Abstract
Nanoporous silicon particles (pSi), with a pore size in the range of 20~60 nm, were modified with polyethyleimine (PEI) to yield pSi-PEI particles, which were subsequently complexed with siRNA. Thus, pSi-PEI/siRNA particles were fabricated, with the PEI/siRNA nanocomplexes mainly anchored inside the nanopore of the pSi particles. These hybrid particles were used as carriers to deliver siRNA to human breast cancer cells. Due to the gradual degradation of the pSi matrix under physiological conditions, the PEI/siRNA nanocomplexes were released from the pore interior in a sustained manner. Physicochemical characterization revealed that the released PEI/siRNA nanocomplexes exhibited well-defined spherical shape and narrow particle size distribution between 15 and 30 nm. Gene knockdown against the ataxia telangiectasia mutated (ATM) cancer gene showed dramatic gene silencing efficacy. Moreover, comprehensive biocompatibility studies were performed for the pSi-PEI/siRNA particles both in vitro and in vivo and demonstrated that the pSi-PEI particles exhibited significantly enhanced biocompatibility. As a consequence, PEI-modified porous silicon particles may have substantial potential as safe and effective siRNA delivery systems.
Keywords: non-viral gene delivery, polyplex, siRNA, porous silicon particles, breast cancer
1. Introduction
In order to reap the full benefits of RNA interference (RNAi)-based therapy, effective siRNA delivery systems are highly desireable. It has been evidenced that non-viral siRNA delivery systems are superior to their viral counterparts, due to easy preparation, lower cost, enhanced biocompatibility, and improved biosafety [1–9]. In particular, mesoporous silica nanoparticles (MSNs), with a typical pore size in the range of 2~10 nm, have demonstrated promise as carriers for nanomedicine, including nucleic acid, among others [10–12]. Nano-constructs, typically formed via self-assembly between oppositely charged species, i.e., a nucleic acid and a cationic polymer, are installed or adsorbed outside the mesopore as “nanogate” to prevent small molecules (i.e., anti-cancer agent) entrapped inside the mesopore from leaking out. Upon application of external stimuli, the nanogate dissociates releasing the small-molecule cargo inside the mesopore. It is recognized that the small pore of MSNs may hinder the efficient loading of larger biomolecules (i.e., nucleic acid or protein). Therefore, most of the studies, in which MSNs were used for nucleic acid delivery, have been focused on surface coating of the MSNs with a cationic polymer (i.e., PEI), allowing for complexation with anionic nucleic acid [11–12]. It should be noted that since the polymer/nucleic acid complexes were adsorbed or anchored on the outer surface of the MSNs, these nano-constructs, which were self-assembled via electrostatic interaction, might be vulnerable to enzymatic degradation or even compromised upon injection in the systemic circulation.
In an effort to address such challenges induced by the small pore size, Na et al. recently managed to expand the pore size of the as-prepared small-pore MSNs, i.e., increased from 5 nm to 23 nm, by means of post-synthesis treatment of the small-pore MSNs [13]. Comparisons were made in terms of their applications as siRNA delivery systems between the two types of MSNs with different pore sizes. Results showed that increasing the pore size of MSNs could be a useful strategy towards the improvement of MSN-based siRNA delivery for in vivo applications.
Over the past few years, we have developed a series of nanoporous silicon particles (pSi) with a much larger pore size (i.e., with an average diameter of 20~60 nm) and have utilized such particles as multi-stage vectors (MSVs) for systemic delivery of therapeutic or diagnostic agents, including siRNA [14–21]. Due to the bigger pore size, nanoconstructs packaged with therapeutics could be readily loaded inside the pore interior of the pSi particles to achieve sustained delivery to tumor tissues. In a typical MSV approach, charged nanoliposomes packaged with small molecule drugs or therapeutic siRNA are loaded into the pore interior of the pSi particles via electrostatic interaction and capillary force. Once inside the body, the pSi particles (or stage 1 particles) are gradually degraded and nanoliposomes (or stage 2 particles) are released from the pSi particles, thus achieving multi-stage release. This delivery system has such advantages as enhanced loading efficiency and easy tunablity in particle shape and size, allowing for efficient encapsulation of nano-sized species into the MSV in order to shield them from contacting with the unintended organs or cells, which leads to minimal toxicity and enhanced efficacy.
Moreover, investigating the effects of shape and size on the biological properties both in vitro and in vivo showed that in comparison to hemi-spherical pSi particles [22], discoidal pSi particles exhibited enhanced properties towards their applications as effective carriers in cancer therapy, as evidenced from their increased surface area, improved biodistribution in multiple animal tumor models, among others. In view of the complex biological environment, i.e., presence of numerous charged species in the plasma and in tumor interstitium, it would be very useful to develop a loading strategy in which the siRNA-containing nanocomplexes are anchored inside the nanopores of pSi particles in order to minimize the interaction between the siRNA-containing nanocomplexes and the charged biological species upon systemic administration of the resultant pSi particles. Upon gradual degradation of the pSi matrix, the siRNA-containing nanocomplexes can be released in a sustained manner such that favourable pharmacokinetics could be achieved. Additional feature rendered by such a delivery system is its versatility for multiple therapies, which is of dramatic clinical significance [23].
Herein, we describe a platform, in which a cationic polymer, namely polyethyleneimine (PEI), is readily conjugated to the pore interior of pSi particles via straightforward chemistry, followed by electrostatic complexation with anionic siRNA to form PEI/siRNA nanoparticles. PEI has been widely used as non-viral delivery systems for nucleic acids [24, 25]. Upon gradual degradation of the pSi matrix under physiological conditions, PEI/siRNA nanoparticles are released from the nanopore confinement. The resulting nanoparticles are subsequently internalized into cells leading to gene silencing. The ataxia telangiectasia mutated (ATM) gene was chosen as the target gene to test this delivery system. We and others have previously shown that ATM plays an important role in cancer therapy [17, 26].
