Abstract
Soluble CD23 (sCD23) plays a role in the positive regulation of an IgE response. Engagement of the beta-2 adrenergic receptor (β2AR) on a B cell is known to enhance the level of both sCD23 and IgE, although the mechanism by which this occurs is not completely understood. We report herein that, in comparison to a CD40L/IL-4-primed murine B cell alone, β2AR engagement on a primed B cell increased gene expression of ADAM10, which is the primary sheddase of CD23, and protein expression of both CD23 and ADAM10, in a PKA- and p38 MAPK-dependent manner, and promoted the localization of these proteins to exosomes as early as two days after priming, as determined by both Western blot and flow cytometry, and confirmed by electron microscopy. In comparison to isolated exosomes released from primed B cells alone, the transfer of exosomes released from β2AR agonist-exposed primed B cells to cultures of recipient primed B cells resulted in an increase in the level of IgE produced per cell, without affecting the number of cells producing IgE, as determined by ELISPOT. These effects still occurred when a β2AR antagonist was added along with the transfer to block residual agonist, and failed to occur when exosomes were isolated from β2AR-deficient B cells. These findings suggest that the mechanism responsible for mediating the β2AR-induced increase in IgE involves a shuttling of the β2AR-induced increase in CD23 and ADAM10 proteins to exosomes that subsequently mediate an increase in IgE.
Introduction
IgE is proposed to play a role in the pathogenesis of allergy and allergic asthma in humans and mice (1). It is known that the level of IgE produced by a B cell is regulated by CD23 (2-10), which is the low affinity receptor for IgE (FcεRII), and which is expressed as a homotrimer on not only the cell surface of B cells (11,12), but also other immune cells, such as macrophages (13). CD23 negatively regulates the level of IgE produced by a B cell when soluble IgE binds to it (2-4), but positively regulates the level of IgE when CD23 is cleaved to a soluble form (5-10), soluble CD23 (sCD23)2 that subsequently binds to CD19/CD21 on a human B cell (6,7). Recently, the expression of CD23 on B cell-derived exosomes has been reported (14,15). Exosomes are cell-derived, cholesterol-rich vesicles that are released by cells, including B cells primed with either LPS or anti-CD40 in the presence of IL-4 (14-16). B cell-derived exosomes also express other proteins, such as MHCII and CD86 (14,15) and contain microRNAs (17). The importance of these molecules being expressed on exosomes is that released exosomes are able to strategically regulate immune cell activity in either an autocrine or paracrine manner at locations far removed from the exosome source (18-20). To date, most studies have focused on the regulation of CD23 cleavage on the B cell surface plasma membrane (12,21). However, recent studies demonstrated that both CD23 and A Disintegrin And Metalloproteinase 10 (ADAM10)2, the primary sheddase for CD23 in a primed B cell (12,22), form a unique interaction intracellularly that results in their packaging into exosomes that are subsequently released from the cell (14), and that CD23 cleavage on exosomes is ADAM10-dependent (14). ADAM10-mediated cleavage of substrates other than CD23 from monocytes, neuroblastoma cell lines, and lymphoma cell lines is also promoted by ADAM10 localization to membrane regions outside of lipid raft domains, as was shown by an increase in ADAM10-mediated cleavage when cholesterol-rich lipid raft microdomains were disrupted by cholesterol depletion or cholesterol-lowering agents (23-26). Because cholesterol-rich lipid raft microdomains are plentiful in exosomes (27,28), the ADAM10 expressed on exosomes is in an ideal membrane environment in which to regulate the cleavage of CD23. Thus, the mechanisms that are known to regulate the level of IgE produced by a B cell involve well-characterized cellular mechanisms involving CD23, ADAM10, sCD23, and possibly exosomes.
In turn, the level of CD23, sCD23, and IgE are regulated by other physiological factors exogenous to the immune system itself. One of these physiological factors is the neurotransmitter norepinephrine (NE)2, which is released after antigen exposure from nerve endings that terminate in the parenchyma of lymphoid tissues [reviewed in (29)], and also, is synthesized in, and released by, CD4+ T cells (30). The level of IgE, as well as the level of sCD23, in the serum and bronchoalveolar lavage fluid (BALF)2 of immunized NE-depleted mice was found to be lower than the level produced in NE-intact mice (31), suggesting that NE may play a physiological role in regulating the IgE response to antigen. Also, the level of IgE produced by primed CD23-deficient B cells that were exposed in vitro to an agonist to the beta-2 adrenergic receptor (β2AR)2, which is the endogenous receptor for NE, is the same as that produced by primed CD23-deficient B cells alone (31), suggesting that the β2AR on a B cell plays a role in regulating the level of IgE produced by first mediating an effect on CD23. More recently, our laboratory reported that β2AR engagement on a CD40L/IL-4-primed B cell increased the rate of mature IgE transcription and protein production, without affecting class switch recombination (31), through a proximal mechanism that involved a Protein Kinase A-dependent (PKA)2 phosphorylation of hematopoietic protein tyrosine phosphatase (HePTP)2(32), which is a phosphatase that functions to bind all unphosphorylated p38 MAPK inside the cell (33). When HePTP is phosphorylated by PKA, the bound p38 MAPK is released and contributes to the pool of free p38 MAPK, causing an increase in the level of p38 MAPK that is now available for phosphorylation by MAPKK (31,32), which is activated by CD40 engagement (34). The β2AR-activated HePTP/p38 MAPK pathway was linked to the regulation of CD23 expression when an inhibitor to p38 MAPK abrogated the β2AR-induced enhancement in CD23 expression at the mRNA level (31). Likewise, the β2AR-induced enhancement in IgE was also linked to p38 MAPK, indicating a common pathway by which β2AR stimulation enhanced CD23 and IgE production by the B cell (31). Taken together, these findings suggested that NE and the β2AR expressed on a B cell play a role in the positive regulation of IgE through a PKA-, HePTP-, and p38 MAPK-dependent mechanism that appears to regulate the level of CD23 and sCD23. However, the mechanism that links these changes following β2AR engagement on a primed B cell to the increase in IgE remains unknown.
Bronchoconstriction and the respiratory distress that is associated with allergic asthma are treated clinically by the administration of a β2AR agonist, which binds to the β2AR expressed on the surface of bronchiolar smooth muscle cells to reduce their contraction around the bronchioles (35). Of relevance to the present study is the clinical evidence that suggests a role for IgE in mediating an exacerbation of allergic asthma in humans and mice (36-39), as well as a role for the β2AR expressed on B cells in the human lung (40,41) to increase the level of IgE produced during β2AR agonist administration. An understanding of the various molecular links between β2AR stimulation on a B cell and the subsequent increase in IgE, if unique, could potentially be blocked during β2AR agonist inhalation therapy to specifically prevent the increase in IgE that might potentially compromise the beneficial effect of the agonist on bronchiolar smooth muscle relaxation.
The goal of the present study was to identify the mechanism that links the proximal intermediates activated following β2AR stimulation on a primed B cell to the more distal intermediates that induce the increase in sCD23 and IgE. To our knowledge, we report for the first time that β2AR stimulation on a primed B cell, as compared to a primed B cell alone, increases the level of gene expression of ADAM10, and protein expression of CD23 and ADAM10 in a PKA- and p38 MAPK-dependent manner. Moreover, the increased level of CD23 and ADAM10 proteins induced by β2AR stimulation appear to localize to exosomes that are released from the B cell to mediate the β2AR-enhancing effect on IgE.
Materials and Methods
Animals
Mice were housed under pathogen-free conditions and used between 6-12 weeks of age. Female BALB/c mice were purchased from the pathogen-free facility at Taconic Farms (Hudson, NY) at six weeks of age and then housed at The Ohio State University in microisolator cages with autoclaved food and water ad libitum. Congenic BALB/c β2AR-deficient mice were derived from FVB β2AR-deficient mice [kindly provided by Brian Kobilka (Stanford University, CA)], after backcrossing for ten generations (32). All experiments complied with the Animal Welfare Act and the National Institutes of Health (Bethesda, MD) guidelines for the care and use of animals in biomedical research.
Reagents
CD40L-Sf9 cells were prepared as described previously (42). Recombinant murine IL-4 was purchased from eBioscience (San Diego, CA). Terbutaline hemisulfate salt, nadolol, the p38 MAPK inhibitor, SB203580, and the PKA inhibitor, H-89, were purchased from Sigma (St. Louis, MO). INCB3619, which is a selective ADAM10/17 inhibitor (43) was kindly provided by Incyte Corporation (Wilmington, DE). All reagents used for B cell isolation, activation, and pharmacologic treatment tested negative for the presence of endotoxin using E-TOXATE (Sigma).
Antibodies
Western blot antibodies were rabbit polyclonal anti-mouse CD23 (kindly provided by Dr. D. Conrad, Virginia Commonwealth University), rabbit polyclonal anti-mouse ADAM10 (Millipore, Billerica, MA) for cell lysates, rabbit monoclonal anti-mouse ADAM10 (EPR5622 (C-terminus), Abcam, Cambridge, MA) for exosomes, rat monoclonal anti-mouse ADAM10 (Clone 139712 (ectodomain), R'D Systems, Minneapolis, MN), rat monoclonal anti-mouse CD63 (R5G2, MBL International, Woburn, MA) or rabbit monoclonal anti-actin (13E5, Cell Signaling Technology, Beverly, MA). Secondary polyclonal antibodies were HRP-labeled goat anti-rabbit IgG and HRP-labeled chicken anti-rat IgG (Cell Signaling Technology). Proteins were detected using a LumiGlo detection kit (Cell Signaling Technology). Ganglioside M1 (GM1)2 was detected using biotin-labeled cholera toxin B subunit (Sigma) and goat polyclonal affinity purified HRP-labeled anti-biotin (Cell Signaling Technology). ELISA antibodies included rat monoclonal anti-mouse CD23 (coating, clone 2G8, Southern Biotech, Birmingham, AL), rabbit polyclonal anti-mouse CD23 (secondary, kindly provided by D. Conrad, Virginia Commonwealth University), and goat polyclonal alkaline phosphatase-labeled anti-rabbit IgG (detecting, H + L; Southern Biotech). Plates were developed with para-NitroPhenylPhosphate (PNPP) substrate (Sigma) in DEM. Antibodies used for ELISPOT included monoclonal anti-mouse IgE (clone R35-72; BD, Franklin Lakes, NJ) and secondary monoclonal AP-labeled rat anti-mouse IgE (clone 23G3; Southern Biotech). Flow cytometry rat monoclonal anti-mouse antibodies and reagents included ADAM10-FITC (Clone 139712), ADAM10-PE (Clone 139712), and isotype control rat IgG2a kappa-PE from R'D, CD23-FITC (B3B4), B220-APC (RA3-6B2), 7-Amino-Actinomycin D (7-AAD)2, isotype rat IgG2a kappa-FITC, and isotype rat IgG2a kappa-APC from eBioscience.
Resting B cell isolation and cell priming
Resting B cell isolation and priming were performed as described previously (31) using negative selection for CD43-negative B cells using an AutoMACS machine (Miltenyi, Bergisch Gladbach, Germany). B cell purity was greater than 98% as determined by FACS. Purified B cells were primed at a density of .5 × 106 B cells/ml with CD40L-Sf9 (1:10) and IL-4 (1 ng/ml) in the absence or presence of terbutaline (10-6 M) and cultured in complete RPMI [RPMI 1640 medium (Cellgro, Manassas, VA), 10% FBS (Atlas Biologicals, Fort Collins, CO), 20 mM HEPES (Gibco, Carlsbad, CA), 100 U/ml penicillin (Invitrogen, Carlsbad, CA), 100 ug/ml streptomycin(Invitrogen), 2 mM glutamine(Invitrogen), 50 uM 2-ME (Sigma)] at 37°C, 5% CO2. In cell concentration experiments, CD40L-Sf9 (1:10) and B cells were serially diluted from 400,000 B cells/well to 12,500 B cells/well in 200 ul volume, followed by the addition of equal amounts of IL-4 (1 ng/ml) in the absence or presence of terbutaline (10-6 M).
Western blot
Western blot and analysis was performed on equal amounts of protein as described previously (31,44) to detect ADAM10, CD23, CD63, Actin, and GM1. Blots were scanned and inverted to perform densitometry using NIH ImageJ64 1.47m.
Quantitative RT-PCR
Quantitative real-time PCR was performed as described previously (31). The following primers were used: beta-actin, 5′-TACAGCT TCACCACCACAGC-3′ (forward) and 5′-AAGGAAGGCTGGAAAAGAGC-3′(reverse), annealing temperature, 58°C; ADAM10, 5′-CACCAATATTTGGGAAACGG-3′ (forward) and 5′TCCTGAGCTCCTGAGGAAAA-3′ (reverse), annealing temperature, 60°C. All primers were synthesized and purchased from Integrated DNA Technologies (Coralville, IA). PCR was performed using the Rotor Gene Q (Qiagen, Germantown, MD) and software used was Rotor Gene Q Series Software (version 2.0.3).
