Abstract
Background
Antimonials remain the primary antileishmanial drugs in most developing countries. However, drug resistance to these compounds is increasing and our understanding of resistance mechanisms is partial.
Methods/Principal Findings
In the present study, quantitative proteomics using stable isotope labelling of amino acids in cell culture (SILAC) and genome next generation sequencing were used in order to better characterize in vitro generated Leishmania infantum antimony resistant mutant (Sb2000.1). Using the proteomic method, 58 proteins were found to be differentially regulated in Sb2000.1. The ABC transporter MRPA (ABCC3), a known marker of antimony resistance, was observed for the first time in a proteomic screen. Furthermore, transfection of its gene conferred antimony resistance in wild-type cells. Next generation sequencing revealed aneuploidy for 8 chromosomes in Sb2000.1. Moreover, specific amplified regions derived from chromosomes 17 and 23 were observed in Sb2000.1 and a single nucleotide polymorphism (SNP) was detected in a protein kinase (LinJ.33.1810-E629K).
Conclusion/Significance
Our results suggest that differentially expressed proteins, chromosome number variations (CNVs), specific gene amplification and SNPs are important features of antimony resistance in Leishmania.
Introduction
Protozoan parasites of the genus Leishmania cause different clinical manifestations known as leishmaniasis which affect 12 million people worldwide with an estimation of 1.5-2 million new cases each year [1]. Chemotherapy represents the only possible way to fight this disease since no vaccine is available. Pentavalent antimony (SbV) compounds, such as sodium stibogluconate (Pentostam) and meglumine antimoniate (Glucantime), remain the first line of treatment against leishmaniasis especially in developing countries [2]. However, in certain regions, such as Bihar state in India, resistance to SbV is now widespread [3-5]. Other drugs such as paromomycin [6] and the orally administered miltefosine [7] are effective and used nowadays against Leishmania. Nonetheless, despite its high cost limiting its use in developing countries [8], liposomal AmB is now considered as the best drug available against visceral leishmaniasis, the most severe form of the disease [9,10].
The mechanisms of resistance to antimony-based drugs were extensively studied in Leishmania. A decrease in the reduction rate from SbV to SbIII, the active form against the parasite, was observed in Pentostam-resistant amastigotes [11]. A decrease in the expression of the aquaglyceroporin AQP1, responsible for the uptake of SbIII, was also observed in in vitro resistant mutants [12,13] and unresponsive clinical strains [14,15]. The gene coding for the ABC transporter MRPA (ABCC3), involved in the sequestration of SbIII once conjugated with thiols, was amplified on an extrachromosomal circular amplicon in in vitro generated mutants [16-20] and clinical unresponsive strains [21,22]. An increase in the level of intracellular thiols, such as cysteine, glutathione and trypanothione, was observed in numerous laboratory-selected resistant strains [18,19,23-27] as well as in unresponsive clinical strains [14,21,22,28,29]. Other proteins were also linked with the antimony resistance phenotype (reviewed in [30]).
Quantitative proteomics and whole genome sequencing methodologies are now emerging as powerful approaches to study drug resistance in microorganisms. In the parasite Leishmania, proteomic studies have already revealed some resistance mechanisms to antimony [31-35]. Similarly, whole genome sequencing has proven useful to study antimony resistance in clinical L. donovani isolates [36] and in in vitro resistant L. major mutants [37]. In the present study, we used a combination of quantitative proteomics using SILAC and whole genome sequencing to characterize in vitro selected SbIII resistant mutant.
Methods
Cell culture
L. infantum (MHOM/MA/67/ITMAP-263) wild-type (WT) and the in vitro generated resistant mutant Sb2000.1 [20,38-40], which is resistant to 2000 µM of SbIII, were described previously. Promastigotes were grown in RPMI-1640 medium for SILAC (minus L-lysine and L-arginine) (Cambridge Isotope Laboratories) supplemented with 75 µM adenosine (Sigma), 28 mM HEPES (Sigma), 40 µM biotin (Sigma), 1% penicillin-streptomycin (Wisent), 5 mg/L hemin (MP Biomedicals), 10 µM biopterin and 10% heat-inactivated dialysed foetal bovine serum (Cambridge Isotope Laboratories). L. infantum WT light medium (normal isotopic abundance) was also supplemented with 242.26 mg/L L-arginine (Sigma) and 50.03 mg/L L-lysine, while 253.68 mg/L 13C6-15N4-L-arginine and 62.21 mg/L 13C6-15N2-L-lysine were added to the resistant mutant heavy medium. Mutant cells were grown in this medium for at least 7 passages to ensure a minimum of 99% of isotope incorporation. Cells from each WT and mutant cultures were counted using a haemocytometer and the resistant strain was then mixed with WT cells in a 1:1 ratio. The SILAC labeling and subsequent mass spectrometry quantification of labeled peptides was done once.
Sample preparation
The membrane-enriched (ME) fraction was obtained by sonication, ultracentrifugation and further purification by Free flow zone electrophoresis (ZE-FFE) as described previously [41]. The cytosolic protein extraction was performed in 2D lysis buffer as described previously [42]. Proteins were then quantified using the 2D Quant kit (GE Healthcare).
Sodium dodecyl sulfate (SDS)-PAGE (1DE)
Protein samples (30 µg) were mixed with 4x premixed protein sample buffer (BioRad) and β-mercaptoethanol (5% final concentration, Sigma), and heated at 95°C for 5 min. Protein mixtures were then loaded on Precast Criterion XT Bis-Tris gradient gels (4-12% polyacrylamide, BioRad) and the SDS-PAGE separation was performed on a Criterion™ gel electrophoresis cell (BioRad) using a PowerPac 200 BioRad power supply set at 200 V for 50 min. For staining, gels were fixed in a solution of 50% methanol: 7.5% acetic acid for 1 h then incubated overnight with SYPRO Ruby Protein Gel stain (BioRad). The destaining step was performed for 30 min in a solution of 15% methanol: 7.5% acetic acid. Gel images were captured on a PerkinElmer ProExpress Proteomic Imaging system. Each sample lane from the SDS-PAGE gels was cut in 40 fractions (24 fractions above 50 kDa and 16 fractions below 50 kDa) with disposable blade (MEE-1x5) mounted on a One Touch GridCutter (Gel Company Inc.).
Protein in-gel digestion
Bands of interest were extracted from SDS-PAGE gels, placed in 96-well plates and washed extensively with HPLC water. Tryptic digestion was performed on a MassPrep liquid handling robot (Waters) according to the manufacturer’s specifications and to the protocol of Shevchenko et al [43] with modifications [44]. Briefly, proteins were reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide. Trypsin digestion was performed using 126 nM of modified porcine trypsin (Sequencing grade, Promega) at 58 °C for 1 h. Digestion products were then extracted using 1% formic acid and 2% acetonitrile followed by 1% formic acid and 50% acetonitrile. The recovered protein extracts were pooled, vacuum centrifuge dried and resuspended into 10 µL of 0.1% formic acid. Aliquots of 2 (for cytosolic fractions) or 5 (for ME fractions) µL were analyzed by mass spectrometry.