2. Materials and Methods
2.1. Materials
All reagents and medium were obtained from Sigma Aldrich (USA), Lonza, or Promega (USA), and used without further purification. RNase-free H2O was supplied by Fisher Scientific (USA). siRNAs were synthesized by Thermo Scientific. All other chemicals and reagents were of analytical grade and were used as received.
2.2. Preparation of pSi particles
pSi particles were fabricated by electrochemical etching of silicon wafers in the Microelectronics Research Center at The University of Texas at Austin as previously described [27]. The pSi particles were oxidized with H2O2 (30%) at 100°C for 2h to the –OH functionality on the surface. Subsequently, the oxidized pSi particles were reacted with 3-(triethoxysilyl)propyl isocyanate (TEIC) to yield the –NCO functionality [28], which was subjected to conjugation of PEI in anhydrous ethanol. The mixture was rinsed with ethanol and centrifuged twice to remove unreacted chemical or PEI from the pSi particles.
2.3. Particle characterization
Zeta potential measurements and dynamic light scattering (DLS) were carried out using a Zeta Sizer Nano ZS (Malvern Instrument, UK). Particles were dispersed in PB buffer (pH 7.4) at a concentration of 0.5 mg/mL. The samples were mixed well by sonication for 10 s before analysis. Morphological observation was performed using Bruker MultiMode atomic force microscopy (Bruker, USA). Particle density was measured using a Multisizer 4 Coulter Particle Counter (Beckman Coulter, USA). Prior to the analysis, the samples were dispersed in the balanced electrolyte solution (ISOTON® II Diluent, Beckman Coulter, USA) and sonicated for 10 s to ensure a homogenous dispersion. Absorbance and fluorescence measurements were performed with BioTek Synergy H4 hybrid multi-mode microplate reader (BioTek, USA) using a Take3 Micro-Volume plate (BioTek, USA). Morphological studies were performed using scanning electron microscopy (FEI Nova NanoSEM 230) operated at 20 keV or a transmission electron microscopy (JEOL 2010) equipped with a CCD camera and operated at 120 keV.
2.4. In vitro transfection
MDA-MB-231 cells were seeded in 6-well microplates at a density of 2×105 cells/well and allowed to attach overnight in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). After the attachment, the culture medium was replaced with fresh DMEM without FBS. The as-prepared pSi-PEI/siRNA particles were re-dispersed with DMEM to desired concentrations. 100 μL of the diluted sample solution was added to each well, and the cells were incubated at 37 °C for 60 h under 5% CO2 atmosphere. As a control, siRNA transfected with a commercial transfection reagent INTERFERin (Polyplus Transfection, France) was used, by following the manufacturer’s instructions.
2.5. Confocal Laser Scanning Microscopy (CLSM) Observation
Confocal images were acquired at the ACTM Core facility of TMHRI, using a Fluoview 1000 laser scanning fluorescence microscope (Olympus, Japan) equipped with an oil-immersion 100× numerical aperture PlanS Apo objective. The cells were seeded on 35-mm dishes with a cover glass bottom (MatTek Corporation, Ashland, MA). In order to visualize the hybrid particles, Alexa Fluor 555-labeled siRNA was used. Identical protocol was employed for the siRNA transfection as that for the above-mentioned transfection experiments. After transfection for desired time intervals, the cells were washed twice with PBS and fixed with 4% paraformaldehyde in PBS. The nuclei and the endosomes/lysosomes were stained with DAPI and LysoTracker Green, respectively. Excitation wavelengths were 405 nm, 488 nm, and 543 nm, for DAPI, LysoTracker Green, and Alexa Fluor, respectively.
2.6. Western Blot
MDA-MB-231 cells were seeded in 6-well plates (2×105 cells per well). Identical cell culture conditions and siRNA transfection protocols as described above were followed. 60 h after the transfection, the cells was rinsed with PBS and harvested by trypsinization. The treated cells were washed and incubated with lysis buffer containing protease and phosphatase inhibitors. Protein lysates were recovered and the concentration was determined using a BCA assay (Thermo, USA). Then, protein lysates were mixed with SDS loading buffer and heated at 95 °C for 5 min. Samples were separated by electrophoresis with Mini-PROTEAN precast gels (Bio-Rad, USA) and transferred to nitrocellulose membranes (Bio-Rad, USA). Membranes were blocked for 1 h in 5% non-fat milk in Tris-buffered saline with 0.1% Tween-20, and subsequently incubated with desired primary antibody (Cell Signaling Technology, USA) overnight. After wash, they were incubated with HRP-conjugated secondary antibody (Cell Signaling Technology, USA) for 1 h. Membranes were washed and protein bands were detected by enhanced chemiluminescence using a ChemiDoc XRS+ imaging system (Bio-Rad, USA).
2.7. Animal experiments
Female FBV mice (5–6 weeks old, 19–24 g, Charles River Laboratories, USA) were maintained in a VAF-barrier facility. Mice were randomly divided into 5 groups (n= 4) and received a single injection through the tail vein. In the single administration setting, each mouse was injected with 100 μL of aqueous solution containing 15 μg or 75 μg of siRNA. Control mice were administered via i.v. injections of PBS and pSi-PEI complexed with scramble siRNA (pSi-PEI/Scr) at an identical siRNA dosage. Fifteen days after injection, full blood was collected for a complete blood test and biochemistry analysis. All animal work was done in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of Houston Methodist Research Institute in Houston, Texas.
