Abstract
Genetic polymorphisms in TNFB are involved in the regulation of its expression and are found to be associated with various autoimmune diseases. The aim of the present study was to determine whether TNFB +252A/G (rs909253) and exon 3 C/A (rs1041981) polymorphisms are associated with vitiligo susceptibility, and expression of TNFB and ICAM1 affects the disease onset and progression. We have earlier reported the role of TNFA in autoimmune pathogenesis of vitiligo, and we now show the involvement of TNFB in vitiligo pathogenesis. The two polymorphisms investigated in the TNFB were in strong linkage disequilibrium and significantly associated with vitiligo. TNFB and ICAM1 transcripts were significantly increased in patients compared to controls. Active vitiligo patients showed significant increase in TNFB transcripts compared to stable vitiligo. The genotype-phenotype analysis revealed that TNFB expression levels were higher in patients with GG and AA genotypes as compared to controls. Patients with the early age of onset and female patients showed higher TNFB and ICAM1 expression. Overall, our findings suggest that the increased TNFB transcript levels in vitiligo patients could result, at least in part, from variations at the genetic level which in turn leads to increased ICAM1 expression. For the first time, we show that TNFB +252A/G and exon 3 C/A polymorphisms are associated with vitiligo susceptibility and influence the TNFB and ICAM1 expression. Moreover, the study also emphasizes influence of TNFB and ICAM1 on the disease progression, onset and gender bias for developing vitiligo.
Introduction
Vitiligo is an acquired, hypomelanotic disease characterized by circumscribed depigmented macules. The absence of melanocytes from the lesional skin due to their destruction has been suggested to be the key event in the pathogenesis of vitiligo [1]. It is a polygenic, multifactorial disorder involving multiple susceptibility genes and unknown environmental triggers [2]–[4]. It affects approximately 0.5–1% of the world population [5]. The exact etiology of vitiligo remains obscure, but autoimmunity has been strongly implicated in the development of disease as approximately 30% of vitiligo patients are affected with at least one additional autoimmune disorder [6], [7]. Epidemiological studies have shown frequent family clustering of vitiligo cases, with elevated risk of vitiligo in first-degree relatives and high concordance in monozygotic twins [4], [6], suggesting a genetic basis for vitiligo. A number of genes have been implicated in the pathogenesis of vitiligo on the basis of genetic linkage and association studies, including MHC, ACE, CAT, CTLA4, COMT, ESR, MBL2, PTPN22, HLA, NALP1, XBP1, FOXP1 and IL2RA that are involved in the regulation of immunity [8], [9].
Cytokines are important mediators of immunity and there is now convincing evidence that cytokines also have an important role in the pathogenesis of autoimmunity [10]. Morreti et al. [11], [12] have shown cytokine imbalance in the skin of vitiligo patients suggesting their role in autoimmune pathogenesis of vitiligo. Analysis of cytokine gene polymorphisms identifies genetic abnormalities of cytokine regulation, thus allows us to establish genotype-phenotype correlation which may be important in unraveling the disease pathogenesis.
Tumor necrosis factor B (TNFB) also known as lymphotoxin A (LTA), is a close homologue of TNFA. Both these cytokines are recognized by the same widely distributed cellular TNF receptors i.e. TNFR1 and TNFR2, as a consequence, many of their effects are similar [13]. TNFB is a Th1 cytokine, primarily produced by activated T and B lymphocytes. It is a potent mediator of inflammatory and immune responses and it is also involved in the regulation of various biological processes including cell proliferation, differentiation, apoptosis, lipid metabolism, coagulation and neurotransmission [14]. The TNFB gene contains four exons and encodes a primary transcript of 2038 nucleotides. TNFB is a protein of 171 amino acids and secreted as a soluble polypeptide, but can form heterotrimers with lymphotoxin-beta, which effectively anchors the TNFB to the cell surface [15]. TNFB induces cell apoptosis upon binding to TNFR1, but it induces inflammatory responses by activating NF-kB nuclear protein upon binding to TNFR2 [16].