ELISA
ELISA was performed as described previously for sCD23 detection (31). IgE protein in B cell culture supernatants was measured using BD OptEIA mouse IgE kit (BD) and developed with TMB substrate (BD) and acid stop (2N H2SO4, Sigma). Densitometric analysis was performed on a SpectraMax Plus microplate reader (Molecular Devices, Sunnyvale, CA) using Softmax Pro (Molecular Devices), version 5.4.1, at a wavelength of 405 nm for sCD23 and 450 nm for IgE and values were compared to a standard curve where the level of detection for each protein is 1-500 ng for sCD23 and 1-100 ng for IgE.
Exosome isolation and gradient centrifugation
cRPMI was depleted of exosomes by ultracentrifugation at 150,000 × g for fifteen hours at 4°C to pellet exosomes naturally present in FBS in a SW-41 swinging bucket rotor (Beckman Coulter, Brea, CA) in a L-70 ultracentrifuge (Beckman). Ultracentrifuge tubes were sterilized prior to exosome depletion of cRPMI by washing with 10% bleach, 70% ethanol, and a final wash with sterile PBS (Gibco). Ultracentrifuged cRPMI was filtered using a 0.2 uM pore filter (Corning, Corning, NY) prior to cell culture. cRPMI was confirmed to be depleted of exosomes by flow cytometry performed as described below. For the isolation of exosomes released by B cells, exosome-containing supernatant from cultured B cells was fractionated as described previously (14) in a SW-41 swinging bucket rotor in a L-70 ultracentrifuge (Beckman). Prior to any experiments, a characterization of the ultracentrifuged culture supernatant was done to confirm the presence of exosomes by layering of isolated exosomes on an iodixanol (Axis-Shield, Dundee, Scotland) density gradient with densities of 1.00, 1.05, 1.10, 1.15, 1.20, and 1.25 g/ml. The iodixanol density gradient was ultracentrifuged at 100,000 × g overnight at 4°C with no brake. The following day, the various density fractions were collected, diluted in PBS, and then each isolated fraction was ultracentrifuged at 100,000 × g for one hour at 4°C with no brake. Pellets were resuspended in PBS and frozen at -80°C until analysis by Western blot as described above. For most experiments, ultracentrifuged culture supernatant that was not separated by a density gradient was fractionated as described previously (14) and was resuspended in PBS and either frozen at -80°C until analysis by Western blot or stained for flow cytometry as described below.
Flow cytometry
B cells were prepared for FACS as described previously (45) for cell surface detection of CD23, ADAM10, B220 and 7-AAD. Appropriate isotype- and species-matched antibodies were used as controls for gating. Stained cells were detected on a FACSCanto II flow cytometer (BD) and results were analyzed using FlowJo (TreeStar, Ashland, OR). 100,000 events were collected and cells were gated on 7-AAD− and B220+ cells to exclude Sf9-CD40L cells. Additionally, CD23 and ADAM10 staining on Sf9-CD40L cells was below the level of detection.
Flow cytometry of exosomes
Exosomes were isolated as described above, resuspended in PBS, single stained for CD23, ADAM10, or stained with an appropriate species- and isotype-matched control for gating. Because we found that much less antibody was required for the staining of exosomes, in comparison to intact B cells, we titrated all isotype-specific and molecule-specific antibodies to assure that non-specific staining was minimal. Stained exosomes were detected on a FACSCanto II flow cytometer (BD). Polystyrene beads 50-100nm in size (Spherotech, Lake Forest, IL) were used to optimize for exosome size and granularity. Results were analyzed using FlowJo (TreeStar).
Electron Microscopy of exosomes
Exosomes were prepared as described above, fixed, and then analyzed by Transmission Electron Microscopy as described previously (16). Formvar-carbon coated grids (Ted Pella, Redding, CA), onto which the fixed exosomes were placed, were then observed under a transmission electron microscope (Tecnai G2 Spirit, FEI, Hillsboro, OR).
Exosome transfer for functional analysis
Two days after priming in the absence or presence of terbutaline, whole supernatant was either transferred to recipient cultures or fractionated into an exosome-depleted or an exosome-intact fraction. Exosome-intact: Exosomes were isolated under sterile conditions as described above and resuspended in 4 ml of exosome-free media (1X concentration) and added to recipient cultures. Exosome-depleted: The exosome-depleted supernatant that remained after exosome isolation was transferred to a sterile tube and added to recipient cultures. IL-4 was added to all transfer cultures at a concentration of 1 ng/ml. Additionally, some cultures also received Nadolol at a concentration of 10-6 M at the time of transfer. Recipient cultures: Two days after priming in the absence of terbutaline, 96-well plates containing cultured cells were centrifuged at 300 × g to pellet cells. Supernatant was removed and whole, exosome-depleted, or exosome-intact fractions were transferred to recipient cells. Cultures receiving transferred fractions were incubated at 37°C, 5% CO2, for 4 days and then supernatants were removed for analysis of IgE production by ELISA or cells were analyzed by ELISPOT.
ELISPOT
ELISPOT was performed as described previously (31). IgE+ spots were quantified on an Axioplan 2 microscope (Zeiss, Oberkochen, Germany) using KS ELISPOT software (Zeiss, version 4.2.0).
Sucrose Density Gradient Isolation of Lipid Rafts
100 × 106 B cells were activated as described above, lysed, and then fractionated by sucrose density gradient separation as described previously (46). Fractions were collected from the top of the gradient for analysis by SDS-PAGE and Western blot probing of equal volumes for ADAM10, CD23, Actin, and GM1.
Statistics
Data were analyzed by ANOVA, followed by Bonferroni post test when data were normally distributed, to determine whether an overall statistically significant change existed. A Student's t test was used for comparison between two treatment groups. A p value of ≤ 0.05 is designated by *, p ≤ 0.01 by **, and p ≤ 0.001 by ***.
Results
ADAM10 mediates the β2AR-specific enhancing effect on sCD23 and IgE
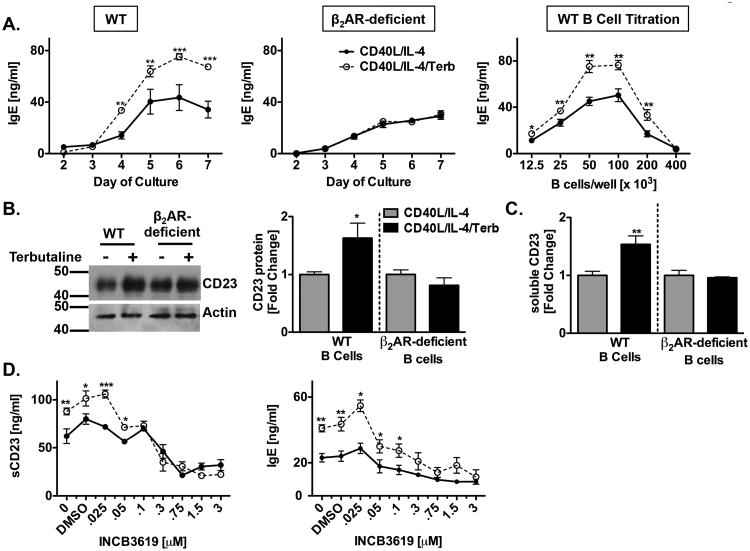
Previous findings showed that IgE, as well as CD23 and sCD23, were increased after the β2AR was engaged on a CD40L/IL-4-primed murine B cell, and that these effects failed to occur in the presence of a β2AR antagonist (31). In the present study, we confirmed that the β2AR was responsible for mediating the IgE-enhancing effect. B cells from WT or β2AR-deficient mice were primed with CD40L/IL-4 in the absence or presence of the selective β2AR agonist, terbutaline, and supernatants were collected on days 2 through 7 for analysis of IgE production by ELISA. The IgE-enhancing effect of β2AR stimulation on primed B cells, in comparison to primed B cells alone, was evident on day 4 and reached a maximal 2-fold increase on day 6 (Figure 1A, left panel); whereas, B cells from β2AR-deficient mice were unable to do so (Figure 1A, middle panel), indicating that the β2AR agonist-induced upregulation in the level of IgE production by primed B cells was specifically a result of β2AR stimulation. It should be noted that we consistently found that β2AR-deficient B cells function similarly to WT B cells for all immune functions, but appear to do so at a lower level of response (31,47). To determine if the β2AR agonist-induced upregulation in the level of IgE production was dependent on cell concentration, as reported previously (48,49), B cells were serially diluted and supernatants were collected on day 6 for analysis of IgE production by ELISA. The IgE-enhancing effect of β2AR stimulation was evident with an input of between 12,500 and 200,000 B cells per well in 200 ul volume, with the maximum β2AR-enhancing effect on IgE exhibited between 50,000 and 100,000 B cells per well (Figure 1A, right panel). We confirmed the cell concentration dependency of IgE production by B cells reported previously, as the addition of 400,000 B cells per well did not exhibit the β2AR-enhancing effect on IgE production, and was significantly reduced in comparison to wells with fewer input B cells, indicating that the number of B cells required to observe the β2AR-enhancing effect was limited by the concentration of B cells per well. β2AR stimulation enhanced CD23 total protein production within 24 hours and continued to increase through day 5 (Figure 1B). Likewise, β2AR stimulation on primed B cells enhanced the level of sCD23 produced, in comparison to primed B cells alone, and this increase was first evident on day 3 (data not shown) and reached a maximal 1.5-fold increase on day 5 (Figure 1C); whereas, B cells from β2AR-deficient mice were unable to increase CD23 or sCD23 in response to the β2AR agonist at any timepoint (Figure 1B-C), also confirming that the agonist-induced increase in CD23 and sCD23 was specifically due to β2AR stimulation on a B cell. Taken together, these findings confirm a role for the β2AR on a B cell in mediating an increase in total CD23 expression, as well as the level of sCD23 and IgE produced.
Figure 1.
ADAM10 modulates the β2AR-mediated enhancing effect on sCD23 and IgE. Resting B cells were isolated from Balb/c (WT) and β2AR-deficient mouse spleens and were primed with CD40L/Sf9 cells (1:10) and IL-4 (1 ng/ml) in the presence (open circles or black bar) or absence (solid circles or grey bar) of the β2AR agonist, terbutaline (Terb, 10-6 M). Total cellular protein and/or culture supernatants were collected for analysis. (A) Left, middle: Six day time course of secreted IgE by WT (left) or β2AR-deficient (middle) B cells using ELISA. Right: Serial dilutions of B cells and CD40L were cultured as above and analyzed for IgE at 6 days using ELISA. Data are presented as mean IgE value (ng/ml) +/- SEM from one representative of two to three independent experiments. (B) CD23 total protein expression at 24 hours using Western blot analysis. One representative blot is shown, with Molecular Weight in kDa, and data are presented as the mean fold difference of total CD23 protein expression in total cellular lysates from WT and β2AR-deficient B cells as standardized to actin +/- SEM from three independent experiments. (C) sCD23 expression at 5 days using ELISA. Data are presented as the mean fold difference of sCD23 from supernatants +/- SEM from three independent experiments. Absolute values of sCD23 were 84 +/- 6 ng/ml (WT) and 61 +/- 5.3 ng/ml (β2AR-deficient). (D) sCD23 (left) and IgE (right) production after five days in the presence of increasing concentrations of the selective ADAM10 inhibitor INCB3619. Data are presented as the mean IgE or sCD23 (ng/ml) +/- SEM from one representative of three independent experiments. Significant differences from priming alone were determined by ANOVA followed by Bonferroni correction or unpaired t-test. *, p = .05, **, p= .01, ***, p=.005.
Based on recent reports that ADAM10 is the primary sheddase of CD23 (12,21,22), it was imperative to determine if the β2AR-induced increase in sCD23 and IgE production was due to a β2AR-induced increase in ADAM10-mediated cleavage of CD23. INCB3619 is a dual ADAM10/ADAM17 inhibitor with an IC50 for various ADAM10 ligands in the range of 0.2-0.7 uM (43). Because ADAM17 is unable to cleave CD23 (12,50), INCB3619 is an excellent compound for the study of ADAM10-specific cleavage of CD23 (12,50). Primed B cells were cultured in the absence or presence of a β2AR agonist with increasing concentrations of INCB3619. The concentration of INCB3619 required to reduce both the control level by half, as well as the β2AR-induced increase of both sCD23 (Figure 1D, left panel) and IgE (Figure 1D, right panel) was between 0.1 and 0.3 uM and occurred independently of changes in cell morphology or cell viability. Concentrations above 2 uM caused a slight reduction in viability (data not shown). This result suggests, for the first time, that ADAM10 plays a role in the β2AR-enhancing effect on sCD23, and further supports that a link exists between the β2AR-mediated increase in sCD23 cleavage and the increase in IgE.