2.8. Whole blood analysis
Blood samples were collected from mice fifteen days after the injection. The samples were analyzed for the number of percentage of total white blood cells (WBC), lymphocytes (LYM), monocytes (MONO), and granulocytes (GRAN). In addition, hematocrit value (HCT), mean corpuscular volume (MCV), red blood cells (RBC), hemoglobin content (HGB), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and red cell distribution width (RDW) were determined.
2.9. Serum biochemistry
Sera were obtained from the blood samples mentioned above. Parameters related to hepatic function (i.e., aspartate aminotransferase (AST), analine aminotransferase (ALT), and alkaline phosphatase (ALKP)) and renal function (i.e., blood urea nitrogen (BUN) and creatinine) were measured.
2.10. Histological evaluation
The tissues were fixed in 10% formalin and embedded in paraffin. Tissue sections (5 μm) were stained with hematoxylin/eosin (H&E). Microscopic analysis was performed. At least five random sections from each slide were examined.
3. Results and Discussion
3.1. Particle modification and physicochemical characterization
Pristine pSi particles, with –OH surface functionality, were modified with 3-(triethoxysilyl)propyl isocyanate (TEIC) to yield isocyanate functionality, which can readily react with the amine-containing PEI polymer (molecular weight: 25 kDa). The resulting positively-charged PEI-conjugated pSi particles (pSi-PEI) were complexed with negatively charged siRNA. Zeta potential measurements confirmed the success of each preparation step (Fig. 1), as evidenced by the results showing that the surface potential of the pSi particles varied in accordance with different surface functionalities.
Figure 1.
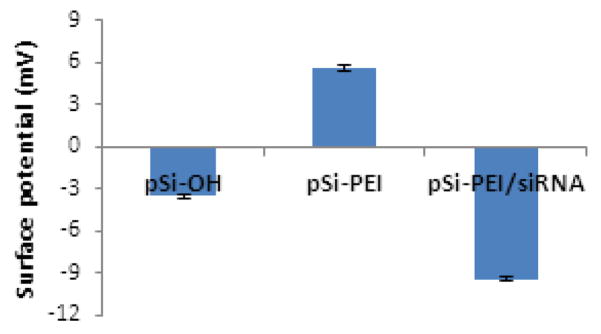
Zeta potential values of pSi particles with various surface functionalities.
SEM analyses were carried out for the pristine pSi, pSi-PEI and pSi-PEI/siRNA particles, as shown in Fig. 2a–c. The nanoporous pSi particles exhibited well-defined disc-like shape, with a diameter of around 1 μm and a height of around 400 nm. The presence of nano-sized pores, with a pore size of between 30 and 60 nm, allowed for efficient encapsulation of siRNA-containing nanocomplexes inside the pore. In addition, TEM observation was performed on the pSi-PEI/siRNA particles (Fig. 2d & 2e). Due to the difference in electron conductivity between the semi-conducting silicon matrix and the insulating polymer layer, the TEM images further revealed that a thin non-conducting layer containing PEI/siRNA nanocomplexes was present in the pore interior as well as on the outer surface of the pSi particles, validating the notion of PEI/siRNA nanocomplexes being anchored onto the pSi matrix.
Figure 2.
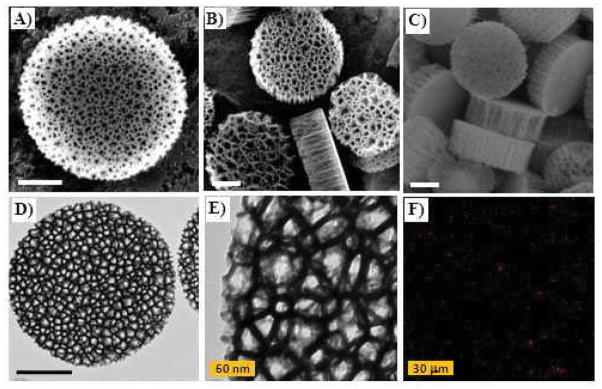
Morphological observations of discoidal pSi particles by SEM (a to c), TEM (d and e), and CLSM (f). SEM images of pSi particles (a), pSi-PEI (b), and pSi-PEI/siRNA (c). TEM images of pSi-PEI/siRNA particles were taken at 20000× (d) and 80000× (e) magnifications, respectively. Confocal image of the pSi-PEI/AF 555-siRNA particles (d). Scale bars represent 300 nm unless indicated.
In addtion to electron microscopies, fluorescence microscopy was used to verify the formation of fluorescent pSi-PEI/siRNA particles. To this end, Alexa Fluor 555-labeled siRNA (AF-siRNA) was used to complex with pSi-PEI to yield pSi-PEI/AF-siRNA particles, which were visualized with a fluorescence microscopy. Fig. 2f shows that when excited with laser, the particles emitted significant red fluorescence, with a size around 1 μm, thus confirming the presence of pSi-PEI/AF-siRNA particles.
3.2. Determination of loading efficiency
It has been recognized that the siRNA loading capacity is critical to the development of practical siRNA delivery systems [2]. To evaluate the loading capacity of the pSi-PEI particles, various amounts of siRNA were used to complex with a given amount of pSi-PEI particles. Centrifugation was performed at 14,000g to ensure the absence of free or unbound siRNA on the resulting pSi-PEI/siRNA particles. Encapsulation efficiency was calculated by the difference in optical density at 260 nm between the supernatant and the pre-loading siRNA solution. Moreover, surface potential measurement was performed to monitor the progress of the complexation between pSi-PEI and siRNA. Fig. 3 shows that the surface potential decreased with increasing siRNA loading until it reached a plateau, suggesting that all the cationic PEI chains were bound to the anionic siRNA. A complete complexation between PEI and siRNA enabled determination of the maximum siRNA loading efficiency for a given pSi-PEI particle. Due to the efficient complexation between the pSi-PEI and siRNA, a maximum of 70 μg of siRNA could be bound to one billion of pSi particles.