Polymorphisms of TNFA and TNFB genes have been shown to affect their production and hence can be associated with several autoimmune diseases [14], [17]. We have earlier reported the crucial role of TNFA in autoimmune pathogenesis of vitiligo [18], and we now show the involvement of TNFB and its downstream effector, intercellular adhesion molecule 1 (ICAM1) in vitiligo pathogenesis. Two well-characterized variants of TNFB are in tight linkage disequilibrium [intron 1 +252A/G (rs909253; IVS1+90 A/G) and exon 3 C/A (rs1041981; Thr26Asn)] are found to influence TNFB expression in vitro [19], [20]. The +252A/G polymorphism was reported to influence TNFB plasma levels. This single nucleotide polymorphism (A/G) affects a phorbol ester-response element and distinguishes the two alleles that have been designated as TNFB1 and TNFB2 [19]. TNFA2 and TNFB2 alleles have much more powerful transcriptional activator binding sites than TNFA1 and TNFB1 alleles, respectively [19]. Other in vitro experiments have shown that TNFB exon 3 C/A (Thr26Asn) polymorphism is associated with a two fold increase in the induction of several cell-adhesion molecules including ICAM1 in the smooth muscle cells of human coronary arteries [21].
It has been suggested that melanocyte death is mediated by apoptosis in the context of autoimmunity, and cytokines such as IFNG, TNFA, TNFB and IL1 can initiate apoptosis [22]. Additionally, IFNG, TNFA and TNFB induce the expression of ICAM1 on the cell-surface of melanocytes [23]. Increased expression of this adhesion molecule on the melanocytes enhances T cell- melanocyte attachment in the skin and may play a role in the destruction of melanocytes in vitiligo [24]. These reports signify that the host's ability to produce cytokines such as TNFA and TNFB may play a crucial role in vitiligo susceptibility.
In the present study, we have made an attempt to understand the role of TNFB and ICAM1 in vitiligo pathogenesis. The objectives of this study were: i.) to determine whether the single nucleotide polymorphisms (SNPs) of TNFB [intron 1 +252A/G (rs909253; IVS1+90 A/G) and exon 3 C/A (rs1041981; Thr26Asn)] are associated with vitiligo susceptibility; ii.) to estimate and compare TNFB and ICAM1 transcript levels in patients and unaffected controls; iii.) to correlate TNFB and ICAM1 transcript levels with onset and progression of the disease.
Materials and Methods
Study subjects
The study group included 524 vitiligo patients comprised of 224 males and 300 females who referred to S.S.G. Hospital, Vadodara and Civil Hospital, Ahmedabad, Gujarat, India (Table 1).
Table 1. Demographic characteristics of vitiligo patients and unaffected controls.
| Characteristic | Vitiligo Patients | Controls |
| n = 524 | n = 592 | |
| Average age(mean age ± SD), yrs | 31.24 ± 12.13 | 27.54 ± 13.26 |
| Sex: Male, No. (%)Female, No. (%) | 224 (42.75)300 (57.25) | 267 (45.10)325 (54.90) |
| Age of onset(mean age ± SD), yrsDuration of disease(mean ± SD), yrs | 21.96 ± 14.908.20 ± 7.11 | NANA |
| Type of vitiligo: | ||
| Generalized, No. (%)Localized, No. (%)Active, No. (%)Stable, No. (%)Family history, No. (%) | 360 (68.70)164 (31.30)393 (75.00)131 (25.00)68 (12.98) | NANANANANA |
‘n’ represents number of patients/controls, SD refers to standard deviation, NA refers to not applicable, yrs refers to years, No. refers to numbers.
The diagnosis of vitiligo was clinically based on characteristic skin depigmentation with typical localization and white colour on the skin lesions under Woods lamp and confirmed as two different types: generalized (including acrofacial vitiligo and vitiligo universalis) and localized by dermatologists. Patients had no other associated autoimmune disease. The patients were divided into two subgroups: active vitiligo (existing lesions were spreading and/or new lesions had appeared within the previous 6 months) and stable vitiligo (no increase in lesion size or number within 6 months or more). Three hundred and ninety three patients were classified with active vitiligo, whereas 131 patients were included in the stable vitiligo group (Table 1).