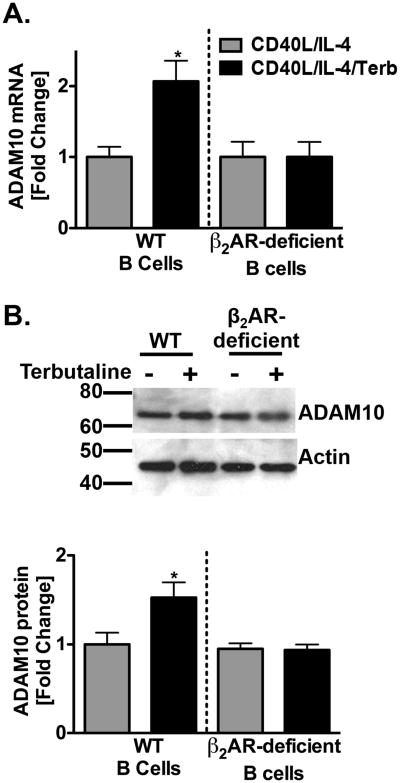
To determine the extent to which β2AR stimulation affected the level of ADAM10 expression, primed B cells from WT or β2AR-deficient B cells were cultured as described above and cell lysates were collected at 1 and 24 hours to measure ADAM10 mRNA and protein expression, respectively, using quantitative RT-PCR and Western blot. The level of ADAM10 mRNA (Figure 2A) and total protein (Figure 2B) were increased 2 and 1.5-fold, respectively, after β2AR stimulation on primed B cells, as compared to primed B cells alone. Conversely, the β2AR enhancement did not occur when β2AR-deficient B cells were used. Taken together with the results from Figure 1, β2AR stimulation on a primed B cell appears to play a role in the upregulation of ADAM10 at the gene level and both CD23 and ADAM10 expression at the protein level.
Figure 2.
ADAM10 gene and protein expression are increased by β2AR stimulation. B cells from WT and β2AR-deficient mice were cultured as described in Figure 1 in the presence (black bars) or absence (grey bars) of terbutaline (Terb). mRNA and total cellular protein, respectively, were collected at one hour and twenty-four hours. (A) Data are presented from three independent experiments as the mean ADAM10 mRNA fold difference as standardized to beta-actin +/- SEM. (B) One representative blot is shown, with Molecular Weight in kDa, and data are presented from three independent experiments as the total ADAM10 protein fold difference as standardized to actin +/- SEM. Significant differences from priming alone were determined by unpaired t-test. *, p = .05.
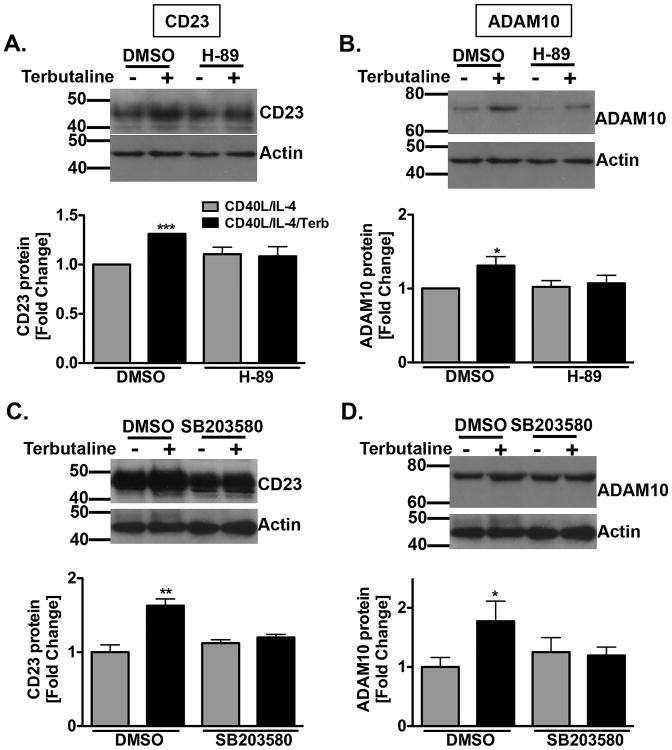
Previous reports had established a proximal link between the β2AR-stimulated and PKA-mediated enhancement of the level of p38 MAPK phosphorylation and IgE (31). To determine if the β2AR-stimulated enhancement of CD23 and ADAM10 was mediated by PKA and/or p38 MAPK, B cells were cultured as described previously in the absence or presence of H-89, a PKA inhibitor, or SB203580, a p38 MAPK inhibitor. After 24 hours, cell lysates were prepared for Western blot analysis. Whereas β2AR stimulation of primed B cells enhanced the level of CD23 and ADAM10, in comparison to primed B cells alone and, also, the PKA inhibitor had no effect on CD23 and ADAM10 in primed B cells alone, B cells exposed to a β2AR agonist in the presence of a PKA or p38 MAPK inhibitor, respectively, were unable to enhance either CD23 (Figure 3A, 3C) or ADAM10 (Figure 3B, 3D) expression. These results indicate that activation of the β2AR/PKA/p38 MAPK pathway is essential for the β2AR-induced enhancement in the level of CD23, ADAM10, and IgE.
Figure 3.
PKA and p38 MAPK inhibition prevents the β2AR-induced enhancement in CD23 and ADAM10 protein. B cells were cultured as described in Figure 1 in the presence (black bar) or absence (grey bar) of terbutaline (Terb). Cell lysates were collected after 24 hours and analyzed by Western blot. A-B. CD23 (A) and ADAM10 (B) expression in the presence of H-89 (.5 uM). C-D. CD23 (C) and ADAM10 (D) expression in the presence of SB203580 (1 uM). One representative blot is shown, with Molecular Weight in kDa, and data are presented from three to four independent experiments as the total protein fold difference compared to priming alone as standardized to actin +/- SEM. Significance based on an unpaired t-test. *, p = .05, **, p= .01, ***, p=.005, as compared to priming alone.
β2AR stimulation does not enhance expression of CD23 and ADAM10 on the B cell surface
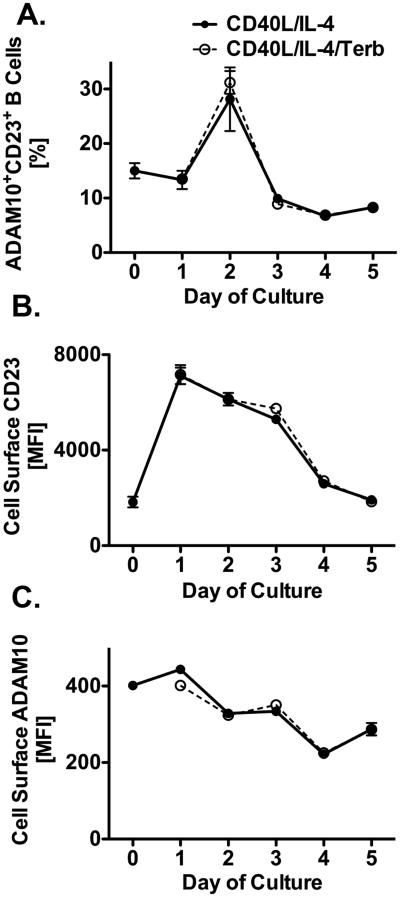
Cleavage of various molecules by ADAM10, such as Notch, FasL, and Amyloid Precursor Protein, has traditionally been thought to occur primarily on the cell surface plasma membrane (12,50,51). Because we showed above that expression of CD23 and ADAM10 were both increased in response to β2AR stimulation on a primed B cell, it was important to determine to what extent this enhancement translated to an increase in expression on the B cell surface. Cultures were set up with B cells, as described previously. FACS analysis of CD23 and ADAM10 expression was done using cells collected on days 0 through 5 after priming. B cells were gated with appropriate antibody isotype controls to include only cells that were both B220+ and viable as determined by 7-AAD exclusion. As shown in Figure 4, the percent of ADAM10+CD23+ cells from both primed and β2AR agonist-exposed conditions increased three-fold on day 2, but went below baseline on day 3 (Figure 4A). Representative histograms of all data are shown in Supplemental Figure 1. We also found that within one day of priming, as determined by MFI, cell surface CD23 increased four-fold in both primed-only B cells and β2AR agonist-exposed primed B cells, and remained elevated until day 3 when levels for both groups began to decrease to baseline levels (Figure 4B). In contrast, the level of ADAM10 on the cell surface slightly increased for both exposure conditions on day 1 then declined to below baseline from days 2 through 5 after priming (Figure 4C). Additionally, almost all B cells were CD23-single positive after CD40L/IL-4 priming, while the same number of B cells that were ADAM10-single positive were also CD23/ADAM10-double positive (Supplemental Figure 1), suggesting that cell surface expression of ADAM10 is linked to CD23 and not vice versa. Also, when using B cells from β2AR-deficient mice, the MFI of cell surface CD23 and ADAM10 after B cell priming in the presence or absence of β2AR stimulation was equivalent (data not shown). Taken together, β2AR stimulation on a B cell enhanced ADAM10 expression at the gene and total protein level, as well as enhanced CD23 expression and cleavage, despite the lack of a change in expression of both CD23 and ADAM10 on the cell surface. These findings suggest a tight regulation of CD23 cleavage by ADAM10, which may occur in a compartment of the B cell, other than the cell surface, that is involved with mediating the β2AR-enhancing effect on IgE production.
Figure 4.
β2AR stimulation does not enhance expression of CD23 and ADAM10 on the B cell surface. B cells from WT mice were cultured as described in Figure 1 in the presence (open circle) or absence (solid circle) of terbutaline (Terb). Cells were collected for FACS analysis from days 1 through 5 after priming. 7-AAD−/B220+/ADAM10+CD23+ Cells Percent Positive (A), MFI of cell surface CD23 (B) and ADAM10 (C). Data are presented as mean percent positive cells (%) or MFI +/- SEM from three independent experiments.
Exosomes released by primed B cells express both CD23 and ADAM10
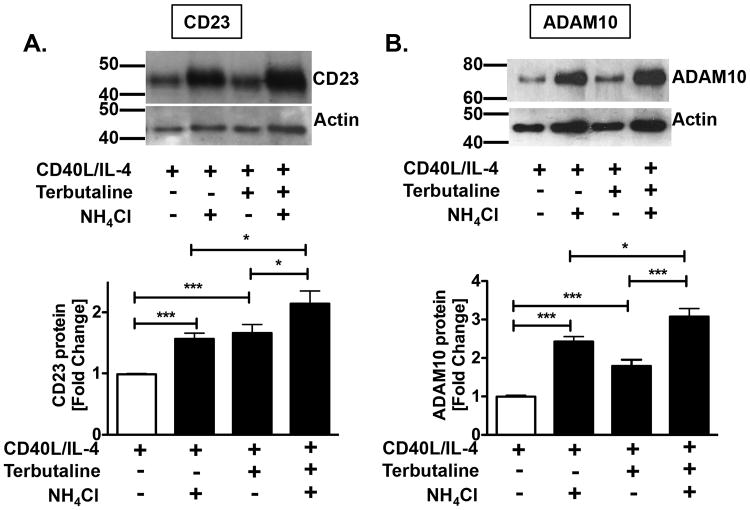
Because the β2AR-induced increase in CD23 and ADAM10 protein did not translate to a change in expression at the cell surface, it became necessary to identify into which cellular compartment the proteins localized to promote CD23 cleavage. Our data thus far did not exclude the possibility that they localized to another membrane within the B cell, as opposed to the cell surface. Data from a recent report showed that, in addition to the cell surface plasma membrane, CD23 and ADAM10 were expressed on vesicles derived from B cells that were characterized as exosomes (14). To determine the extent to which β2AR stimulation on a B cell enhanced CD23 and ADAM10 expression on released exosomes, B cells were cultured as described above in the absence or presence of ammonium chloride (NH4Cl)2 to inhibit exosome release, as has been previously described (14,52). In the absence of NH4Cl, the level of both CD23 (Figure 5A) and ADAM10 (Figure 5B) protein expression, as determined by Western blot analysis of cell lysates, was augmented by β2AR stimulation, in comparison to the level expressed by primed B cells alone. In the presence of NH4Cl, β2AR stimulation on primed B cells increased both CD23 and ADAM10 protein expression 2.2- and 3-fold, respectively, in comparison to primed B cells (Figure 5A-B). These findings suggested that the level of expression of both CD23 and ADAM10 proteins is increased in B cells after β2AR stimulation and that the increase is localized to primarily exosomes.
Figure 5.