Figure 3.
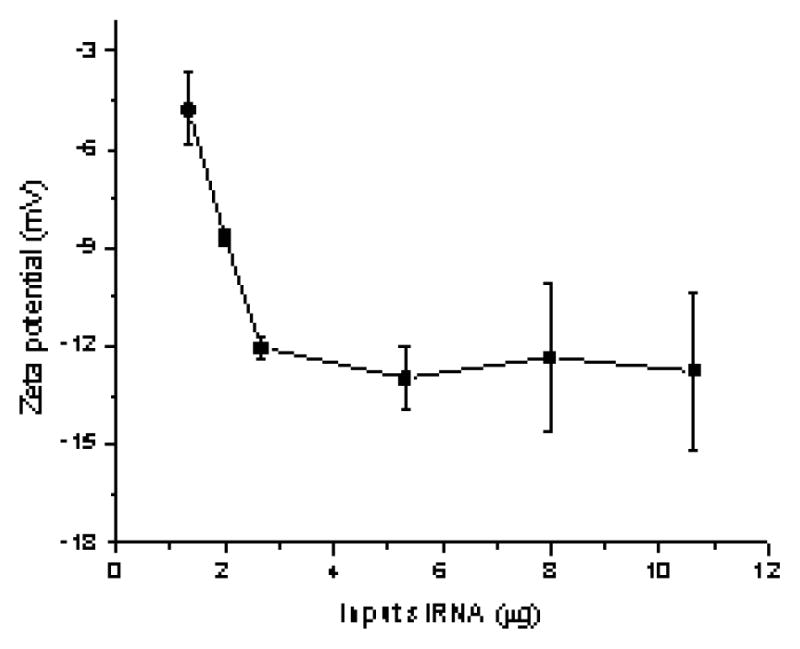
Surface potential changes as a function of input siRNA for a given amount of pSi–PEI particles.
3.3. Degradation of pSi matrix and sustained release of payload
To validate the notion of sustained release of payload anchored inside the nanopores, it was important to verify the structural integrity of the PEI/siRNA nanoparticles during the gradual degradation of the pSi matrix. To this end, the pSi-PEI/siRNA particles were dispersed in PBS and incubated at 37 °C for 24h to ensure complete dissolution of the pSi matrix. Centrifugation was performed and the supernatant was collected for further characterization.
First, morphological observation was performed using atomic force microscopy (AFM). Fig. 4a shows that the resultant PEI/siRNA nanocomplexes exhibited spherical shape with an average particle size ranging from 20 to 40 nm, which is close to the pore size of the pSi particles. The result supports the notion that the particle size of the PEI/siRNA nanocomplexes is governed by the steric confinement of the nanopores. Dynamic light scattering (DLS) measurements (Fig. 4b) reveal that the as-released nanoparticles exhibited a hydrodynamic diameter of around 120 nm, with a very narrow particle size distribution (PDI < 0.1). Note that the difference in the particle size obtained by AFM and by DLS is attributed to the different sample states required by the two characterization techniques, i.e., dry state (ca. 30 nm) vs. hydrated state (120 nm). The results obtianed from DLS measurements are in consistent with those obtained for the PEI/siRNA polyplex nanoparticles reported in the literature [30], even though our particles are somewhat smaller than the latter, likely induced by the confinement effect of the pSi particles. They clearly demonstrate the advantage of a nanopore-templated siRNA delivery system, in which the integrity of the PEI/siRNA nanoparticles anchored inside the nanopores remains intact during the sustained release process.
Figure 4.
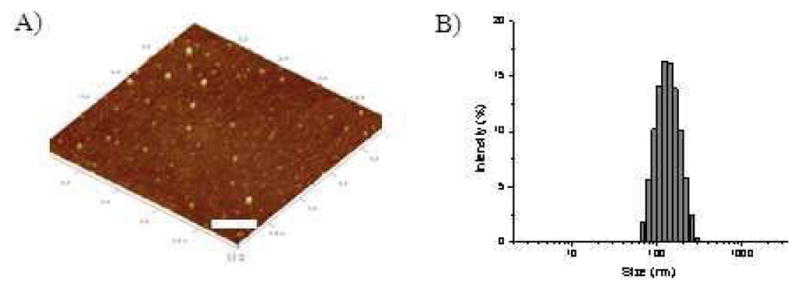
Morphological observation (A) and DLS measurement (B) for PEI/siRNA nanoparticles obtained after complete degradation of the pSi matrix (scale bar: 200 nm).
3.4. Cellular trafficking
In order to gain better insight into how the siRNA-containing particles transport inside the cells, cellular trafficking study using fluorescence microscopy is expected to yield useful information. Cellular trafficking study of hybrid pSi-PEI/Alexa Fluor 555-labeled siRNA particles (or pSi-PEI/AF siRNA) was carried out. These particles were used for transfection with MDA-MB-231 human breast cancer cell line. Confocal laser scanning microscopy (CLSM) was performed 4h and 18h post-transfection. At 4h post-transfection, significant co-localization between the siRNA and the endosomal compartment, as revealed by the overlap between the red Alexa Fluor 555-siRNA and the green staining for the endosomal compartment (Fig. 5a), was observed. The result indicates that particles containing AF-siRNA were taken up by the cells via endocytosis.
Figure 5.
