE11 C-terminus that would explain why levels are low in ATLD1/2. In addition, because mutation of the GAR motif in mice resulted in reduced MRE11 levels, it is formally possible that loss of the C-terminal 76-amino acid residues causes low MRE11 levels by impacting the GAR motif at a distance. Understanding this portion of mammalian MRE11 represents an important future goal for insight into DDRs as well as disease mechanisms of MRN deficiencies.
Although the N-terminal domain of MRE11 is highly conserved, there may be important differences in downstream events under its control. For example, the S. pombe mutation analogous to MRE11ATLD17 (SpMre11 W248R) was not defective in activation of Tel1 (the S. pombe ATM orthologue) despite causing a significant disruption of MRN complex stability (68). Our studies support the notion that mammalian MRE11 dimer formation and NBS1 interaction are mechanistically linked and that this linkage plays an important role in activation of ATM. While a similar linkage may occur between S. pombe Mre11 and Nbs1, communication to the respective DNA damage kinases (ATM versus Tel1) might differ. This makes teleologic sense since simple organisms need only activate checkpoints to provide sufficient time for repair, whereas metazoan cells must decide whether to risk attempting repair, versus initiating senescent or apoptotic programs.
Our studies highlight the notion that crucial differences in disease sequelae among related disorders can result from subtle differences in multi-protein complex stability. In the case of MRN, all inherited disease alleles are hypomorphic owing to embryonic lethality of null alleles. Therefore, this spectrum of disorders provides a superb platform for our greater understanding of inherited syndromes of this sort. Defining the relationship between disease outcomes and specific protein structures and functions has important diagnostic and prognostic implications. With sufficient knowledge, informed decisions can be made regarding long-term medical surveillance and care catered to the specific mutations present. This type of information will also aid in the emerging strategy of developing therapeutics that manipulate protein–protein interactions rather than directly targeting a catalytic site. In the near future, it may be possible to develop compounds that compensate for structural mutations, restoring interactions and catalytic activity.
MATERIALS AND METHODS
DNA construct creation
Select Mre11a mutations were introduced into pEF6-MmMre11a using site-directed mutagenesis (Stratagene) or—for the alternative splice mutant—PCR amplification followed by ligation. These mutants were shuttled into pGBKT7 and pGADT7 (Clontech). MmNbn (GeneCopoeia) were TOPO PCR subcloned (Life Technologies) and shuttled into the Y2H vectors.
MEF engineering and culture
Mre11acond/Δ murine embryonic cell lines (41) were maintained using standard culture conditions. To create clones, cells were transfected (Lipofectamine 2000, Life Technologies) with empty vectors or wild-type MRE11- or mutant MRE11-expressing constructs and clones were isolated and grown under blasticidin (Life Technologies) selection. Prior to each experiment, cells were grown under blasticidin selection for 3 days, split, allowed to recover for a day, treated with replication-defective adeno-cre (University of Michigan Vector Core), allowed to recover for 2 days, split and allowed to recover for 2–3 days after which time experiments were performed. ATM−/− MEFs were as described previously (51). Where IR treatment is indicated, cells were exposed to a 137Cs source.
Mre11a RNA typing
5e5 cells per sample were pelleted, RNA was isolated (AllPrep DNA/RNA Mini Kit, Qiagen), RT-PCR was performed (forward primer: GCAATCTCAACATTTCCATTCC, reverse primer: GTTTCTTCTTGGGCAACTACTG) and amplicons were subjected to Sanger sequencing (sequencing primers: CAGTATTTAGTATCCACGGCAAC, CATCGTCATCATCCTCATCTG, GGAGAAGAGATCAACTTTGGG and CTCTTCCTTGTCCACAAACTC).