Inhibition of exosome release increases CD23 and ADAM10 protein retention within a primed B cell. Resting B cells were cultured as described in Figure 1 in the absence or presence of ammonium chloride (NH4Cl, 10 mM) to inhibit the release of exosomes. Cell lysates were collected after 24 hours and analyzed for CD23 (A) and ADAM10 (B) protein expression by Western blot analysis. Representative blots are shown, with Molecular Weight in kDa, and data were normalized to actin and are presented as the mean fold difference of CD23 and ADAM10 expression +/- SEM from 3-4 independent experiments. Significance based on an unpaired t-test. *, p = .05, **, p= .01, compared to priming in the absence of ammonium chloride alone.
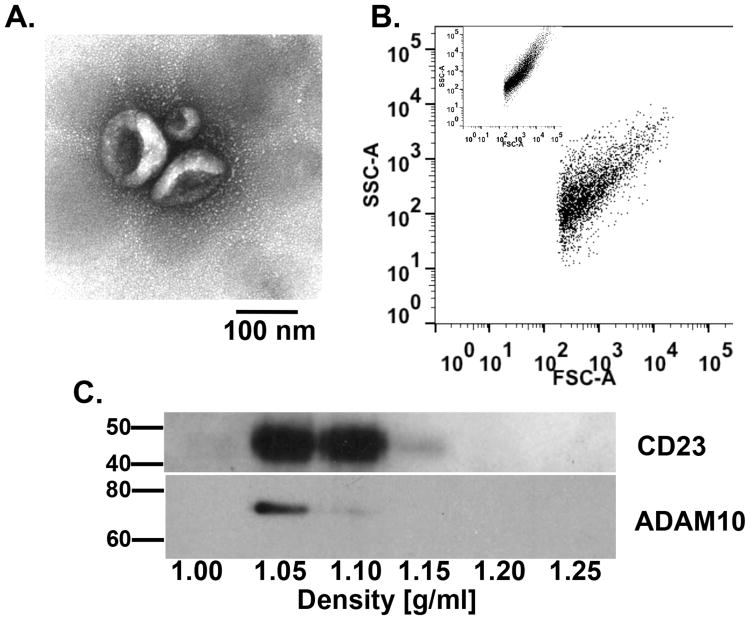
Because several types of vesicles are released from cells (53), it was essential to confirm that the vesicles released by primed B cells in our system satisfied the criteria defined by experts in the field for classifying a microvesicle as an exosome. To this end, B cell-derived vesicles were examined for characteristic features described previously (15), namely, “cup-like” morphology, size between 40-100 nm, and density between 1.08-1.15 g/ml. To evaluate morphology and size, exosomes were isolated from the supernatant of primed B cells cultured for 3 days and fixed and analyzed via TEM. Vesicles isolated on day 3 of priming showed a characteristic cup-like morphology that was less than 100 nm in size (Figure 6A). To confirm size, isolated exosomes were analyzed by flow cytometry. Exosomes measured between 40-100 nm in size (Figure 6B) as compared to ultra-small polystyrene beads to control for accurate size gating (Figure 6B, inset). To measure the density of released exosomes, an iodixanol density gradient centrifugation was used to fractionate supernatant-derived exosomes on day 3, which was the earliest time point at which a large number of exosomes could be isolated. Exosomes expressed both CD23 and ADAM10 in the fractions isolated between 1.05 and 1.10 g/ml (Figure 6C), which is in agreement with previous reports (15). Additionally, the fractions between 1.05 and 1.15 g/ml were the only fractions that contained exosomes as detected by Transmission Electron Microscopy (data not shown). Therefore, these data show that the vesicles released from primed B cells in our model system fulfill the accepted definition required to classify them as exosomes.
Figure 6.
Exosomes are released by primed B cells that express both CD23 and ADAM10. B cells were cultured as described in Figure 1 and B cell-derived exosomes were isolated after 72 hours. (A) Exosomes were fixed and prepared for TEM. Micrograph is representative of three independent experiments. Scale bar represents 100 nm. (B) Exosomes were analyzed by FACS and compared to polystyrene beads of similar size (50-100 nm, inset). Data are presented as Forward Scatter (FSC) versus Side Scatter (SSC) and are representative of three independent experiments. (C) Isolated exosomes were separated by iodixanol density gradient and equal volumes of fractions of 1.00, 1.05, 1.10, 1.15, 1.20, and 1.25 g/ml density were analyzed by Western blot for the expression of CD23 and ADAM10. One representative blot from three independent experiments is shown, with Molecular Weight in kDa.
β2AR stimulation enhances CD23 and ADAM10 expression on exosomes early after priming
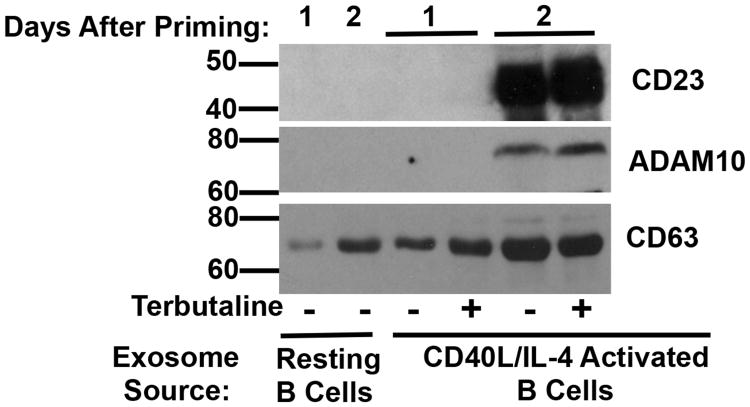
Because inhibition of exosome release with ammonium chloride suggested that the β2AR-induced enhancement in CD23 and ADAM10 expression might be localized to exosomes, we sought to confirm these findings using Western blot analysis of isolated exosomes. To determine if CD23 and ADAM10 were expressed on exosomes released from resting B cells, or if priming was required for CD23 and ADAM10 exosome expression, resting and primed B cells were cultured as described above and exosomes were isolated after 1 through 2 days. Initial analysis of exosome-associated ADAM10 by Western blot with a polyclonal antibody directed at the C-terminal domain of ADAM10 revealed two bands around 65 kDa that corresponded to mature ADAM10 (Supplementary Figure 2, upper panel), which was in contrast to a single band detected by the same antibody around the same molecular weight in cellular lysates (Supplementary Figure 2, lower panel). To determine if this band was non-specific, both cellular lysates and exosomes were examined using monoclonal antibodies directed at either the C-terminal domain or the ectodomain of ADAM10. In exosomes, we found that both monoclonal antibodies detected a single band of ADAM10 (Supplementary Figure 2, upper panel), which corresponded to the lower band detected by the polyclonal ADAM10 antibody, indicating that the upper band detected by the polyclonal antibody was non-specific. A very faint upper band was detected by the ectodomain-specific ADAM10 monoclonal antibody. Thus, all further analyses of ADAM10 expression of exosomes by Western blot were performed with the monoclonal antibody directed at the C terminal domain of ADAM10. Furthermore, in cellular lysates, the C-terminal domain-specific monoclonal ADAM10 antibody detected a higher molecular weight form, around 95 kDa, of ADAM10 (Supplementary Figure 2, lower panel), likely the pro form of ADAM10, a form that was not found in exosomes (Supplementary Figure 2, upper panel), indicating the possibility that only mature ADAM10 is present in exosomes. Exosomes derived from resting B cells did not express CD23 or ADAM10, while exosomes derived 2 days after priming alone expressed CD23 and ADAM10, which was enhanced by β2AR stimulation (Figure 7). As a positive control, a marker for exosomes, CD63, was measured (17), even though CD63 is not a housekeeping gene or loading control, but serves only as an exosome marker. The level of CD63 expression was increased on exosomes derived from B cells exposed to a β2AR agonist at 1 day after priming (Figure 7), but since the function of CD63 is unknown, it was used in this study as purely an exosome marker. The absence of CD23 on released exosomes until 2 days after priming indicates that there is a delay between the synthesis of CD23, which appears in cell lysates and on the cell surface 1 day after priming, and the shuttling of CD23 to exosomes. Furthermore, since ADAM10 is a constitutively expressed protein, the delay in the appearance of ADAM10 in exosomes until 2 days after priming indicates the possibility that CD23 and ADAM10 form an association which allows for shuttling into exosomes, as has been shown previously (14).
Figure 7.
β2AR stimulation enhances CD23 and ADAM10 expression on exosomes at early timepoints after priming. Resting B cells and CD40L/IL-4 primed B cells were cultured as described in Figure 1 and B cell-derived exosomes were isolated from supernatants after 1 through 2 days. Equal amounts of protein were analyzed by Western blot for CD23, ADAM10 and CD63 for each timepoint. One representative blot from three independent experiments is shown, with Molecular Weight in kDa.
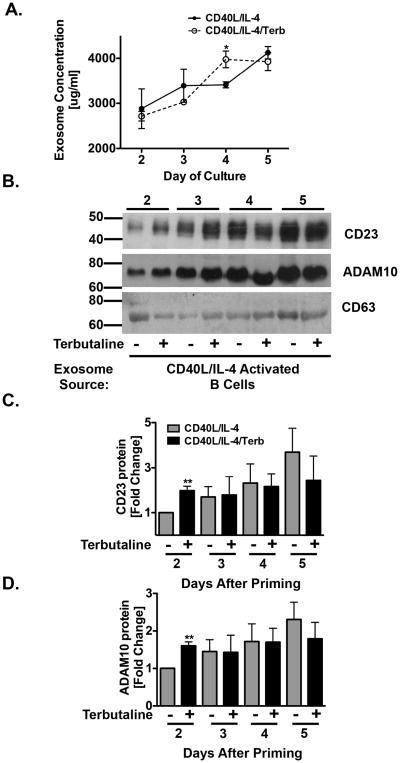
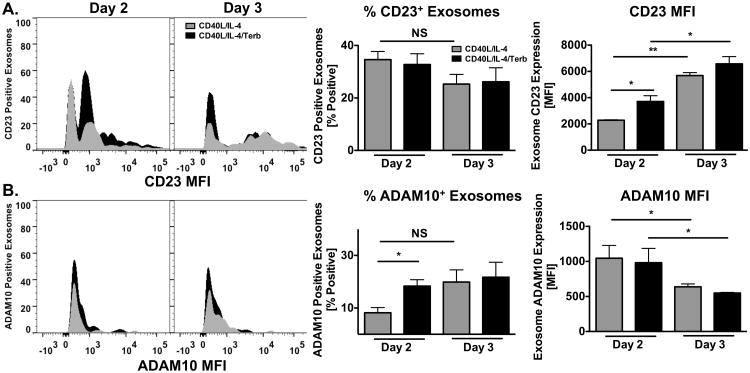
Because the β2AR-enhancement for exosome-associated CD23 and ADAM10 occurred within 2 days of priming, it was important to determine if this enhancement occurred at any later timepoints. Therefore, primed B cells were cultured as described above and exosomes were isolated after 2 through 5 days and analyzed by Western blot. The overall protein level of isolated exosomes was enhanced by β2AR stimulation of primed B cells in comparison to primed alone B cells on day 4, but not on day 2,3, or 5. However, there was an increase over time in the level of protein from isolated exosomes (Figure 8A). The positive control for exosomes, CD63, was increased by β2AR stimulation 3 through 4 days after priming in comparison to exosomes isolated from primed B cells alone. Yet, because the level of protein did not correlate with CD63 expression, we concluded that CD63 is not a proper loading control, although it is routinely used currently in exosome analyses. Until a better loading control is determined, CD63 continues to be used, but the caveat associated with its use is noted. The level of both CD23 and ADAM10 released on exosomes by primed B cells was enhanced by β2AR stimulation 2 days after priming, but remained equal 3 through 5 days after priming (Figure 8B). The positive control for exosomes, CD63, was increased by β2AR stimulation 3 through 4 days after priming. Further analysis by densitometry confirmed that CD23 (Figure 8C) and ADAM10 (Figure 8D) on exosomes released from primed B cells were enhanced by β2AR stimulation 2 days after priming, but not 3 through 5 days after priming, indicating that there is a tight regulation of CD23 and ADAM10 expression on exosomes. Collectively, these results indicate for the first time that CD23 and ADAM10 expression appears on exosomes only after 2 days of B cell priming, and that levels expressed of both proteins increases after β2AR stimulation on primed B cells.
Figure 8.
β2AR stimulation enhances CD23 and ADAM10 expression on exosomes after priming. CD40L/IL-4 primed B cells were cultured as described in Figure 1 and B cell-derived exosomes were isolated from supernatants after 2 through 5 days. Equal amounts of protein were analyzed by Western blot for CD23, ADAM10 and CD63 for each timepoint. (A) BCA analysis of isolated exosomes. Data are presented as mean amount of protein (mg/ml) +/- SEM from 2-3 independent experiments. B-D. B) One representative blot from three independent experiments is shown, with Molecular Weight in kDa, and data are presented as the mean fold difference of CD23 (C) and ADAM10 (D) expression +/- SEM from 3 independent experiments. Significance based on an unpaired t-test. **, p= .01, compared to priming alone.