A total of 592 ethnically age- and sex-matched unaffected individuals (267 males and 325 females) were included in the study (Table 1). None of the healthy individuals or their relatives had any evidence of vitiligo and autoimmune disease. The study plan was approved by the Institutional Ethics Committee for Human Research (IECHR), Faculty of Science, The Maharaja Sayajirao University of Baroda, Vadodara, Gujarat, India. The importance of the study was explained to all participants and written consent was obtained from all subjects before performing the studies. The written consent was also obtained from the guardians on behalf of the children participants involved in the study. IECHR approved the written consent procedure for patient and control participants.
Genotyping of TNFB +252A/G polymorphism
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was used to genotype TNFB +252A/G polymorphism. Five ml venous blood was collected from the patients and healthy subjects in K3EDTA coated vacutainers (BD, Franklin Lakes, NJ 07417, USA). Genomic DNA was extracted from whole blood using ‘QIAamp DNA Blood Kit’ (QIAGEN Inc., Valencia, CA 91355, USA) according to the manufacturer's instructions. The primers used for polymerase chain reaction are mentioned in Table S1 in File S1. The reaction mixture of the total volume of 20 µL included 5 µL (100 ng) of genomic DNA, 10 µL nuclease-free H2O, 2.0 µL 10x PCR buffer, 2 µL 2 mM dNTPs (SIGMA Chemical Co, St. Louis, Missouri, USA), 0.3 µL of 10 µM corresponding forward and reverse primers (Eurofins, Bangalore, India), and 0.3 µL (5 U/µL) Taq Polymerase (Bangalore Genei, Bangalore, India). Amplification was performed using a PTC-100 thermal cycler (MJ Research, Inc., Watertown, Massachusetts, USA) according to the protocol: 95°C for 10 minutes followed by 35 cycles of 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds. The amplified products were checked by electrophoresis on a 2.0% agarose gel stained with ethidium bromide.
Restriction enzyme NcoI (New England Biolabs, Beverly, MA) was used for digesting amplicons of TNFB +252 A/G (Table S1 in File S1). 5 µL of the amplified products were digested with 5 U of the restriction enzyme in a total reaction volume of 25 µL as per the manufacturer's instruction. The digested products with 100 base pair DNA ladder (Bioron, Ludwigshafen am Rhein, Germany) were resolved on 2.0% agarose gels stained with ethidium bromide and visualized under UV transilluminator. A 417 bp undigested product corresponding to A allele and 284 bp and 133 bp digested products corresponding to G allele were identified and three different genotypes were classified as: GG homozygous, AG heterozygous and AA homozygous (Fig. S1 in File S2). More than 10% of the samples were randomly selected for confirmation and the results were 100% concordant (analysis of the chosen samples was repeated by two researchers independently).
Genotyping of TNFB Thr26Asn C/A polymorphism
The genotyping of Thr26Asn C/A SNP of TNFB was carried out by dual colour hydrolysis probes labeled with FAM (for ‘A’ allele) and VIC (for ‘C’ allele) using LightCycler® 480Real-Time PCR protocol with background corrected end point fluorescence analysis in a TaqMan SNP genotyping assay (Assay ID: C_7514870_20; Life Technologies Corp., California, USA) which yielded the three genotypes (AA homozygous, CA heterozygous and CC homozygous) as identified on scattered plot (Fig. S2 in File S2). Real-time PCR was performed in 10 µl volume using LightCycler®480 Probes Master (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer's instructions. A no-template control (NTC) was used with the SNP genotyping assay.
Determination of TNFB, ICAM1 and GAPDH mRNA expression
TNFB, ICAM1 and GAPDH mRNA levels were measured in whole blood of 166 patients and 175 controls by Real-time PCR.
RNA extraction and cDNA synthesis
Total RNA from whole blood was isolated and purified using the RibopureTM - blood Kit (Ambion inc. Texas, USA) following the manufacturer's protocol. RNA integrity was verified by 1.5% agarose gel electrophoresis, RNA yield and purity was determined spectrophotometrically at 260/280 nm. RNA was treated with DNase I (Ambion inc. Texas, USA) before cDNA synthesis to avoid DNA contamination. cDNA synthesis was performed using 1 µg of total RNA by RevertAid First Strand cDNA Synthesis Kit (Fermentase, Vilnius, Lithuania) according to the manufacturer's instructions in the MJ Research Thermal Cycler (Model PTC-200, Watertown, MA, USA).