Mass spectrometry for ME fraction
SILAC experiments for ME fraction were performed on a QSTAR XL QqTOF mass spectrometer equipped with a nanospray II ion source (ABSciex) coupled to an Agilent 1100 HPLC. Five microliters of each protein sample were injected by the Agilent 1100 autosampler onto a 0.075 mm (internal diameter) self-packed IntegraFrit column (New Objective) packed with an isopropanol slurry of 5 µm Jupiter C18 (Phenomenex) stationary phase using a pressure vessel set at 700 psi. The length of the column was 12 cm. Samples were run using a 75 min gradient from 10-40% solvent B (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile) at a flow rate of 250 nL/min. An information-dependant acquisition (IDA) method was set up with the MS survey range set between 400 amu and 1600 amu (1 s) followed by dependent MS/MS scans with a mass range set between 100 and 1600 amu (3 s) of the 3 most intense ions with the enhanced all mode activated. Dynamic exclusion was set for a period of 15 s and a tolerance of 100 ppm.
Mass spectrometry for cytosolic fraction
SILAC experim*ents for cytosolic protein fraction were performed on a TripleTOF 5600 mass spectrometer equipped with a nanospray III ion source (ABSciex) coupled to an Agilent 1200 HPLC. Two microliter samples were injected by the Agilent 1200 autosampler onto a 0.075 mm (internal diameter) self-packed PicoFrit column (New Objective) packed with an isopropanol slurry of 5 µm Jupiter C18 (Phenomenex) stationary phase using a pressure vessel set at 700 psi. The length of the column was 15 cm. Samples were run using a 65 min gradient from 5-35% solvent B (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile) at a flow rate of 300 nL/min. Data were acquired using an ion spray voltage of 2.4 kV, curtain gas of 30 psi, nebulizer gas of 8 psi and an interface heater temperature of 125 °C. An information-dependant acquisition (IDA) method was set up with the MS survey range set between 400 amu and 1250 amu (250 ms) followed by dependent MS/MS scans with a mass range set between 100 and 1800 amu (50 ms) of the 20 most intense ions in the high sensitivity mode with a 2+ to 5+ charge state. Dynamic exclusion was set for a period of 3 s and a tolerance of 100 ppm.
Interpretation of tandem mass spectra and protein identification
Raw data files (n=40 for each sample) were submitted for simultaneous searches using the Protein Pilot version 4 software (ABSciex) utilizing the Paragon and Progroup algorithms [45]. The Protein Pilot program was set up to search the L. infantum proteins in the TriTrypDB LeishPEP database (http://tritrypdb.org/common/downloads/release-4.0/Linfantum/fasta/LinfantumAnnotatedProteins_TriTrypDB-4.0.fasta) with carbamidomethyl (C) as a fixed modification and standard SILAC (Lys +8, Arg +10) settings for QSTAR or TripleTof 5600 instruments. Proteins for which at least two fully trypsin-digested light (L) and heavy (H) peptides were detected at > 99% confidence and quantitative p-value lower than 0.05 were used for subsequent comparative quantitative analysis.
Whole genome sequencing
Genomic DNAs were prepared from mid-log phase cultures of clonal populations of L. infantum 263 WT and Sb2000.1 as described previously [46]. Their sequences were determined by Illumina MiSeq 150-nucleotides paired-end reads which assembled into 3197 and 2920 contigs of at least 500 nucleotides for L. infantum 263 WT and Sb2000.1, respectively. Sequence reads from each strain were aligned to the reference genome L. infantum JPCM5 [47] available at TriTrypDB (version 4.0) [48] using the software bwa (bwa aln, version 0.5.9) with default parameters [49]. The maximum number of mismatches was 4, the seed length was 32 and 2 mismatches were allowed within the seed. The detection of single nucleotide polymorphisms (SNPs) was performed using SAMtools (version 0.1.18), bcftools (distributed with SAMtools) and vcfutils.pl (distributed with SAMtools) [50], with a minimum of three reads to call a potential variation prior to further analysis. Sequencing data are available at the EMBL-EBI European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession number ERP001815 with samples ERS179382 and ERS176090 corresponding to L. infantum 263 WT and Sb2000.1, respectively. Several python (version 2.4.3) and bash (version 3.2) scripts were created to further analyze the data. The quality assessment software SAMStat (v1.08) was used to generate quality reports [51]. Putative SNPs detected by whole genome sequencing were verified by conventional PCR amplification and DNA sequencing. Growth curves were obtained by measuring absorbance at 600 nm of 72-hour cultures with different drug concentrations using an automated microplate reader.
DNA constructs
The gene LinJ.33.1810 was amplified from genomic DNA derived from L. infantum 263 WT. The PCR fragment was purified and digested with both XbaI and HindIII before being cloned into the Leishmania expression vector pSP72αNEOα [52] digested with the same enzymes. The integrity of the cloned open reading frame was confirmed by conventional sequencing before being used for transfection.
Results
Proteomic analysis of the antimony resistant mutant
The L. infantum Sb2000.1 mutant resistant to SbIII was grown in the presence of 13C6-15N4-L-arginine and 13C6-15N2-L-lysine while the parental L. infantum WT strain was maintained in medium containing normal isotopic abundance amino acids. Cells were counted and mutant was mixed with WT cells in a 1:1 ratio. ME fraction and cytosolic proteins were independently extracted from mixed populations and subjected separately to SDS-PAGE separation. Sample lines were further fractionated into 40 pieces of gel, proteins were gel extracted, trypsin digested and peptides were identified and quantified by mass spectrometry and isotopic quantification. Only significant protein identifications with difference in their expression level greater than 1.2-fold between mutant and WT were considered. Thus, 60 differentially expressed proteins were observed in cytosolic or ME fractions of Sb2000.1 when compared to the WT strain (Table 1). Among them, 42 (70%) were up-regulated and 18 (30%) were down-regulated in Sb2000.1. Two proteins were identified independently in both fractions, but varied in the same way (LinJ.21.0310 and LinJ.24.1700) (Table 1). All the identified cytosolic proteins were predicted to be devoid of transmembrane domains (TMDs) according to the TMHMM v2.0 algorithm, whereas 40% of the proteins differentially expressed in ME fractions encoded at least one TMD (Table 1). The latter result is consistent with a previous study using the same method to obtain ME fractions from promastigote parasites [41].
Table 1. MS/MS identifications of differentially expressed proteins in Sb2000.1 compared to WT in SILAC experimentsa.