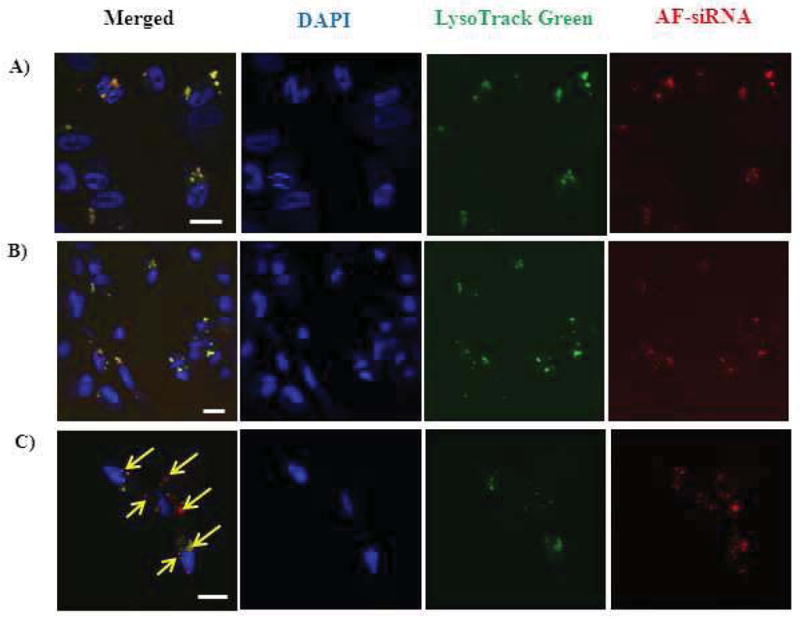
Cellular trafficking study of pSi-PEI/siRNA (a) and PEI/siRNA nanoparticles (b & c). Internalization of particles was performed at 4h (a–b) and 18 h post-transfection (scale bar: 20 μm).
Furthermore, cellular trafficking study by CLSM was performed for the as-released nanoparticles which were recovered after the degradation of pSi-PEI/AF-siRNA hybrid particles under identical experimental conditions as employed above. At 4 h post-transfection, confocal images were acquired by following the identical procedures for the prior CLSM observation. Fig. 5b demonstrates significant co-localization between the siRNA and the endosomal compartments, as seen by from the overlap in fluorescence signals respectively contributed from the labeled siRNA and the endosomal compartment. The results are indicative of cellular internalization of the PEI/AF-siRNA nanoparticles released from the pSi matrix. At a longer time scale (i.e., 18h), lesser degree of overlap between the labeled siRNA and endosome/lysosome compartments was detected, accompanied by the presence of two separate fluorescence signals attributed to the individual species (Fig. 5c). This data indicates that siRNA-containing particles may escape from the late endosomal/lysosomal compartments, thus exerting gene silencing effect.
3.5. Gene knockdown against ATM cancer gene by Western blot
Next, we investigated whether the internalized siRNA-containing particles could result in significant gene knockdown against the target cancer gene, ATM. RNAi efficacy was evaluated for the pSi-PEI/siRNA hybrid particles by performing Western blot analysis on MDA-MB-231 cells. Cells transfected with pSi-PEI particles complexed with scrambled siRNA (pSi-PEI/siSCR), PEI(25k)/siATM nanocomplexes, and siATM using a commercially available transfection reagent (i.e., INTERFERin) were used as controls. As shown in Fig. 6, pSi particles containing scrambled siRNA did not induce any RNAi effect, demonstrating that gene silencing observed for the ATM gene occurred in a sequence-specific manner. In addition, concentration-dependent gene silencing effect was carried out. Dramatic gene knockdown was achieved at an siRNA concentration of 70 nM. The results clearly demonstrate the advantage of utilizing PEI-conjugated pSi particles as an effective carrier to deliver siRNA to human breast cancer cells.
Figure 6.
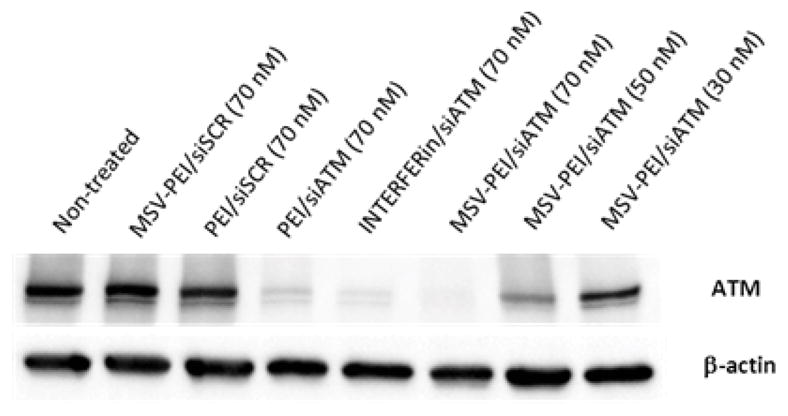
Gene silencing effect against ATM cancer gene at 60 h post-transfection.
3.6. In vitro biocompatibility evaluation
One of the major advantages of using a non-viral siRNA delivery system as opposed to a viral delivery system is its potentially superior biological safety. It has also been recognized that low or minimal toxicity is highly desirable for clinical applications [21]. The cytotoxicity of pSi-PEI/siRNA particles was assessed on MDA-MB-231 cells. MTT assay was performed 48h and 72h after the cells were exposed to the particles. The as-prepared hybrid particles exhibited no cytotoxicity for siRNA concentrations as high as 100 nM (data not shown), indicating significant biocompatibility in vitro. Taking into account the fact that almost complete gene knockdown was achieved at an siRNA concentration of 70 nM, the siRNA delivery system based on pSi-PEI particles is a promising candidate for the enhanced delivery of siRNA to cancer cells in general, and to breast cancer cells in particular.