Immunoblots
Cells were lysed in Laemmli buffer (BioRad) and heated at 100°C for 10 min. Protein concentrations were ascertained by BCA assay (Thermo Scientific). Proteins were resolved by SDS–PAGE, transferred to PVDF membranes (Immobilon) and blocked in 5% milk TBST. Primary antibodies used were as follows: MRE11 (Cell Signaling), RAD50 (Bethyl), NBS1 (Novus), pATM S1987 (Rockland), ATM (Cell Signaling), pKAP1 (Bethyl), KAP1 (Cell Signaling), pCHK1 S345 (Cell Signaling), CHK1 (Cell Signaling), GAPDH (Abcam), HA-tag (Cell Signaling), MYC-tag (Cell Signaling) and tubulin (Pierce). Either fluorophore (Li-Cor)- or peroxidase (Jackson Immunolabs)-conjugated secondary antibodies were used. Quantitation was performed using a Li-Cor Odyssey infrared imaging system.
G2/M checkpoint
7.5e5 cells were plated per 10 cm dish, grown for 48 h, treated with 10 Gy IR from a 137Cs source or mock treated, allowed to recover for an hour and fixed. Cells were probed for the mitotic marker p-histone H3 S10 (56) using Cell Signaling primary antibody and FITC conjugated secondary antibody (BD Pharmingen). Flow cytometry (Accuri C6, BD Biosciences) was performed as described previously (69).
Immunoprecipitation
Ten to twenty million cells were lysed with 50 mm Tris-Cl, 300 mm NaCl, 10% glycerol and 1% NP-40. Lysates were precleared with protein A agarose beads (Roche); lysate protein concentrations were measured by BCA assay (Thermo Scientific); 20 µg/µl protein solutions were made; beads were incubated with either anti-MRE11 antibody (Cell Signaling), anti-RAD50 antibody (Bethyl) or anti-NBS1 antibody (Novus); and 0.5 mg protein was added along with phosphatase and protease inhibitors (Roche). After an overnight incubation, beads were washed four times. Proteins were eluted from the beads with Laemmli buffer (BioRad), and extracts were heated at 95°C for 10 min.
Yeast two-hybrid
Y2HGold (Clontech) were cotransformed using the Yeastmaker Yeast Transformation system (Clontech) per the manufacturer's protocol. To select for cotransformed cells, transformation reactions were plated onto SD-L-W agar (Clontech) plates, and colonies were picked and streaked onto SD-L-W agar plates. To test for interaction by colorimetric assay, the restreaked yeast were picked, grown in SD-L-W (Clontech) overnight, assessed for their density (by OD600nm) and briefly centrifuged. α-Galactosidase activity was visualized as conversion of p-nitrophenyl-α-d-galactopyranoside (colorless) to p-nitrophenoxide (yellow, λmax = 410 nm). Sixteen microliters of supernatant was aliquoted per reaction, 48 µl assay buffer (2 volumes 0.5 m NaOAc, pH 4.5 (aq) and 1 volume 100 mm p-nitrophenyl-α-d-galactopyranoside (Sigma-Aldrich) (aq)) was added and the reactions were incubated at least overnight. Each reaction was quenched with 136 µl 1 Na2CO3 (aq), and OD410 nm readings were taken. Colony growth was assessed by culturing yeast overnight, normalizing yeast density by OD600 nm and plating five 10X serial dilutions onto SD-L-W (loading control) and SD-L-W-H-ade (interaction test) agar plates.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement: None declared.
FUNDING
This work was supported by the National Institutes of Health (R01–HL079118 to D.O.F., T32 GM007863 and T32 CA009676 to J.A.R. and the University of Michigan Cancer Center Support Grant 5-P30-CA46592) and the Leukemia and Lymphoma Society (1074-10 to D.O.F.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Drs JoAnn M. Sekiguchi, Eric Fearon, Mats Ljungman and Xiaochun Yu for helpful discussions regarding the manuscript and project, and Drs Kishore K Chiruvella and Tom Wilson for assistance with the yeast two-hybrid system.