| Systematic IDs | Putative protein name |
Unique peptides
b
(% seq. coverage) |
Ratio Sb2000.1/WT
|
P-value
|
Error Factor
|
TMDs
c
|
Fraction
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transport | ||||||||||||||
| LinJ.05.1140 | vacuolar ATPase subunit-like protein | 2 (5.88) | 1,24 | 0,0583 | 1,26 | 0 | M | |||||||
| LinJ.10.0400 | FT1 folate/biopterin transporter, putative | 3 (5.97) | -2,22 | 0,0450 | 2,15 | 11 | M | |||||||
| LinJ.10.0420 | FT5 folate/biopterin transporter, putative | 6 (6.99) | -1,65 | 0,0618 | 1,76 | 11 | M | |||||||
| LinJ.11.1050 | pretranslocation protein, alpha subunit, putative, SEC61-like (pretranslocation process) protein, putative | 2 (2.26) | 1,46 | 0,0595 | 1,50 | 9 | M | |||||||
| LinJ.23.0290 | MRPA (ABCC3) ABC-thiol transporter | 13 (9.37) | 3,58 | 0,0002 | 1,76 | 8 | M | |||||||
| LinJ.31.1240 | vacuolar-type proton translocating pyrophosphatase 1, putative | 9 (10.86) | -1,36 | 0,0374 | 1,33 | 14 | M | |||||||
| LinJ.32.2150 | cop-coated vesicle membrane protein p24 precursor, putative,ER--golgi transport protein p24, putative | 3 (24.38) | 1,63 | 0,0267 | 1,42 | 1 | M | |||||||
| LinJ.35.1840 | COP-coated vesicle membrane protein gp25L precursor, putative,ER--golgi transport protein gp25L, putative | 4 (18.69) | 1,42 | 0,0160 | 1,25 | 2 | M | |||||||
| Surface | ||||||||||||||
| LinJ.10.0500 | GP63, leishmanolysin,metallo-peptidase, Clan MA(M), Family M8 | 23 (26.04) | -1,91 | 1,257E-07 | 1,17 | 1 | M | |||||||
| LinJ.30.0920 | surface protein amastin, putative | 5 (26.26) | -1,73 | 0,0196 | 1,52 | 4 | M | |||||||
| Cytoskeleton | ||||||||||||||
| LinJ.16.1550 | kinesin, putative | 4 (65.24) | 2,75 | 0,0010 | 1,28 | 0 | M | |||||||
| LinJ.23.0720 | kinesin, putative | 3 (3.51) | 1,67 | 0,0013 | 1,19 | 0 | M | |||||||
| Metabolism | ||||||||||||||
| LinJ.01.0510 | long chain fatty acid CoA ligase, putative | 4 (6.45) | 1,47 | 0,0078 | 1,26 | 0 | M | |||||||
| LinJ.06.1340 | protoporphyrinogen oxidase-like protein | 2 (14.29) | -2,95 | 0,0084 | 1,74 | 1 | M | |||||||
| LinJ.10.0310 | isocitrate dehydrogenase [NADP], mitochondrial precursor, putative | 3 (6.21) | 1,27 | 0,0722 | 1,31 | 0 | C | |||||||
| LinJ.11.1000 | pyruvate phosphate dikinase, putative | 2 (1.20) | 1,38 | 0,0028 | 1,16 | 0 | M | |||||||
| LinJ.11.1100 | sterol 14-alpha-demethylase | 6 (10.63) | 1,45 | 0,0085 | 1,28 | 0 | M | |||||||
| LinJ.13.0960 | NADH-cytochrome B5 reductase, putative | 6 (17.86) | 1,47 | 0,0255 | 1,36 | 1 | M | |||||||
| LinJ.15.1140 | tryparedoxin peroxidase | 12 (42.71) | 1,38 | 0,0623 | 1,44 | 0 | C | |||||||
| LinJ.21.0310 | hexokinase, putative | 2 (6.16) | 1,40 | 0,0182 | 1,26 | 0 | C | |||||||
| LinJ.21.0310 | hexokinase, putative | 10 (16.35) | 1,53 | 0,0034 | 1,26 | 0 | M | |||||||
| LinJ.24.0310 | fumarate hydratase, putative | 2 (4.55) | 1,25 | 0,0766 | 1,31 | 0 | C | |||||||
| LinJ.24.1700 | succinate dehydrogenase flavoprotein, putative | 2 (4.12) | 1,20 | 0,0701 | 1,22 | 0 | C | |||||||
| LinJ.24.1700 | succinate dehydrogenase flavoprotein, putative | 8 (10.05) | 1,37 | 0,0771 | 1,44 | 1 | M | |||||||
| LinJ.30.2590 | formate--tetrahydrofolate ligase | 1 (2.25) | 1,57 | 0,0408 | 1,50 | 0 | C | |||||||
| LinJ.30.2920 | aldehyde dehydrogenase, putative | 3 (3.59) | 1,55 | 0,0089 | 1,25 | 0 | M | |||||||
| LinJ.30.3000 | glyceraldehyde 3-phosphate dehydrogenase, glycosomal | 2 (3.88) | 1,39 | 0,0778 | 1,47 | 0 | M | |||||||
| LinJ.32.3510 | GCVL-2 dihydrolipoamide dehydrogenase, putative | 5 (5.25) | 1,23 | 0,0017 | 1,11 | 0 | M | |||||||
| LinJ.36.3630 | 2-oxoglutarate dehydrogenase E1 component, putative | 3 (3.56) | 1,38 | 0,0840 | 1,46 | 0 | M | |||||||
| LinJ.36.6960 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 10 (21.16) | 1,26 | 0,0471 | 1,25 | 0 | C | |||||||
| Protein folding | ||||||||||||||
| LinJ.27.2350 | heat shock protein DNAJ, putative | 5 (16.92) | -1,25 | 0,0708 | 1,28 | 0 | C | |||||||
| LinJ.28.3060 | heat-shock protein hsp70, putative | 5 (6.79) | 1,43 | 0,0018 | 1,17 | 0 | M | |||||||
| LinJ.30.2540 | heat shock 70-related protein 1, mitochondrial precursor, putative | 3 (4.85) | 1,25 | 0,0373 | 1,23 | 0 | M | |||||||
| LinJ.36.2140 | chaperonin Hsp60, mitochondrial precursor | 40 (52.49) | 1,32 | 0,0008 | 1,14 | 0 | C | |||||||
| Proteolysis | ||||||||||||||
| LinJ.14.0920 | calpain-like cysteine peptidase, putative,cysteine peptidase, Clan CA, family C2, putative | 2 (17.39) | -1,44 | 0,0570 | 1,48 | 0 | C | |||||||
| LinJ.20.1320 | calpain-like cysteine peptidase, putative,calpain-like cysteine peptidase, Clan CA, family C2 | 6 (52.32) | -2,55 | 0,0023 | 1,64 | 0 | M | |||||||
| LinJ.27.0500 | calpain-like cysteine peptidase, putative,cysteine peptidase, Clan CA, family C2, putative | 2 (6.17) | 1,72 | 0,0060 | 1,28 | 0 | M | |||||||
| LinJ.36.1730 | proteasome beta 5 subunit, putative | 3 (11.92) | 1,31 | 0,0345 | 1,27 | 0 | C | |||||||
| Transcription, Translation | ||||||||||||||
| LinJ.17.0110 | elongation factor 1-alpha | 11 (21.87) | 1,26 | 0,0003 | 1,11 | 0 | M | |||||||
| LinJ.29.1920 | 40S ribosomal protein S15A, putative | 7 (25.38) | 1,24 | 0,0576 | 1,25 | 0 | M | |||||||
| LinJ.29.2580 | 60S ribosomal protein L13, putative | 4 (12.27) | -1,26 | 0,0515 | 1,26 | 0 | M | |||||||
| LinJ.30.3650 | 40S ribosomal protein S14 | 4 (20.14) | -1,59 | 0,0021 | 1,20 | 0 | C | |||||||
| LinJ.36.0020 | histone H4 | 6 (34.00) | 1,41 | 0,0569 | 1,43 | 0 | M | |||||||
| Others | ||||||||||||||
| LinJ.20.1350 | SMP-1 small myristoylated protein-1, putative | 10 (19.85) | 1,28 | 0,0941 | 1,42 | 0 | M | |||||||