After the hybrid pSi particles are administered into the bloodstream, they encounter a complex biological environment of plasma proteins and immune cells. Particle uptake by immune cells in circulation, such as macrophages, monocytes, leukocytes and dendritic cells, takes place through various pathways. Thus, it is important to evaluate the extent of response of macrophages to external stimuli. Evaluation of cytokine release was performed on RAW 264.7 mouse macrophage cells. Cells treated with polyinosinic-polycytidylic acid (Poly(I:C)) or PBS were used as control. Levels of two typical pro-inflammatory cytokines (i.e., TNF-α and IL-6), indicative of an inflammatory process, were quantified 24 h after the cells were exposed to various pSi particles at three particle/cell ratios, such as 50:1, 100:1, and 200:1, which respectively corresponds to siRNA dosages of 25 nM, 50 nM, and 100 nM. Fig. 7a and 7b revealed that all three types of pSi particles, i.e., pSi, pSi-PEI and pSi-PEI/siRNA, exhibited similar level of cytokine release to the cells treated with PBS, suggesting that they did not induce any significant increase in cytokine release for the particle/cell ratios employed. This data further confirms the remarkable biocompatibility of the pSi particles.
Figure 7.
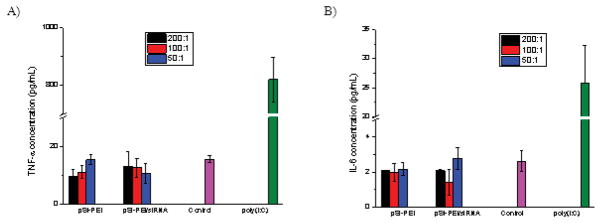
In vitro biocompatability evaluation by determination of pro-inflammatory cytokines secretion, including: a) TNF-α and b) IL-6, using ELISA analysis (n=3).
The above-mentioned toxicity data was unexpected because PEI is known to induce cytotoxicity [24, 31], specifically due to strong interaction between the cationic PEI and the anionic cell membrane. The lack of cytotoxicity from the hybrid delivery system may be related to the superior biocompatibility of the pSi matrix, which confers a “stealth” property for the PEI/siRNA nanoparticles anchored inside the pore. In accordance with those observations for the PEGylated PEI [29], the stealth property indeed dramatically diminished or eliminated the direct contact between PEI and the cells when pSi-PEI/siRNA particles were applied. Only when the pSi matrix was degraded gradually, the PEI/siRNA nanoparticles could be released in a sustained manner.
It should be pointed out that such findings are in contrast to those observed for conventional PEI-based gene delivery systems, in which a large portion of free PEI chains indeed contribute to apparent cytotoxicity [31]. However, in the pSi-PEI conjugation approach described in this study, the free PEI was separated very efficiently and thus removed from the pSi-PEI particles by means of centrifugation, which in turn led to minimal toxicity. One additional advantage rendered by the pSi-PEI conjugation approach was the remarkable reduction of the initial release of PEI/siRNA nanocomplexes from the pore interior, in contrast to the traditional PEI-based siRNA delivery systems, which also accounts for decreased toxicity.
3.7. In vivo biocompatibility evaluation
In vivo biocompatibility evaluation was carried out by treating FBV mice with pSi-PEI particles complexed with siRNA (including siSCR or siATM) at a therapeutic dosage of 15 μg or a supra-therapeutic dosage of 75 μg per mouse. As shown in Fig. 8, there was no body weight change or behavior change during a 15-day treatment period.
Figure 8.
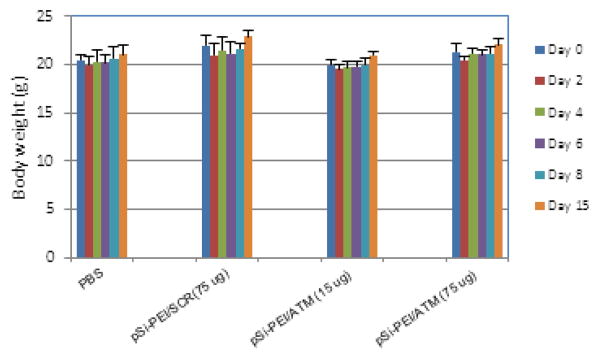
Change of body weight during the period of treatment.