REFERENCES
- 1.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. doi:10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stracker T.H., Petrini J.H. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. doi:10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falck J., Coates J., Jackson S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. doi:10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.H., Paull T.T. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. doi:10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.H., Paull T.T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. doi:10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 6.Paull T.T., Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. doi:10.1016/S1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 7.Paull T.T., Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. doi:10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. doi:10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W., et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. doi:10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadrado M., Martinez-Pastor B., Murga M., Toledo L.I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 2006;203:297–303. doi: 10.1084/jem.20051923. doi:10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S., et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. doi:10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 12.Varon R., Vissinga C., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. doi:10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 13.Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R., 3rd, Hays L., Morgan W.F., Petrini J.H. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. doi:10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 14.McKinnon P.J. ATM and ataxia telangiectasia. EMBO Rep. 2004;5:772–776. doi: 10.1038/sj.embor.7400210. doi:10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demuth I., Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. 2007;26:7792–7798. doi: 10.1038/sj.onc.1210876. doi:10.1038/sj.onc.1210876. [DOI] [PubMed] [Google Scholar]
- 16.Athma P., Rappaport R., Swift M. Molecular genotyping shows that ataxia-telangiectasia heterozygotes are predisposed to breast cancer. Cancer Genet. Cytogenet. 1996;92:130–134. doi: 10.1016/s0165-4608(96)00328-7. doi:10.1016/S0165-4608(96)00328-7. [DOI] [PubMed] [Google Scholar]
- 17.Cybulski C., Wokolorczyk D., Kluzniak W., Jakubowska A., Gorski B., Gronwald J., Huzarski T., Kashyap A., Byrski T., Debniak T., et al. An inherited NBN mutation is associated with poor prognosis prostate cancer. Br. J. Cancer. 2013;108:461–468. doi: 10.1038/bjc.2012.486. doi:10.1038/bjc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuhlke K.A., Johnson A.M., Okoth L.A., Stoffel E.M., Robbins C.M., Tembe W.A., Salinas C.A., Zheng S.L., Xu J., Carpten J.D., et al. Identification of a novel NBN truncating mutation in a family with hereditary prostate cancer. Fam. Cancer. 2012;11:595–600. doi: 10.1007/s10689-012-9555-1. doi:10.1007/s10689-012-9555-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciara E., Piekutowska-Abramczuk D., Popowska E., Grajkowska W., Barszcz S., Perek D., Dembowska-Baginska B., Perek-Polnik M., Kowalewska E., Czajnska A., et al. Heterozygous germ-line mutations in the NBN gene predispose to medulloblastoma in pediatric patients. Acta Neuropathol. 2010;119:325–334. doi: 10.1007/s00401-009-0608-y. doi:10.1007/s00401-009-0608-y. [DOI] [PubMed] [Google Scholar]
- 20.Seemanova E., Jarolim P., Seeman P., Varon R., Digweed M., Swift M., Sperling K. Cancer risk of heterozygotes with the NBN founder mutation. J. Natl Cancer Inst. 2007;99:1875–1880. doi: 10.1093/jnci/djm251. doi:10.1093/jnci/djm251. [DOI] [PubMed] [Google Scholar]
- 21.Swift M., Reitnauer P.J., Morrell D., Chase C.L. Breast and other cancers in families with ataxia-telangiectasia. N. Engl. J. Med. 1987;316:1289–1294. doi: 10.1056/NEJM198705213162101. doi:10.1056/NEJM198705213162101. [DOI] [PubMed] [Google Scholar]
- 22.Thompson D., Duedal S., Kirner J., McGuffog L., Last J., Reiman A., Byrd P., Taylor M., Easton D.F. Cancer risks and mortality in heterozygous ATM mutation carriers. J. Natl Cancer Inst. 2005;97:813–822. doi: 10.1093/jnci/dji141. doi:10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 23.Renwick A., Thompson D., Seal S., Kelly P., Chagtai T., Ahmed M., North B., Jayatilake H., Barfoot R., Spanova K., et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat. Genet. 2006;38:873–875. doi: 10.1038/ng1837. doi:10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 24.Fletcher O., Johnson N., dos Santos Silva I., Orr N., Ashworth A., Nevanlinna H., Heikkinen T., Aittomaki K., Blomqvist C., Burwinkel B., et al. Missense variants in ATM in 26,101 breast cancer cases and 29,842 controls. Cancer Epidemiol. Biomarkers Prev. 2010;19:2143–2151. doi: 10.1158/1055-9965.EPI-10-0374. doi:10.1158/1055-9965.EPI-10-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J., Petersen S., Tessarollo L., Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. doi:10.1016/S0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 26.Dumon-Jones V., Frappart P.O., Tong W.M., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263–7269. [PubMed] [Google Scholar]