| LinJ.28.2430 | glycosomal membrane protein, putative | 3 (11.71) | 1,62 | 0,0007 | 1,20 | 0 | M | |||||||
| LinJ.36.1420 | Transitional endoplasmic reticulum ATPase, putative,valosin-containing protein homolog | 8 (14.78) | -1,26 | 0,0774 | 1,30 | 0 | C | |||||||
| Hypothetical | ||||||||||||||
| LinJ.06.0640 | hypothetical protein, conserved | 4 (5.05) | -1,29 | 0,0242 | 1,22 | 0 | C | |||||||
| LinJ.08.1010 | hypothetical protein, conserved | 4 (3.01) | 1,34 | 0,0688 | 1,42 | 0 | M | |||||||
| LinJ.14.0460 | hypothetical protein, conserved | 2 (12.11) | -1,40 | 0,0808 | 1,51 | 0 | C | |||||||
| LinJ.17.0010 | hypothetical protein, conserved | 8 (7.62) | 1,26 | 0,0252 | 1,21 | 0 | M | |||||||
| LinJ.24.2250 | hypothetical protein, conserved | 3 (7.48) | 1,47 | 0,0123 | 1,25 | 0 | M | |||||||
| LinJ.26.1020 | hypothetical protein, conserved | 3 (4.19) | 1,32 | 0,0125 | 1,19 | 1 | M | |||||||
| LinJ.26.1960 | hypothetical protein, conserved | 4 (4.48) | 1,29 | 0,0030 | 1,15 | 0 | M | |||||||
| LinJ.27.0720 | hypothetical protein, conserved | 3 (2.92) | -1,94 | 0,0969 | 3,62 | 0 | C | |||||||
| LinJ.29.1600 | hypothetical protein, conserved | 3 (5.06) | -2,69 | 0,0004 | 1,28 | 14 | M | |||||||
| LinJ.30.3190 | hypothetical protein, conserved | 6 (15.57) | 1,26 | 0,0554 | 1,27 | 1 | M | |||||||
| LinJ.34.3960 | hypothetical protein, conserved | 4 (3.68) | 1,24 | 0,0616 | 1,26 | 1 | M | |||||||
| LinJ.35.0140 | hypothetical protein, conserved | 3 (17.65) | 1,51 | 0,0758 | 1,68 | 1 | M | |||||||
| LinJ.35.1010 | hypothetical protein, conserved | 2 (3.71) | -2,11 | 0,0992 | 2,98 | 0 | C | |||||||
| LinJ.36.5310 | hypothetical protein, conserved | 1 (1.44) | -1,37 | 0,0491 | 1,36 | 0 | C | |||||||
aTMDs, transmembrane domains; M, membrane-enriched fraction; C, cytosolic fraction. bPeptide identifications were accepted if they reached greater then 95% probability as specified by the Peptide Prophet algorithm. cTMDs were predicted using TMHMM v2.0.
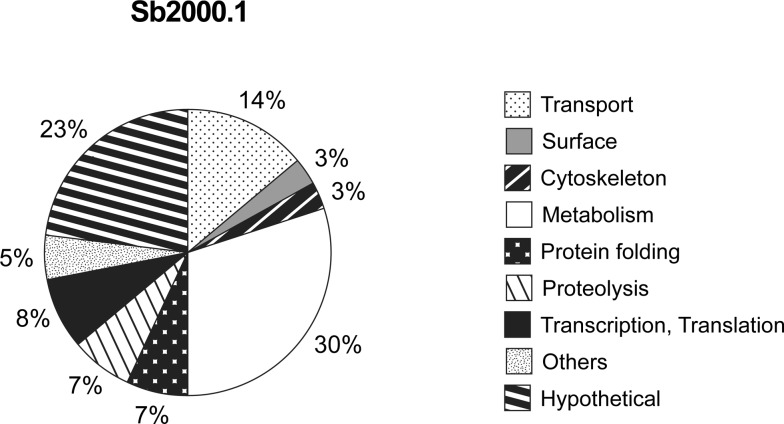
Differentially expressed proteins with statistical significance were sorted into functional classes according to GeneDB annotations and Gene Ontology. Among the proteins harbouring domains enabling functional assignment, about one third (30%) of the differentially expressed proteins were functionally ascribed to the metabolism group (Figure 1). Hypothetical proteins represented approximately a quarter (23%) of the identified proteins. The third group in importance regrouped proteins involved in transport (14%). The other functional classes (less than 10% each) represented by the proteins identified were transcription/translation, protein folding, proteolysis, cytoskeleton, surface and others (Figure 1).
Figure 1. Functional assignment of proteins found differentially expressed by SILAC between L. infantum WT and Sb2000.1 mutant.
Protein functional classification was based on GeneDB annotations and Gene Ontology.
Among the metabolic pathways present in our dataset, glycolysis and TCA cycle seemed to be increased in the Sb2000.1 mutant compared to the WT cells (Table 1). Indeed, the glycolytic enzymes hexokinase (LinJ.21.0310), glyceraldehyde 3-phosphate dehydrogenase (LinJ.30.3000) and 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (LinJ.36.6960) were found to be up-regulated in Sb2000.1, whereas the TCA cycle up-regulated enzymes included the NADP-dependant isocitrate dehydrogenase (LinJ.10.0310), the 2-oxoglutarate dehydrogenase E1 component (LinJ.36.3630), GCVL-2 dihydrolipoamide dehydrogenase (LinJ.32.3510), succinate dehydrogenase flavoprotein (LinJ.24.1700) and the fumarate hydratase (LinJ.24.0310).
The genes coding for candidate proteins being detected either as up- or down-regulated in our proteomic study were cloned into an expression vector and electroporated in L. infantum WT and Sb2000.1 cells, respectively (Table S1). The genes of candidate proteins up-regulated in Sb2000.1 were also transfected in a partial revertant strain (Sb2000.1REV) [20] (Table S1). Transfection of the ABC transporter MRPA gene (LinJ.23.0290), whose gene product was highlighted for the first time here by a proteomic approach, conferred antimony resistance in WT and partial revertant (Table S1). However, transfection of the 14 other candidate genes failed to show a direct link with SbIII resistance (Table S1).
Aneuploidy and amplicon formation lead to drug resistance
Comparative whole genome sequencing allowed the detection of genomic loci amplification and chromosomal aneuploidies in the Sb2000.1 mutant (Figure S1). Indeed, when compared to the WT strain, aneuploidy was observed for 8 chromosomes in Sb2000.1 (chromosomes 9, 11, 12, 20, 21, 23, 26 and 31). It is interesting to note that the copy number of chromosomes 9 and 31 in the mutant appear to be decreased compared to the WT cells, but not constantly across the whole chromosome and not at a level that would reflect the complete deletion of one chromosomal copy in the parasite population (Figure S1). This phenomenon suggests a mosaic aneuploidy (reviewed in [53]) for chromosome 9 in the mutant populations where a proportion of the cells have the same number of chromosomes than the WT and the other proportion have a deleted copy of the chromosome. For chromosome 31 however, approximately two times more reads were obtained in comparison to the other chromosomes in Sb2000.1, which suggests a tetraploid state, and the small decrease in copy number for this chromosome in the mutant could thus result from the loss of one of the four alleles.