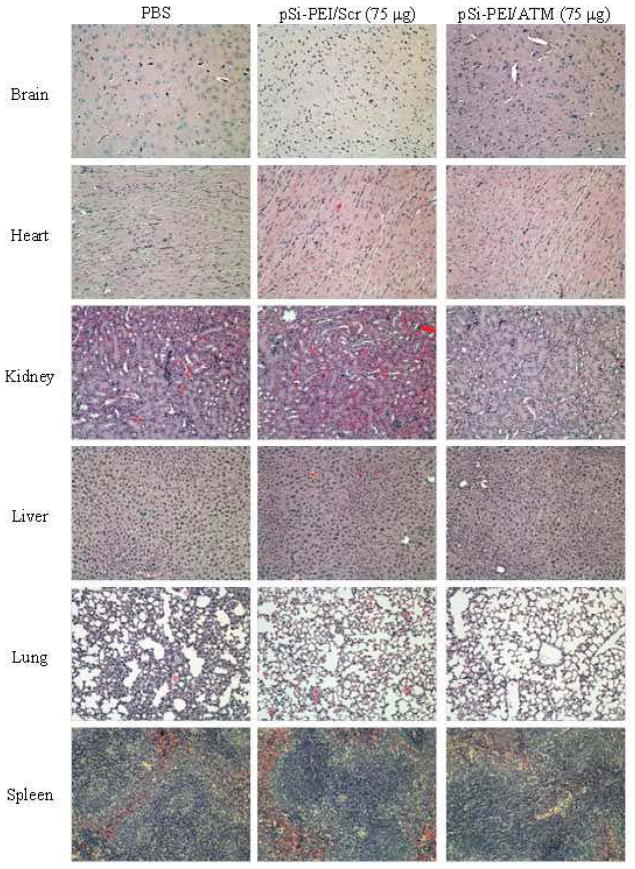
Fifteen days after the treatments, blood samples were collected for biochemical and hematological analyses in order to assess potential damage to major organs (i.e., liver or renal function) (Fig. 9). In comparison to the PBS-treated group, there was no significant difference in biomarker activity or concentration, such as aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALKP) for liver function (Fig. 9a), and blood urea nitrogen (BUN) and creatinine for renal function (Fig. 9b). Likewise, hematological analysis, including white blood cell (WBC), lymphocyte (LYMPH), granulocyte (GRAN), and monocyte (MONO), did not show significant difference between the control and the siRNA-treated groups (Fig. 9c). These in vivo biocompatibility results are consistent with the in vitro results mentioned previously. In additon, the significant biocompatibility of the pSi-PEI particles as siRNA delivery system was demonstrated with the histological examination of the major tissues such as brain, heart, kidney, liver, lung, and spleen of the control and treatment groups. As shown in Fig. 10, no apparent morphological changes were observed. Overall, this data is demonstrative of superior biocompatibility of utilizing pSi-PEI particles as effective siRNA carriers.
Figure 9.
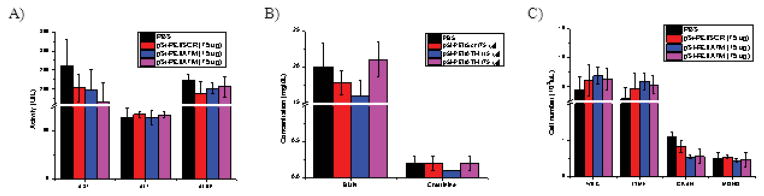
Blood chemistry analysis. Serum samples were collected from mice 15 days after treatment. Levels of a) hepatic enzymes such as aspartate aminotransferase (AST), analine aminotransferase (ALT), and alkaline phosphatase (ALKP); b) renal function biomarkers, including blood urea nitrogen (BUN) and creatinine; and c) hematological paremeters analysis, including white blood cells (WBC), lymphocytes (LYM), granulocytes (GRAN), and monocytes (MONO). Each value represents the mean±SD (n=4).
Figure 10.
Histological examination. Major organs were collected 15 days after injection, and evaluated for potential tissue damages by H&E staining. Representative images (10×) of brain, heart, kidney, liver, lung, and spleen from mice in the control groups (PBS and pSi-PEI/Scr 75 μg) and the treatment groups (pSi-PEI/ATM 75 μg) are shown.
4. Conclusion
We have demonstrated that the PEI-conjugated pSi particles are promising candidates for the facile preparation of a non-toxic delivery system and enable sustained delivery of siRNA to human breast cancer cells. In vitro experiments showed that the resultant siRNA-containing particles could be internalized into cells and effectively silence the ATM cancer gene. Furthermore, remarkable biocompatibility was observed both in vitro and in vivo. In addition to significantly simplifying the manufacturing process for the RNAi-based therapeutics, one important implication of this innovative strategy is that such delivery platforms can be easily adapted for a multiptude of clinical applications by merely adjusting the siRNA of concern. Therefore, such a universal the versatility of this siRNA delivery platform is expected to have great potential in personalized cancer therapy.
Scheme 1.
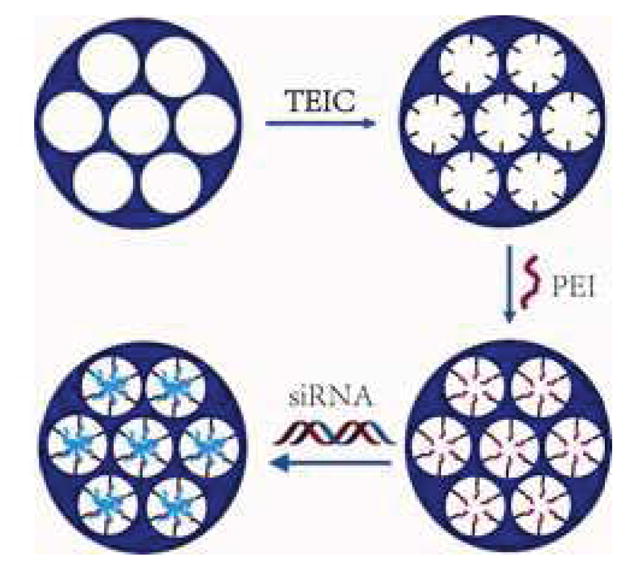
Synthesis of pSi-PEI/siRNA hybrid particles.