To extend the Western blot findings to a quantitative level, FACS analysis of individually isolated exosomes was performed to indicate the number of exosomes expressing CD23 and ADAM10, as well as the levels expressed. B cells from WT and β2AR-deficient mice were prepared as described previously and exosomes were isolated on days 2 through 5 after priming. Because the Western blot results in Figure 7 indicated an absence of CD23 and ADAM10 expression on exosomes prior to day 2, FACS analysis was performed starting on day 2. CD23 and ADAM10 were analyzed individually on exosomes because there was a concern that the use of both detecting antibodies at the same time might provide an inaccurate profile due to the small size of exosomes, and because the number of samples available for gating controls was limited. The total number of exosomes isolated from primed B cells alone and those primed B cells exposed to a β2AR agonist increased from 2 through 5 days after priming, as compared to the previous day (Supplemental Figure 3), and was not increased by β2AR stimulation on primed B cells, in comparison to primed-only B cells at any time point. Likewise, the number of CD23+ exosomes remained constant throughout days 1 through 5 (Figure 9A). However, after 2 days of priming, a 1.8-fold increase in the level of CD23, as determined by MFI, was evident on exosomes derived from primed B cells exposed to a β2AR agonist (Figure 9A). This difference in CD23 expression was not evident on days 3 through 5 after priming, and was not evident on exosomes derived from β2AR-deficient B cells at any time point (data not shown). In contrast to CD23, the number of ADAM10+ exosomes was augmented 1.9-fold by β2AR stimulation 2 days after priming, but returned to a control number on days 3 through 5 after priming and was not evident on exosomes derived from β2AR-deficient B cells (Figure 9B). Also, in contrast to CD23, the expression of ADAM10 on exosomes, as determined by MFI, was not augmented by β2AR stimulation after days 2 through 5 of priming, but remained the same as exosomes from primed B cells alone. Together, these data indicate that β2AR stimulation induced changes in exosome phenotype only after 2 days of priming that included an increase in the level of CD23 expression on, and the number of ADAM10+, exosomes.
Figure 9.
β2AR stimulation enhances the level of CD23 expression and the percent of ADAM10+ exosomes at early timepoints. B cells were cultured as described in Figure 1 in the presence (black histogram, bar) or absence (grey histogram, bar) of terbutaline (Terb). Exosomes were isolated between 2 and 3 days and analyzed by FACS for CD23 and ADAM10 expression. Data are presented as one representative histogram, mean Percent, or MFI +/- SEM from 2-3 independent experiments. (A) CD23 expression on exosomes. Percent CD23+ and MFI of CD23+ exosomes. (B) ADAM10 expression on exosomes. Percent ADAM10+ and MFI of ADAM10+ exosomes. Significant differences were determined by unpaired t-test. *, p = .05, **, p= .01.
B cell-derived exosomes directly enhance IgE production
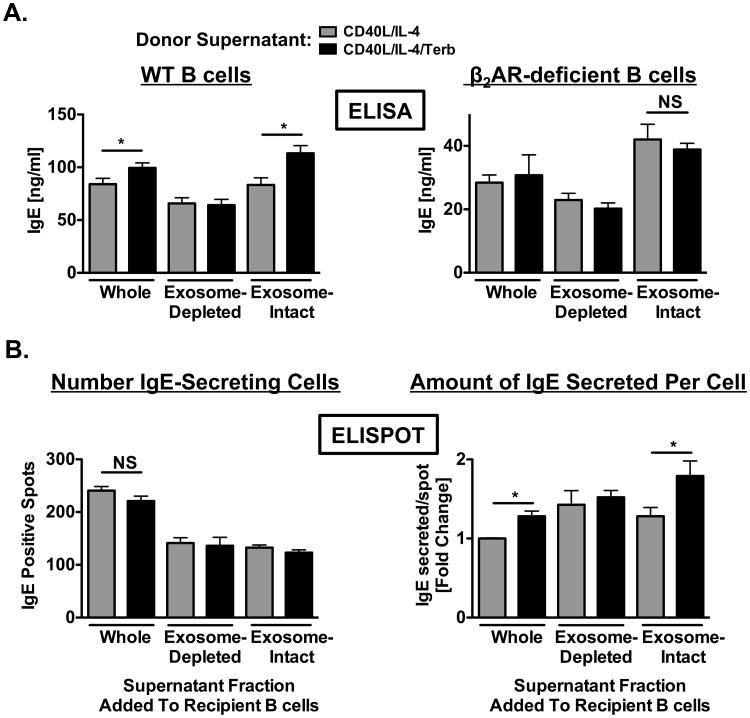
What remained unknown was the extent to which the exosomes derived from β2AR agonist-exposed primed B cells, in comparison to those from primed B cells alone, were able to mediate an increase in IgE. To address this unknown, B cells from WT or β2AR-deficient mice were primed in the absence or presence of the β2AR agonist, terbutaline, for 2 days, at which time either the whole supernatant was removed and transferred directly to another culture of 2-day primed recipient B cells, or the supernatant was first fractionated by ultracentrifugation into exosome-intact and exosome-depleted components that were then transferred to 2-day primed recipient B cells. The recipient B cells were then cultured for another 4 days, at which time supernatants were removed and analyzed for the level of IgE. The transfer of either the whole unfractionated supernatant or the isolated exosome-intact fraction from β2AR agonist-exposed primed B cells, in comparison to those fractions from primed B cells alone, increased the level of IgE produced by recipient cells by 1.2- or 1.3-fold, respectively, while the transfer of the exosome-depleted fraction did not (Fig 10A, left panel). In contrast, the transfer of exosome fractions from β2AR-deficient B cells exposed to a β2AR agonist did not increase IgE (Figure 10A, right panel). Because previous work from our laboratory indicated that β2AR stimulation on a B cell did not affect class switch recombination, but did induce an increase in the amount of IgE secreted per cell, as opposed to an increase in the number of B cells secreting IgE (31), we tested to what extent the transfer of exosomes induced the same effect. The number of IgE-secreting cells, as determined by ELISPOT, was similar in the recipient cultures, regardless of which condition the supernatant fraction was transferred from, although the number was higher when recipient B cells were exposed to whole supernatant fractions from either primed B cells alone or those primed B cells exposed to a β2AR agonist (Figure 10B, left panel). As expected, B cell-derived exosomes with a β2AR-induced increase in CD23 and ADAM10 expression mediated an increase in the level of IgE produced on a per cell basis in the recipient cultures by 1.3- to 1.4-fold (Figure 10B, right panel). These findings suggest that the exosomes released from primed B cells exposed to a β2AR agonist not only express a higher level of CD23 and a greater percentage of ADAM10+ exosomes, as compared to exosomes from primed B cells alone, but also function to directly mediate the β2AR-induced increase in the level of IgE.
Figure 10.
Transfer of exosomes derived from primed B cells exposed to a β2AR agonist directly enhance IgE production by recipient B cells. Donor supernatants: resting B cells were cultured as described in Figure 1 in the presence (black) or absence (grey) of terbutaline. Supernatants containing B cell-derived exosomes were removed after two days and the supernatant was fractionated by ultracentrifugation into exosome-intact and exosome-depleted components. Recipient B cells: resting B cells were cultured as described in Figure 1 in the absence of terbutaline. Whole, exosome-intact, or exosome-depleted supernatants were transferred in the presence of IL-4 (1 ng/ml) to recipient cells that were primed for 2 days in the absence of terbutaline and which had supernatants removed prior to donor supernatant transfers. (A) Supernatant fractions were transferred from donor cultures containing either WT (left panel) or β2AR-deficient (right panel) B cells. IgE produced by recipient cells was measured by ELISA. Data are presented as mean IgE values (ng/ml) +/- SEM from one representative independent experiment of three from at least three replicate measurements. (B) Comparison of the number of recipient IgE-secreting cells (left panel) and the amount of IgE secreted by these cells (right panel) using the ELISPOT assay. Data are presented as the number of IgE-positive spots/well or as the fold change compared to whole supernatant transfer in IgE secreted on a per spot basis +/- SEM from one representative independent experiment of three from at least three replicate measurements. Significance is based on an unpaired t-test. *, p = .05, as compared to B cells exposed to fractions obtained from primed B cells alone.
A number of parameters were tested to establish the effectiveness and specificity of the exosome-mediated effect. First, we found that recipient B cells produced a very low level of IgE unless IL-4 was added to the recipient culture along with the transferred fractions (data not shown). We also observed that the exosome-depleted supernatant was capable of enhancing the level of IgE produced in recipient B cells alone, in comparison to the transfer of medium alone with IL-4 (data not shown), indicating that some soluble factor present in the exosome-depleted supernatant obtained from the primed B cells alone was able to transfer a slight enhancement themselves to the recipient B cells above baseline. Also, a further 1.3-fold enhancement of IgE was observed in the level of IgE produced by recipient B cells with the transfer of either exosome-intact or whole supernatant fractions that was above that produced by exosome-depleted supernatant fractions (Figure 10A) and, significantly, the same fractions from primed B cells exposed to β2AR stimulation were further able to enhance IgE production (Figure 10A), indicating that the β2AR-enhancing effect on IgE could be transferred via exosomes. Second, to neutralize any effect that might have been transferred due to any residual β2AR agonist, the β2AR antagonist nadolol was added to recipient cell cultures along with the transferred supernatant fractions. Under this condition, the β2AR-enhancing effect on the level of IgE produced by the recipient cells remained when the donor supernatants were exosome-intact and whole supernatants (Supplemental Figure 4A), indicating that any residual terbutaline did not alter the β2AR-enhanced exosome effect on IgE. Third, to determine the extent to which the timing of exosome transfer affected the transfer of the enhancing effect, exosome-intact supernatant fractions from day 3 donor cultures were transferred onto day 3 primed-only recipient B cells. The level of IgE produced by recipient B cells was 4-fold less, in comparison to day 2 transfers, and the primed β2AR-exposed donor fractions were unable to enhance the level of IgE produced, in comparison to fractions from primed alone B cells (Supplemental Figure 4B). This finding suggested that exosomes released by day 2 from a primed B cell are able to function maximally. Finally, to determine the extent to which the concentration of exosomes transferred affected the level of the enhancing effect, day 2 exosomes were resuspended to a 1X or 2X concentration prior to transfer to recipient B cells. The 1X and 2X concentrations of exosomes derived from B cells exposed to a β2AR agonist enhanced IgE production, in comparison to those derived from primed B cells alone (Supplemental Figure 4C), indicating that the β2AR-enhancing effect was unaffected by exosome concentration. Taken together, these findings established the conditions under which the exosome transfer conditions were optimal for them to mediate the β2AR agonist-induced enhancing effect on IgE, demonstrating that the β2AR-induced increase in CD23 and ADAM10 on B cell-derived exosomes is functionally relevant.
ADAM10 and CD23 organize into membrane microdomains
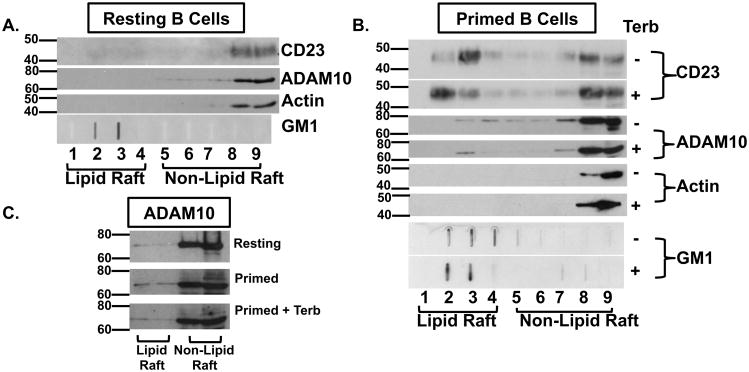
The question remained as to the membrane region into which the increased level of β2AR-induced CD23 and ADAM10 was compartmentalized, which would influence where CD23 cleavage occurred. Recent reports showed that ADAM10-mediated cleavage of various substrates within a membrane was enhanced upon cholesterol depletion, which resulted in a disruption of lipid rafts (23-26), suggesting that ADAM10 activity was greater outside of a cholesterol-rich lipid raft region of a membrane. To determine if membrane localization within B cell membranes was affected by β2AR stimulation, B cells were primed and cultured as described previously and cell lysates were fractionated on days 0 through 5 after priming via sucrose density gradient centrifugation. Equal volumes of each fraction were analyzed by Western blot for the presence of CD23, ADAM10, actin, which is a protein found in non-lipid raft fractions (54), and GM1, which is a sphingolipid enriched in membrane lipid raft fractions (55). In resting cells, CD23 and ADAM10 were primarily located in non-lipid raft regions (Figure 11A) and, after priming, there was a change in distribution of CD23 and ADAM10 to both fractions. Two days after priming and β2AR stimulation, an increased localization of CD23, but not ADAM10, to non-lipid raft fractions was observed, as compared to primed B cells alone (Figure 11B), and persisted throughout day 3 (data not shown). Because the resting level of ADAM10 differed in non-lipid raft fractions derived from resting as compared to primed B cells, equal volumes from resting fractions that were three times the volume used for activated fractions and corresponded to equal protein in the non-lipid raft fractions were compared from the lipid and non-lipid raft fractions, fractions 3-4 and 8-9, respectively, in order to determine if ADAM10 was present in lipid raft fractions of resting cells. Low amounts of ADAM10 were detected in lipid raft fractions from resting B cells, and ADAM10 expression in these fractions increased marginally upon priming of the B cell (Figure 11C). The relatively small amount of change in localization of ADAM10 versus CD23 may be explained by the effect that priming has on the expression of CD23 in comparison to ADAM10. Whereas, ADAM10 is constitutively expressed and it's surface expression is relatively unaffected by B cell priming, priming of the B cell substantially enhances the surface expression of CD23 in comparison to resting cells (Figure 4). Thus, whereas the resting lipid raft fractions contained very little CD23, priming of the B cell enhanced CD23 expression to the extent that a large amount of CD23 was detected in both fractions. In comparison, the absence of an upregulation of ADAM10 expression in response to priming alone translated to a very subtle increase in ADAM10 expression in the lipid raft fractions. These findings suggested that B cell membranes, be it on the cell surface or released exosomes, express CD23 and ADAM10 in regions where cleavage is optimal, and that this pattern of localization is the same in primed B cells alone and after β2AR stimulation.