Sequencing of Sb2000.1 revealed two amplicons. The first one is derived from chromosome 23 (Figure S1) and encodes the ABC transporter MRPA, hence corroborating our proteomic data where MRPA was shown to be 3.58 times more abundant in Sb2000.1 than in WT cells (Table 1). The second identified amplicon originated from chromosome 17 and contained 12 genes mostly coding for hypothetical proteins (Figure S1). However, we could not link the genes present on this amplicon with either our proteomic results or known resistance genes already described in the literature. Interestingly, we also observed a deleted region at the end of chromosome 6 in Sb2000.1 (Figure S1). In our WT strain, an amplicon originated from this region is naturally present (Ubeda et al., manuscript submitted) and thus the sequencing data support the notion that this amplicon is lost in the antimony mutant (Figure S1). This correlates with our proteomic results where the protein coded by LinJ.06.1340 (Table 1), a gene present on that particular locus of chromosome 6, is 2.95 times less abundant in the Sb2000.1 mutant compared to the WT. Another striking observation made from sequencing data is the apparent deletion in the mutant cells of a region on chromosome 26, which has also been observed in other analysed Leishmania resistant mutants (Figure S1). This region contains no gene that we could relate to a general drug resistance mechanism and, since it was observed in many contexts, it may represent a dispensable highly rearranged locus.
Single nucleotide polymorphisms implicated in antimony resistance
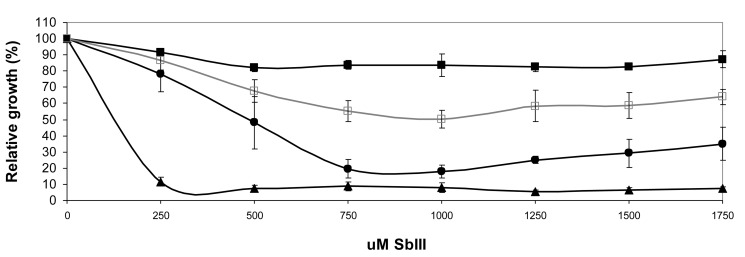
The L. infantum Sb2000.1 mutant remains considerably more resistant to SbIII than its wild-type parent when cultured in the absence of drug pressure for several passages leading to the loss of MRPA amplicons (L. infantum Sb2000.1REV in Figure 2) which suggested a role for a stable mutation in resistance. Only few single nucleotide polymorphisms (SNPs) were identified by whole genome sequencing of the antimony resistant mutant, corroborating data obtained on L. major [37]. Furthermore, most of the SNPs tested by PCR revealed the presence of polymorphic alleles in the WT, one of them already carrying the resistant mutant allele. These SNPs were not further analysed. However, one homozygous SNP in a gene coding for a protein kinase (LinJ.33.1810) was confirmed by conventional PCR and DNA resequencing. This mutation consists of a guanine to adenine transition at gene position 1885 giving rise to a change from a glutamic acid to a lysine at position 629 in the protein. It is interesting to note that the glutamic acid at this position is conserved in all other sequenced Leishmania species. To assess the role of this SNP in the resistance phenotype, we cloned the WT version of LinJ.33.1810 and transfected it in either L. infantum WT or Sb2000.1 cells. No phenotype was observed when the gene was overexpressed in WT cells (data not shown), but transfection of the WT copy of this protein kinase coding gene sensitized moderately but significantly the Sb2000.1 mutant to SbIII (Figure 2), hence confirming the link with the antimony resistance phenotype.
Figure 2. Antimony susceptibilities in Sb2000.1 and Sb2000.1REV transfected with LinJ.33.1810-WT.
Growth curves in the presence of increasing drug concentrations were performed by monitoring 72-hour cultures by OD measurement at 600 nm. The average of at least three independent experiments is shown. L.infantum WT (filled triangles); Sb2000.1 transfected with pSP72-αNEOα (filled squares); Sb2000.1 transfected with pSP72-αNEOα-LinJ.33.1810WT (open squares); Sb2000.1REV (filled circles).
Discussion
Pentavalent antimonials (SbV) have been the first-line drugs for the treatment of leishmaniasis for almost the last seven decades but clinical resistance to these drugs has emerged as an obstacle to successful treatment, especially in India [3-5]. Using whole genome sequencing and SILAC methodologies, we probed the genomic and proteomic changes induced by SbIII in an in vitro generated L. infantum resistant mutant. Since membrane proteins are often underrepresented in proteomic studies, we chose to analyse SILAC-labeled ME and cytosolic proteins separately in order to increase the proteome coverage. Many of the identified proteins were hypothetical and thus will need further investigation of their potential role in drug resistance. Proteomic analysis of membrane proteins has allowed the detection of MRPA, a known protein in SbIII resistance phenotype [16-22]. As previously shown [20], transfection of this gene in WT cells gave rise to an intermediary SbIII resistance phenotype (Table S1). However, transfection of 14 other candidate genes coding for proteins differentially expressed failed to show a direct involvement in drug resistance (Table S1). This suggests that the proteins identified here are either involved in drug resistance by an indirect mean or that a combination of some of these proteins is required to obtain a resistance phenotype. It is also possible that some of the differences between these isogenic parasites are not related to antimony resistance and were due to the mutagenic nature of the drug. One possible approach to decrease the false-negative discovery rate would be to study more than one mutant and concentrate on recurrent mutations in independent mutants.
The surface metallopeptidase GP63 had a net decrease in its expression in the Sb2000.1 mutant (Table 1). This protein has already been shown to be down-regulated in tunicamycin-resistant L. chagasi [54], but up-regulated in sinefungin-resistant L. donovani and L. tropica [55]. Similarly, the highly immunogenic amastin surface glycoproteins were previously reported as being modified at the parasite cell surface in several clinical Leishmania strains isolated from unresponsive patients to antimonials [36]. Our SILAC data indicated a decrease in the protein levels of one amastin (LinJ.30.0920) (Table 1). Histone H4 (LinJ.36.0020), that was more abundant in Sb2000.1 at the protein level, has previously been shown to be overexpressed at the RNA level in antimony-resistant L. donovani field isolates [56].
The levels of heat-shock proteins and chaperonins are also known to be increased in the presence of antimony and its related metal arsenite [57]. The heat-shock protein hsp70, which is already known to increase antimony tolerance [58], was indeed up-regulated in the resistant mutant as well as the HSP60 chaperonin (Table 1). A number of other heat-shock proteins and chaperonins were also detected as overproduced in L. infantum Sb2000.1 (Table 1). While not experimentally tested, these proteins are more likely to be implicated in a more general drug/stress response mechanism.
Key energy metabolism pathways were up-regulated in the Sb2000.1 mutant (Table 1). The observed modulation of glycolytic enzymes is supported by a previous report [34] and may correspond to a general stress response mechanism. TCA cycle is also up-regulated in the SbIII resistant mutant (Table 1).
Some evidences have shown that antimony kills the parasite by a caspase-independent apoptosis-like process involving DNA fragmentation [59-61]. In line with these observations, an interesting protein down-regulated in our SbIII mutant was the calpain-like cysteine peptidase SKCRP14.1 (LinJ.14.0920). This protein has already been shown to be down-regulated in a L. donovani field strain resistant to SbV and has been proposed to be a regulator of drug-mediated programmed cell death [31].