- 27.Kang J., Bronson R.T., Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 2002;21:1447–1455. doi: 10.1093/emboj/21.6.1447. doi:10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. doi:10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 29.Spring K., Ahangari F., Scott S.P., Waring P., Purdie D.M., Chen P.C., Hourigan K., Ramsay J., McKinnon P.J., Swift M., et al. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nat. Genet. 2002;32:185–190. doi: 10.1038/ng958. doi:10.1038/ng958. [DOI] [PubMed] [Google Scholar]
- 30.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. doi:10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 31.Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. doi:10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 32.Stewart G.S., Maser R.S., Stankovic T., Bressan D.A., Kaplan M.I., Jaspers N.G., Raams A., Byrd P.J., Petrini J.H., Taylor A.M. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. doi:10.1016/S0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 33.Delia D., Piane M., Buscemi G., Savio C., Palmeri S., Lulli P., Carlessi L., Fontanella E., Chessa L. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum. Mol. Genet. 2004;13:2155–2163. doi: 10.1093/hmg/ddh221. doi:10.1093/hmg/ddh221. [DOI] [PubMed] [Google Scholar]
- 34.Fernet M., Gribaa M., Salih M.A., Seidahmed M.Z., Hall J., Koenig M. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Hum. Mol. Genet. 2005;14:307–318. doi: 10.1093/hmg/ddi027. doi:10.1093/hmg/ddi027. [DOI] [PubMed] [Google Scholar]
- 35.Uchisaka N., Takahashi N., Sato M., Kikuchi A., Mochizuki S., Imai K., Nonoyama S., Ohara O., Watanabe F., Mizutani S., et al. Two brothers with ataxia-telangiectasia-like disorder with lung adenocarcinoma. J. Pediatr. 2009;155:435–438. doi: 10.1016/j.jpeds.2009.02.037. doi:10.1016/j.jpeds.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 36.Chaki M., Airik R., Ghosh A.K., Giles R.H., Chen R., Slaats G.G., Wang H., Hurd T.W., Zhou W., Cluckey A., et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. doi:10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitts S.A., Kullar H.S., Stankovic T., Stewart G.S., Last J.I., Bedenham T., Armstrong S.J., Piane M., Chessa L., Taylor A.M., et al. hMRE11: genomic structure and a null mutation identified in a transcript protected from nonsense-mediated mRNA decay. Hum. Mol. Genet. 2001;10:1155–1162. doi: 10.1093/hmg/10.11.1155. doi:10.1093/hmg/10.11.1155. [DOI] [PubMed] [Google Scholar]
- 38.Theunissen J.W., Kaplan M.I., Hunt P.A., Williams B.R., Ferguson D.O., Alt F.W., Petrini J.H. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. doi:10.1016/S1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- 39.Bartkova J., Tommiska J., Oplustilova L., Aaltonen K., Tamminen A., Heikkinen T., Mistrik M., Aittomaki K., Blomqvist C., Heikkila P., et al. Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene. Mol. Oncol. 2008;2:296–316. doi: 10.1016/j.molonc.2008.09.007. doi:10.1016/j.molonc.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo G., Yao M.S., Bender C.F., Mills M., Bladl A.R., Bradley A., Petrini J.H. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. doi:10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. doi:10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda T., Sumiyoshi T., Takahashi M., Kataoka T., Asahara T., Inui H., Watatani M., Yasutomi M., Kamada N., Miyagawa K. Alterations of the double-strand break repair gene MRE11 in cancer. Cancer Res. 2001;61:23–26. [PubMed] [Google Scholar]
- 43.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer. J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. doi:10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 44.Oba D., Hayashi M., Minamitani M., Hamano S., Uchisaka N., Kikuchi A., Kishimoto H., Takagi M., Morio T., Mizutani S. Autopsy study of cerebellar degeneration in siblings with ataxia-telangiectasia-like disorder. Acta Neuropathol. 2010;119:513–520. doi: 10.1007/s00401-010-0639-4. doi:10.1007/s00401-010-0639-4. [DOI] [PubMed] [Google Scholar]
- 45.Hopfner K.P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. doi:10.1016/S0092-8674(01)00335-X. [DOI] [PubMed] [Google Scholar]