Figure 11.
CD23 and ADAM10 localization to non-lipid raft domains within primed B cell membranes increases after β2AR stimulation. Resting B cells (A) or CD40L/IL-4-primed B cells (B) were cultured as described in Figure 1 in the presence (+) or absence (-) of terbutaline (Terb). Whole cell lysate fractions were isolated from a sucrose density gradient after 2 days. Fractions 1-4 represent less dense lipid raft fractions and Fractions 5-9 represent more dense non-lipid raft fractions. Equal volumes of each cell lysate fraction was analyzed by Western blot for the presence of CD23, ADAM10, and Actin. Equal volumes were also loaded via slot blot and analyzed for the presence of Ganglioside 1 (GM1), a sphingolipid that is enriched in lipid raft fractions. (C) Equal volumes corresponding to equal amounts of protein in the non-lipid raft fractions were loaded from Fractions 3-4 (Lipid Raft) and Fractions 8-9 (Non-lipid Raft) and analyzed for ADAM10. One representative blot from three independent experiments is shown, with Molecular Weight in kDa.
Discussion
A critical role for CD23 and sCD23 in regulating the level of IgE produced by a B cell is well known. However, the role of the β2AR expressed on a B cell in regulating the level of CD23, sCD23, and IgE is less well studied, even though this form of regulation may be relevant both physiologically, when NE is released in the vicinity of B cells after antigen exposure, and clinically, when a β2AR agonist is used therapeutically to treat bronchoconstriction. The goal of the present study was to identify the mechanism that links β2AR engagement on a CD40L/IL-4-primed B cell to an increase in the level of CD23, sCD23 and IgE. We report in the present study that three mechanisms appear to be involved. The first mechanism involves a β2AR-induced increase in both CD23 and ADAM10 expression at the mRNA and protein level. The second mechanism involves localization of the β2AR-induced increase in CD23 and ADAM10 to B cell-derived exosomes and their release from the cell. And finally, the third mechanism involves the ability of the released exosomes to directly mediate the β2AR-induced increase in IgE.
One of the more unexpected findings in the present study was that the β2AR-induced increase in CD23 and ADAM10 expression in a B cell did not translate to an increase in expression on the plasma membrane, which is where the cleavage of CD23 was originally proposed to occur (12), but instead, preferentially increased on B cell-derived exosomes. The presence of CD23 and ADAM10 on exosomes confirmed recent reports using Western blot analysis showing that B cell priming with anti-CD40 and IL-4 alone induced the expression of CD23 on B cell-derived exosomes (15), and that B cell priming with LPS and IL-4 induced both CD23 and ADAM10 (14). The enhancement of CD23 and ADAM10 expression on B cell-derived exosomes, as opposed to the cell surface, following β2AR engagement on a primed B cell, suggested three possible means by which the β2AR-induced increase in CD23 and ADAM10 protein expression might occur. First, an increase in cell surface expression failed to occur because CD23 was cleaved at the cell surface and released immediately by ADAM10 to produce sCD23, as was proposed previously (12) and, thus, a transient increase at the cell surface might occur, but would be undetectable using FACS analysis. Second, any increase of CD23 and ADAM10 proteins at the B cell surface might preferentially endocytose together, as was proposed previously (14), to then fuse with intracellular multivesicular bodies (MVB)2(19,56) that would subsequently fuse with the plasma membrane for release from the cell (19,56,57). In support of this possibility is the report that the cell surface expression of ADAM10 on the surface of Human Embryonic Kidney 293 (HEK293) cells is regulated by endocytosis, as evidenced when using an inhibitor to the endocytic protein dynamin (58), suggesting that any ADAM10 expression at the plasma membrane was cleared from the surface via endocytosis. Support for the co-endocytosis of CD23 with ADAM10 is provided by a previous report that the stalk region of CD23 binds to ADAM10 in a protease-independent manner, and that ADAM10 is essential for the inclusion of CD23 on exosomes (14), indicating that a unique interaction occurs between CD23 and ADAM10 that directs their preferential shuttling into exosomes. Furthermore, the present findings showed that all ADAM10+ B cells were also CD23+, but not vice versa, suggesting that CD23 may be required for cell surface expression of ADAM10 on B cells. Finally, the β2AR-induced increase in newly synthesized CD23 and ADAM10 proteins may shuttle directly from the Golgi into MVB's for fusion with the plasma membrane and subsequent release from the cell, as shown for ADAM10 in breast tumor cell lines (51,57). Support for the immediate shuttling of these proteins to exosomes was that specific inhibition of exosome release by ammonium chloride, as defined previously (14,52), caused retention of exosomes intracellulary, which was measured as an increase in the CD23 and ADAM10 content of cellular lysates. This finding indicated that the proteins were preferentially localized to exosomes that were unable to leave the cell in the presence of ammonium chloride. Thus, the means by which CD23 and ADAM10 localize to exosomes are not mutually exclusive, but the present results suggest that the β2AR-induced increase in CD23 and ADAM10 is preferentially shuttled to exosomes for release from the cell and subsequent cleavage of CD23, instead of shuttled to the cell surface plasma membrane.
We show that exosomes released by donor cells exposed to a β2AR agonist expressed a higher level of both CD23 and ADAM10, suggesting that the exosomes provided a source for a higher level of sCD23 to positively regulate the IgE response of a recipient primed B cell that had already undergone class switch recombination. We propose that the transfer of the β2AR-enhancing effect on IgE was due to the exosomes released from the donor cells themselves, and not due to something inherent to the recipient B cells, for the following reasons. First, the recipient B cells received equal numbers of exosomes from both the agonist-exposed and –unexposed donor cells, ruling out any effect due to a difference in exosome number, and suggesting that the effect was mediated by the higher levels of CD23 and ADAM10 expressed on the transferred exosomes. Second, the transfer of any residual β2AR agonist, which might have affected recipient cells, was unlikely because the addition of a β2AR antagonist along with the donor exosomes was unable to prevent the enhancing effect. Also, the half-life of terbutaline in culture is 7 hours (59), and exerts an enhancing effect only within the first 24 hours of priming, as reported previously (60), suggesting that any effect on a B cell 48 hours after priming was unlikely. And, finally, because the transfer of the enhancing effect occurred only with exosomes collected on the peak day of the β2AR-induced increase in CD23 and ADAM10 expression (day 2), we propose that day 2 might also be the peak time of sCD23 production to exert a positive regulation of the IgE response. To confirm this proposal, future experiments will be done to confirm the level of sCD23 produced by the transferred exosomes, although such measurements may be difficult to perform due to the extensive ultracentrifugation steps required to isolate exosomes, which may result in a loss of sCD23 during processing of samples, as well as the difficulty in differentiating between sCD23 and the CD23 expressed on exosomes by ELISA.
Previous work focused on regulation of ADAM10 activity by membrane localization to lipid versus non-lipid raft fractions (23-26). It has been reported that lipid rafts inhibit the ability of ADAM10 to cleave substrates, such as amyloid precursor protein, IL-6 receptor, and CD30, and that the disruption of lipid rafts permits ADAM10 to exert sheddase activity on these substrates (23-26). In the present study, CD40L/IL-4 priming of the B cell promoted the localization of CD23 and ADAM10 from exclusively non-lipid raft regions in a resting cell, to both non-lipid and lipid raft regions. It is possible that the presence of CD23 and ADAM10 in non-lipid raft regions and the apparent β2AR-induced increase in CD23 localization to non-lipid raft regions indicates that non-lipid raft fractions are the site of CD23 cleavage to produce enhanced sCD23. This finding also suggests that redistribution to lipid raft regions is a means by which the cell controls the extent of ADAM10-mediated cleavage. Thus, the localization within non-lipid raft regions assures that cleavage of CD23 will occur, while the localization to lipid raft regions assures that ADAM10 activity will be controlled. The presence of lipid rafts within exosomes has previously been noted [reviewed by (28)], and exosomes are derived from the lipid-rich lipid raft regions of cellular membranes (61) where the interaction of CD23 and ADAM10 within this region allows for exosome formation within MVB's that are released from the cell (14). The fact that both exosomes and lipid rafts have similar densities when isolated by sucrose and iodixanol density centrifugation (15,62,63), suggests that the lipid raft measurements made in the present study detect both exosome and plasma membrane lipid raft-associated CD23 and ADAM10. Because we were able to isolate lipid raft and non-lipid raft fractions expressing CD23 and ADAM10 from specifically the exosome fraction of an iodixanol density gradient (data not shown), the possibility exists that ADAM10-mediated cleavage of CD23 also occurs on lipid-rich exosomes that is also regulated by localization to non-lipid raft domains. Future studies will focus on the role that lipid raft and non-lipid raft domains play in the β2AR-induced increase in ADAM10-mediated cleavage of CD23, and will also identify the extent to which preferential localization occurs on an exosome, as opposed to a plasma membrane.
Whereas the signaling intermediates that were responsible for mediating the β2AR-induced increase in CD23 and ADAM10 are mostly unknown, a number of reports that address CD23 and ADAM10 regulation have provided some clues. The upregulation of CD23 expression on a B cell occurs in both human and mouse B cells following exposure to either IL-4 (64,65), anti-CD40 antibody (66), or a Th2 clone in the presence of antigen (67). We reported previously that β2AR stimulation on a CD40L/IL-4-primed B cell enhanced CD23 expression beyond the level induced by CD40L/IL-4 alone (31), and in the present report, we confirm that this increase was mediated by the β2AR specifically and in a PKA- and p38 MAPK-dependent manner. The signaling intermediates responsible for the β2AR-induced enhancement in CD23 may also be linked to the known role of STAT6 in regulating the IL-4–induced increase in CD23 (68) via p38 MAPK regulation of the transactivation domain of STAT6 (69). Because the pathway by which β2AR engagement enhances IgE is via a β2AR-induced, PKA-dependent phosphorylation of HePTP (32), which releases bound p38 MAPK for phosphorylation by the MAPKK pathway activated following CD40 engagement (34), we propose that the β2AR-induced increase in CD23 expression is likely due to the p38 MAPK-dependent enhancement of IL-4-mediated STAT6 activation via interaction with the transactivation domain, which ultimately leads to an increase in the level of CD23 expression.
In contrast to CD23 regulation, signaling intermediates that upregulate ADAM10 expression are largely unknown. B cells express ADAM10 constitutively (70,71), as was confirmed in the present study, and which is in contrast to CD23 expression that is negligible in resting B cells (65). In the present report, β2AR engagement on a primed B cell induced an increase in ADAM10 expression at both the gene and protein level, which was associated with an increase in CD23 cleavage, and which was blocked with an ADAM10 inhibitor. The reported signaling intermediates that regulate ADAM10 expression appear to function through the activation of p38 MAPK(72-74). Promoter analysis of the ADAM10 gene reveals binding sites for the Retinoic Acid Receptor alpha and NF-κB (72), supporting a previous finding that ADAM10 protein was upregulated by activation of the retinoic acid pathway in a neuroblastoma cell line (73). Additionally, ADAM10 gene and protein expression were also found to be upregulated by activation of the p38 MAPK/ERK1/2 pathway in primary cortical cultures (74), while other reports showed that p38 MAPK was able to phosphorylate both Retinoic Acid Receptor alpha and NF-κB when using Mouse Embryonic Fibroblasts and NIH 3T3 cells (75,76). In the present report, we show that the β2AR-enhanced expression of ADAM10 is also mediated by p38 MAPK, and is likely mediated by the aforementioned β2AR-activated PKA/HePTP/p38 MAPK signaling pathway described for CD23. Taken together, both CD23 and ADAM10 expression can be regulated by p38 MAPK, and the β2AR on a B cell can further contribute to this regulation through the same signaling intermediate, indicating that a common mechanism exists by which β2AR engagement increases two proteins important for IgE regulation, which is also linked to increased p38 MAPK activation.