While aneuploidy is a well-known drug resistance mechanism in cancer cells (reviewed in [62]) and pathogenic yeasts [63], as well as in Leishmania [20,64], we could not link our observed CNVs by whole genome sequencing with our proteomic results. This is nonetheless consistent with a previous study in yeast where it was shown that an extra copy of a chromosome did not necessarily lead to an increase in proteins encoded by the genes present on this specific chromosome [65]. In yeast, either the extra mRNA molecules are not translated or the protein products are degraded shortly after synthesis [65]. The same principle may apply for Leishmania, thus explaining the non-concordance of CNVs and protein levels detected in Sb2000.1 parasites.
Two amplicons respectively derived from chromosomes 17 and 23 were detected from our sequencing data in the SbIII resistant mutant (Figure S1). The resistance phenotype was partially reversible in Sb2000.1 after only 30 passages in the absence of antimony, mainly due to the loss of the amplicon derived from chromosome 23 coding for the ABC transporter MRPA [20].
Our sequencing approach identified a low level of SNPs in the resistant mutant and only one SNP was confirmed when tested by PCR and conventional DNA sequencing. Transfection of the WT version of the protein kinase coded by LinJ.33.1810 in Sb2000.1 conferred a modest but reproducible and significant sensitization of the mutant (Figure 2). Antimony is a known protein phosphatase inhibitor [66], and a mutation in a protein kinase may compensate for the inhibition of a phosphatase. Also, downregulation of protein kinase has recently been linked [67] and associated [68] to resistance to antimony in Leishmania field isolates and it is possible that the mutation reported here serves similar purposes. While mutations are usually stable, the LinJ.33.1810-E629K allele was not observed in Sb2000.1REV possibly by the selection of a minor subpopulation within the mutant whole population. This mutation ‘reversion’, along with the loss of the MRPA amplicon, may explain the decrease in resistance for the revertant line.
In conclusion, our study has revealed that CNVs, amplicons, point mutations and changes in protein expression are associated with drug resistance. Several proteins were identified as being significantly modulated in the Sb2000.1 mutant and our proteomic study has reinforced the impact of SbIII resistance on several important metabolic pathways in Leishmania. This study has also reinforced the role of MRPA, identified for the first time in a proteomic study, as a key player in the antimony resistance. Furthermore, aneuploidy for 8 chromosomes and a SNP in a gene coding for a protein kinase have been correlated to antimony resistance.
Supporting Information
Chomosomal copy number variations in Sb2000.1 compared to WT. Chromosomes were divided into genomic windows of 5kb and the number of reads mapping to each windows determined in L. infantum Sb2000.1 and WT. The Sb2000.1/WT log2 ratios of read counts were then plotted for each genomic window on a per chromosome basis.
(PDF)
Genes whose expression of the protein product was significantly modulated in L. infantum Sb2000.1 compared to L. infantum WT in the SILAC experiments were functionally characterized for their role in resistance by gene transfection experiments.
(XLS)
Acknowledgments
We would like to thank Maxim Isabelle for the help in planning and setting up of SILAC experiments. We also would like to thank Sandra Breuils-Bonnet and Benjamin Nehmé for sample preparation prior to MS/MS and Frédéric Raymond and Sébastien Boisvert for their help with the whole genome sequencing and subsequent bioinformatics analyses.
Funding Statement
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and by the Ministère du Développement Economique, Innovation et Exportation (MDEIE) grants to AD and by CIHR grants to GGP (MOP209278) and MO (MOP13233). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Murray HW, Berman JD, Davies CR, Saravia NG (2005) Advances in leishmaniasis. Lancet 366: 1561-1577. doi: 10.1016/S0140-6736(05)67629-5. PubMed: 16257344. [DOI] [PubMed] [Google Scholar]
- 2. Alvar J, Croft S, Olliaro P (2006) Chemotherapy in the treatment and control of leishmaniasis. Adv Parasitol 61: 223-274. doi: 10.1016/S0065-308X(05)61006-8. PubMed: 16735166. [DOI] [PubMed] [Google Scholar]
- 3. Lira R, Sundar S, Makharia A, Kenney R, Gam A et al. (1999) Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J Infect Dis 180: 564-567. doi: 10.1086/314896. PubMed: 10395884. [DOI] [PubMed] [Google Scholar]
- 4. Sundar S (2001) Drug resistance in Indian visceral leishmaniasis. Trop Med Int Health 6: 849-854. doi: 10.1046/j.1365-3156.2001.00778.x. PubMed: 11703838. [DOI] [PubMed] [Google Scholar]
- 5. Thakur CP, Dedet JP, Narain S, Pratlong F (2001) Leishmania species, drug unresponsiveness and visceral leishmaniasis in Bihar, India. Trans R Soc Trop Med Hyg 95: 187-189. [DOI] [PubMed] [Google Scholar]
- 6. Sundar S, Jha TK, Thakur CP, Sinha PK, Bhattacharya SK (2007) Injectable paromomycin for Visceral leishmaniasis in India. N Engl J Med 356: 2571-2581. doi: 10.1056/NEJMoa066536. PubMed: 17582067. [DOI] [PubMed] [Google Scholar]
- 7. Sundar S, Rosenkaimer F, Makharia MK, Goyal AK, Mandal AK et al. (1998) Trial of oral miltefosine for visceral leishmaniasis. Lancet 352: 1821-1823. doi: 10.1016/S0140-6736(98)04367-0. PubMed: 9851383. [DOI] [PubMed] [Google Scholar]
- 8. Olliaro P, Sundar S (2009) Anthropometrically derived dosing and drug costing calculations for treating visceral leishmaniasis in Bihar, India. Trop Med Int Health 14: 88-92. doi: 10.1111/j.1365-3156.2008.02195.x. PubMed: 19121150. [DOI] [PubMed] [Google Scholar]
- 9. Bern C, Adler-Moore J, Berenguer J, Boelaert M, den Boer M et al. (2006) Liposomal amphotericin B for the treatment of visceral leishmaniasis. Clin Infect Dis 43: 917-924. doi: 10.1086/507530. PubMed: 16941377. [DOI] [PubMed] [Google Scholar]
- 10. Chappuis F, Sundar S, Hailu A, Ghalib H, Rijal S et al. (2007) Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol 5: 873-882. PubMed: 17938629. [DOI] [PubMed] [Google Scholar]
- 11. Shaked-Mishan P, Ulrich N, Ephros M, Zilberstein D (2001) Novel Intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J Biol Chem 276: 3971-3976. doi: 10.1074/jbc.M005423200. PubMed: 11110784. [DOI] [PubMed] [Google Scholar]
- 12. Gourbal B, Sonuc N, Bhattacharjee H, Legare D, Sundar S et al. (2004) Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem 279: 31010-31017. doi: 10.1074/jbc.M403959200. PubMed: 15138256. [DOI] [PubMed] [Google Scholar]
- 13. Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M (2005) Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol 57: 1690-1699. doi: 10.1111/j.1365-2958.2005.04782.x. PubMed: 16135234. [DOI] [PubMed] [Google Scholar]