Acknowledgments
The authors acknowledge Asad Moten, a visiting doctoral student at the Department of Primary Care Health Sciences and Nuffield Department of Clinical Medicine at the University of Oxford (UK) for his help in editing the manuscript. This work was supported by Department of Defense grants W81XWH-09-1-0212 and W81XWH-12-1-0414, National Institute of Health grants U54CA143837 and U54CA151668, the State of Texas CPRIT grant RP121071, and the Ernest Cockrell Jr. Distinguished Endowed Chair. Financial support from the NSFC (grant number: 51173035) is also acknowledged (M.Z.). The authors do not disclose any potential conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5(3):161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 2.Nishiyama N, Kataoka K. Current state, achievements, and future prospects of polymeric micelles as nanocarriers for drug and gene delivery. Pharmacol Therapeut. 2006;112(3):630–648. doi: 10.1016/j.pharmthera.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Kataoka K. Nano-structured composites based on calcium phosphate for cellular delivery of therapeutic and diagnostic agents. Nano Today. 2009;4(6):508–517. [Google Scholar]
- 4.Miyata K, Nishiyama N, Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem Soc Rev. 2012;41(7):2562–2574. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 5.Ogris M, Wagner E. Targeting tumors with non-viral gene delivery systems. Drug Discov Today. 2002;7(8):479–485. doi: 10.1016/s1359-6446(02)02243-2. [DOI] [PubMed] [Google Scholar]
- 6.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Deliv Rev. 2002;54(5):715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 7.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13(18):1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 8.Felber AE, Dufresne M-H, Leroux J-C. pH-sensitive vesicles, polymeric micelles, and nanospheres prepared with polycarboxylates. Adv Drug Deliv Rev. 2012;64(11):979–992. doi: 10.1016/j.addr.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Upponi JR, Torchilin VP. Design of multifunctional non-viral gene vectors to overcome physiological barriers: dilemmas and strategies. Int J Pharm. 2012;427(1):3–20. doi: 10.1016/j.ijpharm.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Lee JE, Lee N, Kim T, Kim J, Hyeon T. Multifunctional mesoporous silica nanocomposite nanoparticles for theranostic applications. Accounts Chem Res. 2011;44(10):893–902. doi: 10.1021/ar2000259. [DOI] [PubMed] [Google Scholar]
- 11.Xia T, Kovochich M, Liong M, Meng H, Kabehie S, George S, et al. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano. 2009;3(10):3273–3286. doi: 10.1021/nn900918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Chen Y, Wang M, Ma Y, Xia W, Gu H. A mesoporous silica nanoparticle-PEI-fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials. 2013;34(4):1391–1401. doi: 10.1016/j.biomaterials.2012.10.072. [DOI] [PubMed] [Google Scholar]
- 13.Na H-K, Kim M-H, Park K, Ryoo S-R, Lee KE, Jeon H, et al. Efficient functional delivery of siRNA using mesoporous silica nanoparticles with ultralarge pores. Small. 2012;8(11):1752–1761. doi: 10.1002/smll.201200028. [DOI] [PubMed] [Google Scholar]
- 14.Godin B, Tasciotti E, Liu X, Serda RE, Ferrari M. Multistage nanovectors: from concept to novel imaging contrast agents and therapeutics. Accounts Chem Res. 2011;44(10):979–989. doi: 10.1021/ar200077p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen H, You J, Zhang G, Ziemys A, Li Q, Bai L, et al. Cooperative, nanoparticle-enabled thermal therapy of breast cancer. Adv Healthc Mater. 2012;1(1):84–89. doi: 10.1002/adhm.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen H, Sun T, Ferrari M. Nanovector delivery of siRNA for cancer therapy. Cancer Gene Ther. 2012;19(6):367–373. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu R, Huang Y, Mai JH, Zhang G, Guo X, Xia XJ, et al. Multistage vectored siRNA targeting Ataxia-Telangiectasia Mutated for breast cancer therapy. Small. 2013;9(9–10):1799–1808. doi: 10.1002/smll.201201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen H, Rodriguez-Aguayo C, Xu R, Gonzalez-Villasana V, Mai J, Huang Y, et al. Enhancing chemotherapy response with sustained EphA2 silencing using multistage vector delivery. Clin Cancer Res. 2013;19(7):1806–1815. doi: 10.1158/1078-0432.CCR-12-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E, et al. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 2009;70(9):3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ananta JS, Godin B, Sethi R, Moriggi L, Liu X, Serda RE, et al. Geometrical confinement of gadolinium-based contrast agents in nanoporous particles enhances T-1 contrast. Nat Nanotechnol. 2010;5(11):815–821. doi: 10.1038/nnano.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasciotti E, Liu X, Bhavane R, Plant K, Leonard AD, Price BK, et al. Mesoporous silicon particles as a multistage delivery system for imaging and therapeutic applications. Nat Nanotechnol. 2008;3(3):151–157. doi: 10.1038/nnano.2008.34. [DOI] [PubMed] [Google Scholar]
- 22.Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, et al. Size and shape effects in the biodistribution of intravascularly injected particles. J Control Release. 2010;141(3):320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer D, Li YX, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 25.Russ V, Guenther M, Halama A, Ogris M, Wagner E. Oligoethylenimine-grafted polypropylenimine dendrimers as degradable and biocompatible synthetic vectors for gene delivery. J Control Release. 2008;132(2):131–140. doi: 10.1016/j.jconrel.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Canman CE, Lim DS. The role of ATM in DNA damage responses and cancer. Oncogene. 1998;17(25):3301–3308. doi: 10.1038/sj.onc.1202577. [DOI] [PubMed] [Google Scholar]
- 27.Chiappini C, Tasciotti E, Fakhoury JR, Fine D, Pullan L, Wang Y-C, et al. Tailored porous silicon microparticles: fabrication and properties. Chem Phys Chem. 2010;11(5):1029–1035. doi: 10.1002/cphc.200900914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VSY. A polyamidoamine dendrimer-capped mesoporous silica nanosphere-based gene transfection reagent. J Am Chem Soc. 2004;126(41):13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 29.Petersen H, Fechner PM, Martin AL, Kunath K, Stolnik S, Roberts CJ, et al. Polyethylenimine-graft-poly(ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjug Chem. 2002;13(4):845–854. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- 30.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Clamme JP, Azoulay J, Mely Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys J. 2003;84(3):1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]