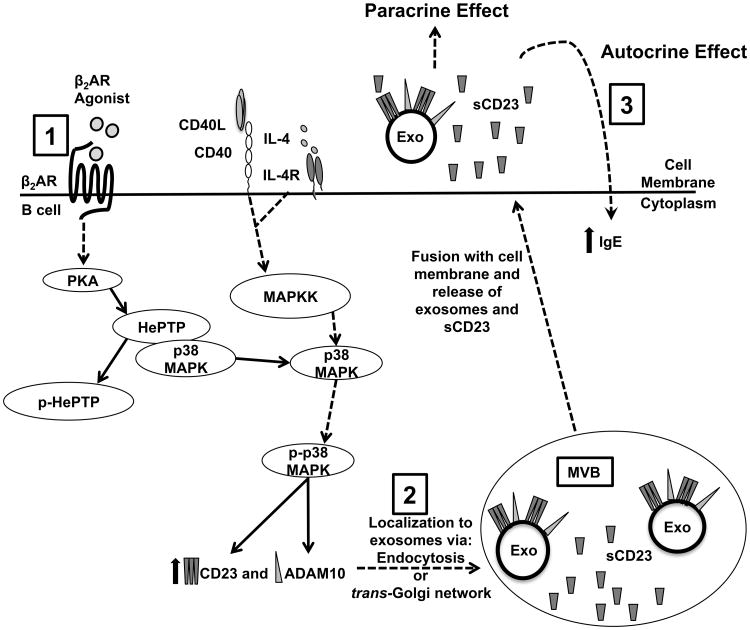
Although we elucidated the kinetics of the β2AR-induced enhancement of CD23 and ADAM10 expression on exosomes, our ultimate goal was to identify the mechanism by which β2AR engagement on a primed B cell enhanced the level of IgE produced. The present finding suggested that exosomes might play an essential role in mediating this effect on IgE, and potentially, play a role in mediating a physiological effect that influences conditions such as allergy and asthma. However, to our knowledge, few studies have addressed the physiological relevance of exosomes derived from mice undergoing an allergic sensitization protocol to promote allergic asthma. Two recent studies measured the tolerizing potential of exosomes derived from the serum and BALF of mice undergoing a tolerizing protocol. Exosomes, also called tolerosomes in these studies, were used prophylactically to prevent the IgE response in a murine allergic asthma model (20,77), suggesting that exosomes from tolerized mice were able to induce tolerance in response to sensitization to the same antigen. The mechanism of the transfer of an anti-inflammatory phenotype in vivo, is suggested by previous findings that B cell-derived exosomes transferred antigen-MHC II complexes to follicular dendritic cells (78), and that CD23+ B cells were required for optimal IgE-antigen presentation to CD11c+ dendritic cells to allow for CD4+ T cell activation (79). Whereas the transfer of a sensitizing phenotype via exosomes offers many intriguing avenues of research, prevention of a potential β2AR-induced increase in IgE in the treatment of allergic asthma would also be helpful. The present findings suggest a mechanism that can be targeted to prevent a β2AR-induced increase in the production of IgE. Early intervention after β2AR agonist inhalation using either ADAM10 inhibitors and/or molecules that would prevent the biogenesis of exosomes might prevent a β2AR-induced increase in IgE production. Additionally, in patients, the expression of CD23 or ADAM10 on exosomes in the blood might be used as a biomarker to predict outcomes for beta agonist therapy in patients, with high levels of these proteins indicating that higher IgE might be produced to exacerbate the clinical condition. In conclusion, and as depicted in Figure 12, the present study demonstrated that β2AR stimulation on a primed B cell, in comparison to primed B cells alone, mediates an increase in the level of IgE, first by enhancing CD23 and ADAM10 gene and protein expression via activation of the PKA/HePTP/p38 MAPK signaling pathway. The β2AR-mediated enhancement in CD23 and ADAM10 protein is found on B cell-derived exosomes that are released by the B cell, and which, upon transfer to recipient primed B cells, mediate the β2AR-enhancing effect on the level of IgE produced per cell. Targeting of any of the mechanisms that induce the increased expression on exosomes or the formation of exosomes may provide a therapeutic approach that can be used to prevent the β2AR-mediated enhancing effect on IgE, without affecting the β2AR-mediated dilating effect on bronchiolar smooth muscle.
Figure 12.
Adrenergic Regulation of IgE Involves Modulation of CD23 and ADAM10 Expression on Exosomes. (1) Wild type B cells primed in the presence of a β2AR agonist enhance CD23 and ADAM10 production through a PKA-dependent phosphorylation of HePTP, which subsequently releases bound p38 MAPK for phosphorylation by the CD40-triggered MAPKK pathway. (2) β2AR stimulation augments both CD23 and ADAM10 expression on exosomes, but not on the cell surface, either through an endocytic pathway starting at the cell surface or through fusion of vesicles containing CD23 and ADAM10 that are derived from the trans-Golgi network with MVB's. (3) Exosomes and sCD23 generated in MVB are released from the cell upon fusion of the MVB with the plasma membrane. The release of sCD23 from exosomes enhances IgE production by acting in either an autocrine or paracrine manner, as suggested previously (14,78,79). The autocrine mechanism may involve a sCD23-mediated enhancement of IgE production by the cell from which the sCD23 was released. The paracrine mechanism may involve the exosomes acting at distal sites where the CD23 on the exosomes will complex with antigen bound to IgE. This complex will be phagocytosed by dendritic cells so that they can present antigen to T cells to activate them to help B cells that have endocytosed the exosome complexes for processing and presentation to the dendritic cell-activated T cell. Taken together, the β2AR-enhanced expression of CD23 and ADAM10 on B cell-derived exosomes may explain the mechanism responsible for mediating the effect of β2AR stimulation on a B cell to increase the level of IgE produced.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical assistance from the Campus Microscopy Imaging Facilities at OSU. We thank Dr. Caroline Whitacre for the use of her flow cytometer and ELISPOT reader. We also thank Dr. Chris Lucas for critical review of the manuscript.
Footnotes
This work was supported by research grants from the National Institute of Allergy and Infectious Disease, R01-AI37326 (to V.M.S), and F31-AI089031 (to C.J.P.).
Abbreviations used in this paper were: sCD23, soluble CD23; NE, Norepinephrine; BALF, Bronchoalveolar Lavage Fluid; ADAM10, A Disintegrin And Metalloproteinase 10; β2AR, Beta-2 Adrenergic Receptor; HePTP, Hematopoietic Protein Tyrosine Phosphatase; PKA, Protein Kinase A; 7-AAD, 7-Amino-Actinomycin D; GM1, Ganglioside M1; NH4Cl, Ammonium Chloride; MVB, Multivesicular Body
References
- 1.Oettgen HC, Geha RS. IgE regulation and roles in asthma pathogenesis. J Allergy Clin Immunol. 2001;107:429–440. doi: 10.1067/mai.2001.113759. [DOI] [PubMed] [Google Scholar]
- 2.Sherr E, Macy E, Kimata H, Gilly M, Saxon A. Binding the low affinity Fc epsilon R on B cells suppresses ongoing human IgE synthesis. J Immunol. 1989;142:481–489. [PubMed] [Google Scholar]
- 3.Nakamura T, Kloetzer WS, Brams P, Hariharan K, Chamat S, Cao X, LaBarre MJ, Chinn PC, Morena RA, Shestowsky WS, Li YP, Chen A, Reff ME. In vitro IgE inhibition in B cells by anti-CD23 monoclonal antibodies is functionally dependent on the immunoglobulin Fc domain. Int J Immunopharmacol. 2000;22:131–141. doi: 10.1016/s0192-0561(99)00068-5. [DOI] [PubMed] [Google Scholar]
- 4.Yabuuchi S, Nakamura T, Kloetzer WS, Reff ME. Anti-CD23 monoclonal antibody inhibits germline Cepsilon transcription in B cells. Int Immunopharmacol. 2002;2:453–461. doi: 10.1016/s1567-5769(01)00187-4. [DOI] [PubMed] [Google Scholar]
- 5.Yu P, Kosco-Vilbois M, Richards M, Kohler G, Lamers MC. Negative feedback regulation of IgE synthesis by murine CD23. Nature. 1994;369:753–756. doi: 10.1038/369753a0. [DOI] [PubMed] [Google Scholar]
- 6.Aubry JP, Pochon S, Graber P, Jansen KU, Bonnefoy JY. CD21 is a ligand for CD23 and regulates IgE production. Nature. 1992;358:505–510. doi: 10.1038/358505a0. [DOI] [PubMed] [Google Scholar]
- 7.Hibbert RG, Teriete P, Grundy GJ, Beavil RL, Reljic R, Holers VM, Hannan JP, Sutton BJ, Gould HJ, McDonnell JM. The structure of human CD23 and its interactions with IgE and CD21. J Exp Med. 2005;202:751–760. doi: 10.1084/jem.20050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnefoy JY, Gauchat JF, Life P, Graber P, Aubry JP, Lecoanet-Henchoz S. Regulation of IgE synthesis by CD23/CD21 interaction. Int Arch Allergy Immunol. 1995;107:40–42. doi: 10.1159/000236924. [DOI] [PubMed] [Google Scholar]
- 9.McCloskey N, Hunt J, Beavil RL, Jutton MR, Grundy GJ, Girardi E, Fabiane SM, Fear DJ, Conrad DH, Sutton BJ, Gould HJ. Soluble CD23 monomers inhibit and oligomers stimulate IGE synthesis in human B cells. J Biol Chem. 2007;282:24083–24091. doi: 10.1074/jbc.M703195200. [DOI] [PubMed] [Google Scholar]
- 10.Cooper AM, Hobson PS, Jutton MR, Kao MW, Drung B, Schmidt B, Fear DJ, Beavil AJ, McDonnell JM, Sutton BJ, Gould HJ. Soluble CD23 controls IgE synthesis and homeostasis in human B cells. J Immunol. 2012;188:3199–3207. doi: 10.4049/jimmunol.1102689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilmon MA, Shelburne AE, Chan-Li Y, Holmes KL, Conrad DH. CD23 trimers are preassociated on the cell surface even in the absence of its ligand, IgE. J Immunol. 2004;172:1065–1073. doi: 10.4049/jimmunol.172.2.1065. [DOI] [PubMed] [Google Scholar]
- 12.Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, Broadway N, Hartmann D, Saftig P, Umland S, Sehara-Fujisawa A, Black RA, Ludwig A, Becherer JD, Conrad DH, Blobel CP. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006;7:1293–1298. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]
- 13.Acharya M, Borland G, Edkins AL, Maclellan LM, Matheson J, Ozanne BW, Cushley W. CD23/FcepsilonRII: molecular multi-tasking. Clin Exp Immunol. 2010;162:12–23. doi: 10.1111/j.1365-2249.2010.04210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews JA, Gibb DR, Chen BH, Scherle P, Conrad DH. CD23 Sheddase A disintegrin and metalloproteinase 10 (ADAM10) is also required for CD23 sorting into B cell-derived exosomes. J Biol Chem. 2010;285:37531–37541. doi: 10.1074/jbc.M110.141556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunderson SC, Schuberth PC, Dunn AC, Miller L, Hock BD, MacKay PA, Koch N, Jack RW, McLellan AD. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J Immunol. 2008;180:8146–8152. doi: 10.4049/jimmunol.180.12.8146. [DOI] [PubMed] [Google Scholar]
- 16.Arita S, Baba E, Shibata Y, Niiro H, Shimoda S, Isobe T, Kusaba H, Nakano S, Harada M. B cell activation regulates exosomal HLA production. Eur J Immunol. 2008;38:1423–1434. doi: 10.1002/eji.200737694. [DOI] [PubMed] [Google Scholar]
- 17.Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81:1171–1182. doi: 10.1016/j.bcp.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Admyre C, Bohle B, Johansson SM, Focke-Tejkl M, Valenta R, Scheynius A, Gabrielsson S. B cell-derived exosomes can present allergen peptides and activate allergen-specific T cells to proliferate and produce TH2-like cytokines. J Allergy Clin Immunol. 2007;120:1418–1424. doi: 10.1016/j.jaci.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 20.Prado N, Marazuela EG, Segura E, Fernandez-Garcia H, Villalba M, Thery C, Rodriguez R, Batanero E. Exosomes from bronchoalveolar fluid of tolerized mice prevent allergic reaction. J Immunol. 2008;181:1519–1525. doi: 10.4049/jimmunol.181.2.1519. [DOI] [PubMed] [Google Scholar]
- 21.Lemieux GA, Blumenkron F, Yeung N, Zhou P, Williams J, Grammer AC, Petrovich R, Lipsky PE, Moss ML, Werb Z. The low affinity IgE receptor (CD23) is cleaved by the metalloproteinase ADAM10. J Biol Chem. 2007;282:14836–14844. doi: 10.1074/jbc.M608414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, Conrad DH. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010;207:623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews V, Schuster B, Schutze S, Bussmeyer I, Ludwig A, Hundhausen C, Sadowski T, Saftig P, Hartmann D, Kallen KJ, Rose-John S. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE) J Biol Chem. 2003;278:38829–38839. doi: 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 24.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojro E, Gimpl G, Lammich S, Marz W, Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc Natl Acad Sci U S A. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Tresckow B, Kallen KJ, von Strandmann EP, Borchmann P, Lange H, Engert A, Hansen HP. Depletion of cellular cholesterol and lipid rafts increases shedding of CD30. J Immunol. 2004;172:4324–4331. doi: 10.4049/jimmunol.172.7.4324. [DOI] [PubMed] [Google Scholar]