- 14. Decuypere S, Rijal S, Yardley V, De Doncker S, Laurent T et al. (2005) Gene expression analysis of the mechanism of natural Sb(V) resistance in Leishmania donovani isolates from Nepal. Antimicrob Agents Chemother 49: 4616-4621. doi: 10.1128/AAC.49.11.4616-4621.2005. PubMed: 16251303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mandal S, Maharjan M, Singh S, Chatterjee M, Madhubala R (2010) Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and -resistant clinical isolates of Leishmania donovani from India. J Antimicrob Chemother 65: 496-507. doi: 10.1093/jac/dkp468. PubMed: 20067981. [DOI] [PubMed] [Google Scholar]
- 16. Ouellette M, Borst P (1991) Drug resistance and P-glycoprotein gene amplification in the protozoan parasite Leishmania. Res Microbiol 142: 737-746. doi: 10.1016/0923-2508(91)90089-S. PubMed: 1961984. [DOI] [PubMed] [Google Scholar]
- 17. Grondin K, Papadopoulou B, Ouellette M (1993) Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res 21: 1895-1901. doi: 10.1093/nar/21.8.1895. PubMed: 8098523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grondin K, Haimeur A, Mukhopadhyay R, Rosen BP, Ouellette M (1997) Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenite-resistant Leishmania tarentolae. EMBO J 16: 3057-3065. doi: 10.1093/emboj/16.11.3057. PubMed: 9214623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haimeur A, Brochu C, Genest P, Papadopoulou B, Ouellette M (2000) Amplification of the ABC transporter gene PGPA and increased trypanothione levels in potassium antimonyl tartrate (SbIII) resistant Leishmania tarentolae. Mol Biochem Parasitol 108: 131-135. doi: 10.1016/S0166-6851(00)00187-0. PubMed: 10802326. [DOI] [PubMed] [Google Scholar]
- 20. Leprohon P, Légaré D, Raymond F, Madore E, Hardiman G et al. (2009) Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res 37: 1387-1399. doi: 10.1093/nar/gkn1069. PubMed: 19129236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mittal MK, Rai S, Ashutosh, Ravinder, Gupta S et al. (2007) Characterization of natural antimony resistance in Leishmania donovani isolates. Am J Trop Med Hyg 76: 681-688. PubMed: 17426170. [PubMed] [Google Scholar]
- 22. Mukherjee A, Padmanabhan PK, Singh S, Roy G, Girard I et al. (2007) Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J Antimicrob Chemother 59: 204-211. PubMed: 17213267. [DOI] [PubMed] [Google Scholar]
- 23. Mukhopadhyay R, Dey S, Xu N, Gage D, Lightbody J et al. (1996) Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci U S A 93: 10383-10387. doi: 10.1073/pnas.93.19.10383. PubMed: 8816809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Légaré D, Papadopoulou B, Roy G, Mukhopadhyay R, Haimeur A et al. (1997) Efflux systems and increased trypanothione levels in arsenite-resistant Leishmania. Exp Parasitol 87: 275-282. doi: 10.1006/expr.1997.4222. PubMed: 9371094. [DOI] [PubMed] [Google Scholar]
- 25. Haimeur A, Guimond C, Pilote S, Mukhopadhyay R, Rosen BP et al. (1999) Elevated levels of polyamines and trypanothione resulting from overexpression of the ornithine decarboxylase gene in arsenite-resistant Leishmania. Mol Microbiol 34: 726-735. doi: 10.1046/j.1365-2958.1999.01634.x. PubMed: 10564512. [DOI] [PubMed] [Google Scholar]
- 26. Guimond C, Trudel N, Brochu C, Marquis N, El Fadili A et al. (2003) Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Res 31: 5886-5896. doi: 10.1093/nar/gkg806. PubMed: 14530437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukherjee A, Roy G, Guimond C, Ouellette M (2009) The gamma-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol Microbiol 74: 914-927. doi: 10.1111/j.1365-2958.2009.06907.x. PubMed: 19818018. [DOI] [PubMed] [Google Scholar]
- 28. Mandal G, Wyllie S, Singh N, Sundar S, Fairlamb AH et al. (2007) Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology 134: 1679-1687. PubMed: 17612420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh N, Almeida R, Kothari H, Kumar P, Mandal G et al. (2007) Differential gene expression analysis in antimony-unresponsive Indian kala azar (visceral leishmaniasis) clinical isolates by DNA microarray. Parasitology 134: 777-787. doi: 10.1017/S0031182007002284. PubMed: 17306059. [DOI] [PubMed] [Google Scholar]
- 30. Ashutosh, Sundar S, Goyal N (2007) Molecular mechanisms of antimony resistance in Leishmania. J Med Microbiol 56: 143-153. doi: 10.1099/jmm.0.46841-0. PubMed: 17244793. [DOI] [PubMed] [Google Scholar]
- 31. Vergnes B, Gourbal B, Girard I, Sundar S, Drummelsmith J et al. (2007) A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol Cell Proteomics 6: 88-101. PubMed: 17050524. [DOI] [PubMed] [Google Scholar]
- 32. El Fadili K, Drummelsmith J, Roy G, Jardim A, Ouellette M (2009) Down regulation of KMP-11 in Leishmania infantum axenic antimony resistant amastigotes as revealed by a proteomic screen. Exp Parasitol 123: 51-57. doi: 10.1016/j.exppara.2009.05.013. PubMed: 19500579. [DOI] [PubMed] [Google Scholar]
- 33. Kumar A, Sisodia B, Misra P, Sundar S, Shasany AK et al. (2010) Proteome mapping of overexpressed membrane-enriched and cytosolic proteins in sodium antimony gluconate (SAG) resistant clinical isolate of Leishmania donovani. Br J Clin Pharmacol 70: 609-617. doi: 10.1111/j.1365-2125.2010.03716.x. PubMed: 20840452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biyani N, Singh AK, Mandal S, Chawla B, Madhubala R (2011) Differential expression of proteins in antimony-susceptible and -resistant isolates of Leishmania donovani. Mol Biochem Parasitol 179: 91-99. doi: 10.1016/j.molbiopara.2011.06.004. PubMed: 21736901. [DOI] [PubMed] [Google Scholar]
- 35. Walker J, Gongora R, Vasquez JJ, Drummelsmith J, Burchmore R et al. (2012) Discovery of factors linked to antimony resistance in Leishmania panamensis through differential proteome analysis. Mol Biochem Parasitol 183: 166-176. doi: 10.1016/j.molbiopara.2012.03.002. PubMed: 22449941. [DOI] [PubMed] [Google Scholar]
- 36. Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH et al. (2011) Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21: 2143-2156. doi: 10.1101/gr.123430.111. PubMed: 22038251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukherjee A, Boisvert S, do Monte-Neto RL, Coelho AC, Raymond F et al. (2013) Telomeric gene deletion and intrachromosomal amplification in antimony resistant Leishmania. Mol Microbiol. [DOI] [PubMed] [Google Scholar]
- 38. El Fadili K, Messier N, Leprohon P, Roy G, Guimond C et al. (2005) Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob Agents Chemother 49: 1988-1993. doi: 10.1128/AAC.49.5.1988-1993.2005. PubMed: 15855523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leprohon P, Légaré D, Ouellette M (2009) Intracellular localization of the ABCC proteins of Leishmania and their role in resistance to antimonials. Antimicrob Agents Chemother 53: 2646-2649. doi: 10.1128/AAC.01474-08. PubMed: 19307364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moreira W, Leprohon P, Ouellette M (2011) Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis 2: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brotherton MC, Racine G, Ouameur AA, Leprohon P, Papadopoulou B et al. (2012) Analysis of Membrane-Enriched and High Molecular Weight Proteins in Leishmania infantum Promastigotes and Axenic Amastigotes. J Proteome Res 11: 3974-3985. doi: 10.1021/pr201248h. PubMed: 22716046. [DOI] [PubMed] [Google Scholar]