- 46.Park Y.B., Chae J., Kim Y.C., Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. doi:10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Boisvert F.M., Dery U., Masson J.Y., Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. doi:10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boisvert F.M., Hendzel M.J., Masson J.Y., Richard S. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle. 2005;4:981–989. doi: 10.4161/cc.4.7.1830. doi:10.4161/cc.4.7.1830. [DOI] [PubMed] [Google Scholar]
- 49.Dery U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., Masson J.Y. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell. Biol. 2008;28:3058–3069. doi: 10.1128/MCB.02025-07. doi:10.1128/MCB.02025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Z., Vogel G., Coulombe Y., Dubeau D., Spehalski E., Hebert J., Ferguson D.O., Masson J.Y., Richard S. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 2012;22:305–320. doi: 10.1038/cr.2011.128. doi:10.1038/cr.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buis J., Stoneham T., Spehalski E., Ferguson D.O. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat. Struct. Mol. Biol. 2012;19:246–252. doi: 10.1038/nsmb.2212. doi:10.1038/nsmb.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakkenist C.J., Kastan M.B. DNA Damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. doi:10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 53.Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. doi:10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 54.Beamish H., Lavin M.F. Radiosensitivity in ataxia-telangiectasia: anomalies in radiation-induced cell cycle delay. Int. J. Radiat. Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. doi:10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- 55.Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 56.Van Hooser A., Goodrich D.W., Allis C.D., Brinkley B.R., Mancini M.A. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell. Sci. 1998;111(Pt 23):3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- 57.Mockel C., Lammens K., Schele A., Hopfner K.P. ATP Driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. doi:10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lammens K., Bemeleit D.J., Mockel C., Clausing E., Schele A., Hartung S., Schiller C.B., Lucas M., Angermuller C., Soding J., et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. doi:10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J.H., Mand M.R., Deshpande R.A., Kinoshita E., Yang S.H., Wyman C., Paull T.T. ATM Kinase activity is regulated by ATP-driven conformational changes in the MRN complex. J. Biol. Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiller C.B., Lammens K., Guerini I., Coordes B., Feldmann H., Schlauderer F., Mockel C., Schele A., Strasser K., Jackson S.P., et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. doi:10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hohl M., Kwon Y., Galvan S.M., Xue X., Tous C., Aguilera A., Sung P., Petrini J.H. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 2011;18:1124–1131. doi: 10.1038/nsmb.2116. doi:10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G., et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. doi:10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das D., Moiani D., Axelrod H.L., Miller M.D., McMullan D., Jin K.K., Abdubek P., Astakhova T., Burra P., Carlton D., et al. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J. Mol. Biol. 2010;397:647–663. doi: 10.1016/j.jmb.2010.01.049. doi:10.1016/j.jmb.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim H.S., Kim J.S., Park Y.B., Gwon G.H., Cho Y. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. doi:10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delia D., Fontanella E., Ferrario C., Chessa L., Mizutani S. DNA damage-induced cell-cycle phase regulation of p53 and p21waf1 in normal and ATM-defective cells. Oncogene. 2003;22:7866–7869. doi: 10.1038/sj.onc.1207086. doi:10.1038/sj.onc.1207086. [DOI] [PubMed] [Google Scholar]
- 66.Williams G.J., Williams R.S., Williams J.S., Moncalian G., Arvai A.S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P., et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. doi:10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maser R.S., Zinkel R., Petrini J.H. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. doi:10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 68.Limbo O., Moiani D., Kertokalio A., Wyman C., Tainer J.A., Russell P. Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair. Nucleic Acids Res. 2012;40:11435–11449. doi: 10.1093/nar/gks954. doi:10.1093/nar/gks954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Theunissen J.W., Petrini J.H. Methods for studying the cellular response to DNA damage: influence of the Mre11 complex on chromosome metabolism. Methods Enzymol. 2006;409:251–284. doi: 10.1016/S0076-6879(05)09015-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.