- 27.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nance DM, Sanders VM. Autonomic innervation and regulation of the immune system (1987-2007) Brain Behav Immun. 2007;21:736–745. doi: 10.1016/j.bbi.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergquist J, Tarkowski A, Ekman R, Ewing A. Discovery of endogenous catecholamines in lymphocytes and evidence for catecholamine regulation of lymphocyte function via an autocrine loop. Proc Natl Acad Sci U S A. 1994;91:12912–12916. doi: 10.1073/pnas.91.26.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pongratz G, McAlees JW, Conrad DH, Erbe RS, Haas KM, Sanders VM. The Level of IgE Produced by a B Cell Is Regulated by Norepinephrine in a p38 MAPK- and CD23-Dependent Manner. J Immunol. 2006;177:2926–2938. doi: 10.4049/jimmunol.177.5.2926. [DOI] [PubMed] [Google Scholar]
- 32.McAlees JW, Sanders VM. Hematopoietic protein tyrosine phosphatase mediates beta2-adrenergic receptor-induced regulation of p38 mitogen-activated protein kinase in Blymphocytes. Mol Cell Biol. 2009;29:675–686. doi: 10.1128/MCB.01466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena M, Williams S, Tasken K, Mustelin T. Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nature cell biology. 1999;1:305–311. doi: 10.1038/13024. [DOI] [PubMed] [Google Scholar]
- 34.Nika K, Hyunh H, Williams S, Paul S, Bottini N, Tasken K, Lombroso PJ, Mustelin T. Haematopoietic protein tyrosine phosphatase (HePTP) phosphorylation by cAMP-dependent protein kinase in T-cells: dynamics and subcellular location. Biochem J. 2004;378:335–342. doi: 10.1042/BJ20031244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker JK, Penn RB, Hanania NA, Dickey BF, Bond RA. New perspectives regarding beta(2) -adrenoceptor ligands in the treatment of asthma. Br J Pharmacol. 2011;163:18–28. doi: 10.1111/j.1476-5381.2010.01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med. 1989;320:271–280. doi: 10.1056/NEJM198902023200502. [DOI] [PubMed] [Google Scholar]
- 37.Borish L, Chipps B, Deniz Y, Gujrathi S, Zheng B, Dolan CM. Total serum IgE levels in a large cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2005;95:247–253. doi: 10.1016/S1081-1206(10)61221-5. [DOI] [PubMed] [Google Scholar]
- 38.Hanania NA. Targeting airway inflammation in asthma: current and future therapies. Chest. 2008;133:989–998. doi: 10.1378/chest.07-0829. [DOI] [PubMed] [Google Scholar]
- 39.Soresi S, Togias A. Mechanisms of action of anti-immunoglobulin E therapy. Allergy Asthma Proc. 2006;27:S15–23. [PubMed] [Google Scholar]
- 40.Davis C, Conolly ME, Greenacre JK. Beta-adrenoceptors in human lung, bronchus and lymphocytes. Br J Clin Pharmacol. 1980;10:425–432. doi: 10.1111/j.1365-2125.1980.tb01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conolly ME, Greenacre JK. The beta-adrenoceptor of the human lymphocyte and human lung parenchyma. Br J Pharmacol. 1977;59:17–23. doi: 10.1111/j.1476-5381.1977.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren WD, Berton MT. Induction of germ-line gamma 1 and epsilon Ig gene expression in murine B cells. IL-4 and the CD40 ligand-CD40 interaction provide distinct but synergistic signals. J Immunol. 1995;155:5637–5646. [PubMed] [Google Scholar]
- 43.Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, Yang G, Li Y, Yao W, Vaddi K, Gazdar AF, Friedman SM, Jablons DM, Newton RC, Fridman JS, Minna JD, Scherle PA. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucas CR, Cordero-Nieves HM, Erbe RS, McAlees JW, Bhatia S, Hodes RJ, Campbell KS, Sanders VM. Prohibitins and the cytoplasmic domain of CD86 cooperate to mediate CD86 signaling in Blymphocytes. J Immunol. 2013;190:723–736. doi: 10.4049/jimmunol.1201646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasprowicz DJ, Kohm AP, Berton MT, Chruscinski AJ, Sharpe AH, Sanders VM. Stimulation of the B cell receptor, CD86 (B7-2), and the beta 2-adrenergic receptor intrinsically modulates the level of IgG1 and IgE produced per B cell. JImmunol. 2000;165:680–690. doi: 10.4049/jimmunol.165.2.680. [DOI] [PubMed] [Google Scholar]
- 46.Kondadasula SV, Roda JM, Parihar R, Yu J, Lehman A, Caligiuri MA, Tridandapani S, Burry RW, Carson WE., 3rd Colocalization of the IL-12 receptor and FcgammaRIIIa to natural killer cell lipid rafts leads to activation of ERK and enhanced production of interferon-gamma. Blood. 2008;111:4173–4183. doi: 10.1182/blood-2007-01-068908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders VM, Kasprowicz DJ, Swanson-Mungerson MA, Podojil JR, Kohm AP. Adaptive immunity in mice lacking the beta(2)-adrenergic receptor. Brain Behav Immun. 2003;17:55–67. doi: 10.1016/s0889-1591(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 48.Caven TH, Shelburne A, Sato J, Chan-Li Y, Becker S, Conrad DH. IL-21 dependent IgE production in human and mouse in vitro culture systems is cell density and cell division dependent and is augmented by IL-10. Cell Immunol. 2005;238:123–134. doi: 10.1016/j.cellimm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Rabah D, Conrad DH. Effect of cell density on in vitro mouse immunoglobulin E production. Immunology. 2002;106:503–510. doi: 10.1046/j.1365-2567.2002.01435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003;17:292–294. doi: 10.1096/fj.02-0430fje. [DOI] [PubMed] [Google Scholar]
- 52.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 54.Gupta N, Wollscheid B, Watts JD, Scheer B, Aebersold R, DeFranco AL. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nat Immunol. 2006;7:625–633. doi: 10.1038/ni1337. [DOI] [PubMed] [Google Scholar]
- 55.Gupta N, DeFranco AL. Visualizing lipid raft dynamics and early signaling events during antigen receptor-mediated B-lymphocyte activation. Mol Biol Cell. 2003;14:432–444. doi: 10.1091/mbc.02-05-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 58.Carey RM, Blusztajn JK, Slack BE. Surface expression and limited proteolysis of ADAM10 are increased by a dominant negative inhibitor of dynamin. BMC cell biology. 2011;12:20. doi: 10.1186/1471-2121-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders VM, Munson AE. Beta adrenoceptor mediation of the enhancing effect of norepinephrine on the murine primary antibody response in vitro. J Pharmacol Exp Ther. 1984;230:183–192. [PubMed] [Google Scholar]
- 60.Sanders VM, Munson AE. Kinetics of the enhancing effect produced by norepinephrine and terbutaline on the murine primary antibody response in vitro. J Pharmacol Exp Ther. 1984;231:527–531. [PubMed] [Google Scholar]
- 61.de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102:4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Morris R, Lawrence MJ, Quinn PJ. The isolation and structure of membrane lipid rafts from rat brain. Biochimie. 2007;89:192–196. doi: 10.1016/j.biochi.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 64.Defrance T, Aubry JP, Rousset F, Vanbervliet B, Bonnefoy JY, Arai N, Takebe Y, Yokota T, Lee F, Arai K, et al. Human recombinant interleukin 4 induces Fc epsilon receptors (CD23) on normal human Blymphocytes. J Exp Med. 1987;165:1459–1467. doi: 10.1084/jem.165.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conrad DH, Keegan AD, Kalli KR, Van Dusen R, Rao M, Levine AD. Superinduction of low affinity IgE receptors on murine Blymphocytes by lipopolysaccharide and IL-4. J Immunol. 1988;141:1091–1097. [PubMed] [Google Scholar]
- 66.Gordon J, Katira A, Strain AJ, Gillis S. Inhibition of interleukin 4-promoted CD23 production in human Blymphocytes by transforming growth factor-beta, interferons or anti-CD19 antibody is overriden on engaging CD40. Eur J Immunol. 1991;21:1917–1922. doi: 10.1002/eji.1830210821. [DOI] [PubMed] [Google Scholar]
- 67.Keegan AD, Conrad DH. The murine lymphocyte receptor for IgE. V. Biosynthesis, transport, and maturation of the B cell Fc epsilon receptor. J Immunol. 1987;139:1199–1205. [PubMed] [Google Scholar]
- 68.Tinnell SB, Jacobs-Helber SM, Sterneck E, Sawyer ST, Conrad DH. STAT6, NF-kappaB and C/EBP in CD23 expression and IgE production. Int Immunol. 1998;10:1529–1538. doi: 10.1093/intimm/10.10.1529. [DOI] [PubMed] [Google Scholar]
- 69.Pesu M, Aittomaki S, Takaluoma K, Lagerstedt A, Silvennoinen O. p38 Mitogen-activated protein kinase regulates interleukin-4-induced gene expression by stimulating STAT6-mediated transcription. J Biol Chem. 2002;277:38254–38261. doi: 10.1074/jbc.M201427200. [DOI] [PubMed] [Google Scholar]
- 70.Howard L, Lu X, Mitchell S, Griffiths S, Glynn P. Molecular cloning of MADM: a catalytically active mammalian disintegrin-metalloprotease expressed in various cell types. Biochemical Journal. 1996;317(Pt 1):45–50. doi: 10.1042/bj3170045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A geneatlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prinzen C, Muller U, Endres K, Fahrenholz F, Postina R. Genomic structure and functional characterization of the human ADAM10 promoter. FASEB J. 2005;19:1522–1524. doi: 10.1096/fj.04-3619fje. [DOI] [PubMed] [Google Scholar]
- 73.Tippmann F, Hundt J, Schneider A, Endres K, Fahrenholz F. Up-regulation of the alpha-secretase ADAM10 by retinoic acid receptors and acitretin. FASEB J. 2009;23:1643–1654. doi: 10.1096/fj.08-121392. [DOI] [PubMed] [Google Scholar]
- 74.Wan XZ, Li B, Li YC, Yang XL, Zhang W, Zhong L, Tang SJ. Activation of NMDA receptors upregulates a disintegrin and metalloproteinase 10 via a Wnt/MAPK signaling pathway. J Neurosci. 2012;32:3910–3916. doi: 10.1523/JNEUROSCI.3916-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bruck N, Vitoux D, Ferry C, Duong V, Bauer A, de H, Rochette-Egly C. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 2009;28:34–47. doi: 10.1038/emboj.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1) EMBO J. 2003;22:1313–1324. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Almqvist N, Lonnqvist A, Hultkrantz S, Rask C, Telemo E. Serum-derived exosomes from antigen-fed mice prevent allergic sensitization in a model of allergic asthma. Immunology. 2008;125:21–27. doi: 10.1111/j.1365-2567.2008.02812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Denzer K, van Eijk M, Kleijmeer MJ, Jakobson E, de Groot C, Geuze HJ. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J Immunol. 2000;165:1259–1265. doi: 10.4049/jimmunol.165.3.1259. [DOI] [PubMed] [Google Scholar]
- 79.Henningsson F, Ding Z, Dahlin JS, Linkevicius M, Carlsson F, Gronvik KO, Hallgren J, Heyman B. IgE-mediated enhancement of CD4+ T cell responses in mice requires antigen presentation by CD11c+ cells and not by B cells. PLoS One. 2011;6:e21760. doi: 10.1371/journal.pone.0021760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.