- 42. Brotherton MC, Racine G, Foucher AL, Drummelsmith J, Papadopoulou B et al. (2010) Analysis of stage-specific expression of basic proteins in Leishmania infantum. J Proteome Res 9: 3842-3853. doi: 10.1021/pr100048m. PubMed: 20583757. [DOI] [PubMed] [Google Scholar]
- 43. Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850-858. doi: 10.1021/ac950914h. PubMed: 8779443. [DOI] [PubMed] [Google Scholar]
- 44. Havlis J, Thomas H, Sebela M, Shevchenko A (2003) Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem 75: 1300-1306. doi: 10.1021/ac026136s. PubMed: 12659189. [DOI] [PubMed] [Google Scholar]
- 45. Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH et al. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6: 1638-1655. doi: 10.1074/mcp.T600050-MCP200. PubMed: 17533153. [DOI] [PubMed] [Google Scholar]
- 46. Raymond F, Boisvert S, Roy G, Ritt JF, Légaré D et al. (2012) Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res 40: 1131-1147. doi: 10.1093/nar/gkr834. PubMed: 21998295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839-847. doi: 10.1038/ng2053. PubMed: 17572675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP et al. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38: D457-D462. doi: 10.1093/nar/gkq373. PubMed: 19843604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754-1760. doi: 10.1093/bioinformatics/btp324. PubMed: 19451168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078-2079. doi: 10.1093/bioinformatics/btp352. PubMed: 19505943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lassmann T, Hayashizaki Y, Daub CO (2011) SAMStat: monitoring biases in next generation sequencing data. Bioinformatics 27: 130-131. doi: 10.1093/bioinformatics/btq614. PubMed: 21088025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Papadopoulou B, Roy G, Ouellette M (1992) A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J 11: 3601-3608. PubMed: 1396560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sterkers Y, Lachaud L, Bourgeois N, Crobu L, Bastien P et al. (2012) Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania. Mol Microbiol 86: 15-23. PubMed: 22857263. [DOI] [PubMed] [Google Scholar]
- 54. Wilson ME, Hardin KK (1990) The major Leishmania donovani chagasi surface glycoprotein in tunicamycin-resistant promastigotes. J Immunol 144: 4825-4834. PubMed: 2191040. [PubMed] [Google Scholar]
- 55. Soteriadou KP, Tzinia AK, Mamalaki A, Phelouzat MA, Lawrence F et al. (1994) Expression of the major surface glycoprotein of Leishmania, gp63, in wild-type and sinefungin-resistant promastigotes. Eur J Biochem 223: 61-68. doi: 10.1111/j.1432-1033.1994.tb18966.x. PubMed: 8033909. [DOI] [PubMed] [Google Scholar]
- 56. Singh R, Kumar D, Duncan RC, Nakhasi HL, Salotra P (2010) Overexpression of histone H2A modulates drug susceptibility in Leishmania parasites. Int J Antimicrob Agents 36: 50-57. doi: 10.1016/j.ijantimicag.2010.03.012. PubMed: 20427152. [DOI] [PubMed] [Google Scholar]
- 57. Lawrence F, Robert-Gero M (1985) Induction of heat shock and stress proteins in promastigotes of three Leishmania species. Proc Natl Acad Sci U S A 82: 4414-4417. doi: 10.1073/pnas.82.13.4414. PubMed: 3859870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brochu C, Haimeur A, Ouellette M (2004) The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite leishmania. Cell Stress Chaperones 9: 294-303. doi: 10.1379/CSC-15R1.1. PubMed: 15544167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sereno D, Holzmuller P, Mangot I, Cuny G, Ouaissi A et al. (2001) Antimonial-mediated DNA fragmentation in Leishmania infantum amastigotes. Antimicrob Agents Chemother 45: 2064-2069. doi: 10.1128/AAC.45.7.2064-2069.2001. PubMed: 11408224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee N, Bertholet S, Debrabant A, Muller J, Duncan R et al. (2002) Programmed cell death in the unicellular protozoan parasite Leishmania. Cell Death Differ 9: 53-64. doi: 10.1038/sj.cdd.4400952. PubMed: 11803374. [DOI] [PubMed] [Google Scholar]
- 61. Sudhandiran G, Shaha C (2003) Antimonial-induced increase in intracellular Ca2+ through non-selective cation channels in the host and the parasite is responsible for apoptosis of intracellular Leishmania donovani amastigotes. J Biol Chem 278: 25120-25132. doi: 10.1074/jbc.M301975200. PubMed: 12707265. [DOI] [PubMed] [Google Scholar]
- 62. Duesberg P, Li R, Sachs R, Fabarius A, Upender MB et al. (2007) Cancer drug resistance: the central role of the karyotype. Drug Resist Updat 10: 51-58. doi: 10.1016/j.drup.2007.02.003. PubMed: 17387035. [DOI] [PubMed] [Google Scholar]
- 63. Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367-370. doi: 10.1126/science.1128242. PubMed: 16857942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ubeda JM, Légaré D, Raymond F, Ouameur AA, Boisvert S et al. (2008) Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol 9: R115. doi: 10.1186/gb-2008-9-7-r115. PubMed: 18638379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M et al. (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916-924. doi: 10.1126/science.1142210. PubMed: 17702937. [DOI] [PubMed] [Google Scholar]
- 66. Pathak MK, Yi T (2001) Sodium stibogluconate is a potent inhibitor of protein tyrosine phosphatases and augments cytokine responses in hemopoietic cell lines. J Immunol 167: 3391-3397. PubMed: 11544330. [DOI] [PubMed] [Google Scholar]
- 67. Kazemi-Rad E, Mohebali M, Khadem-Erfan MB, Saffari M, Raoofian R et al. (2013) Identification of antimony resistance markers in Leishmania tropica field isolates through a cDNA-AFLP approach. Exp Parasitol 135: 344-349. doi: 10.1016/j.exppara.2013.07.018. PubMed: 23928349. [DOI] [PubMed] [Google Scholar]
- 68. Ashutosh, Garg M, Sundar S, Duncan R, Nakhasi HL et al. (2012) Downregulation of mitogen-activated protein kinase 1 of Leishmania donovani field isolates is associated with antimony resistance. Antimicrob Agents Chemother 56: 518-525. doi: 10.1128/AAC.00736-11. PubMed: 22064540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chomosomal copy number variations in Sb2000.1 compared to WT. Chromosomes were divided into genomic windows of 5kb and the number of reads mapping to each windows determined in L. infantum Sb2000.1 and WT. The Sb2000.1/WT log2 ratios of read counts were then plotted for each genomic window on a per chromosome basis.
(PDF)
Genes whose expression of the protein product was significantly modulated in L. infantum Sb2000.1 compared to L. infantum WT in the SILAC experiments were functionally characterized for their role in resistance by gene transfection experiments.
(XLS)