Real-time PCR
The expression of TNFB, ICAM1 and GAPDH transcripts were measured by real-time PCR using gene specific primers (Eurofins, Bangalore, India) as shown in Table S1 in File S1. Expression of the GAPDH gene was used as a reference. Real-time PCR was performed in duplicates in 20 µl volume using LightCycler®480 SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer's instructions. The thermal cycling conditions included an initial activation step at 95°C for 10 min, followed by 45 cycles of denaturation, annealing and amplification (95°C for 10 sec, 65°C for 15 sec, 72°C for 20 sec.). The fluorescence data collection was performed during the extension step. At the end of the amplification phase a melt curve analysis was carried out to check the specificity of the products formed (Fig. S3 in File S2). The PCR cycle at which PCR amplification begins its exponential phase and product fluorescence intensity finally rises above the background and becomes visible was considered as the crossing point (CP) or cycle threshold (CT). The ΔCT or ΔCP value was determined as the difference between the cycle threshold of target genes (TNFB/ICAM1) and reference gene (GAPDH). The difference between the two ΔCP values (ΔCP Controls and ΔCP patients) was considered as ΔΔCP to obtain the value of fold expression (2−ΔΔCp).
Statistical analyses
Evaluation of the Hardy-Weinberg equilibrium (HWE) was performed for both the polymorphisms in patients and controls by comparing the observed and expected frequencies of the genotypes using chi-square analysis. The distribution of the genotypes and allele frequencies of TNFB polymorphisms in patients and control subjects were compared by chi-square test. p-values less than 0.025 were considered as statistically significant due to Bonferroni's correction for multiple testing. Odds ratio (OR) with respective confidence interval (95% CI) for disease susceptibility was also calculated. Age of onset analysis and relative expression of TNFB and ICAM1 in patient and control groups were plotted and analyzed by Mann-Whitney Wilcoxon test. Chi-square and Mann-Whitney Wilcoxon tests were performed using Prism 4 software (Graphpad software Inc; San Diego CA, USA, 2003). The statistical power of detection of the association with the disease at the 0.05 level of significance was determined by using the G* Power software [25].
Results
Analysis of association between TNFB +252A/G and exon 3 C/A (Thr26Asn) polymorphisms and susceptibility to vitiligo
The genotype and allele frequencies of the +252 A/G polymorphism in 524 vitiligo patients and 592 controls are summarized in Table 2. The TNFB +252 A/G polymorphism was found to be in significant association with vitiligo patients compared to controls (p = 0.002; Table 2). The minor allele (G) of +252 A/G was more frequent in the vitiligo group compared to the control group (27.0% versus 21.0%, p = 0.001; OR: 1.424, 95% CI: 1.171–1.732) consistent with a susceptibility effect (Table 2). The distribution of TNFB +252 A/G genotype frequencies was consistent with Hardy-Weinberg expectations in control and patient groups (p>0.05) (Table 2). Moreover, generalized vitiligo (GV) group showed significant association of +252 A/G polymorphism when the genotypes were compared with those of the control group (p<0.0001); however localized vitiligo (LV) group did not show significant association of the polymorphism (p = 0.826) (Table 3). Also, there was significant difference in allele frequencies of the polymorphism for GV group when the alleles were compared with those of the control group (p<0.0001); however, the LV group did not show significant difference in allele frequencies (p = 0.759) (Table 3). In addition, gender based analysis of TNFB +252 A/G polymorphism suggested significant association of minor +252 G allele with female vitligo patients as compared to male patients (30.0% versus 23.0%, p = 0.021); however, genotype frequencies could not achieve significance due to Bonferroni's correction for multiple testing (p = 0.039) (Table S2 in File S1). Similar results were obtained from the association study of TNFB C/A SNP (Tables S3 & S4 in File S1) as the two SNPs studied were in perfect LD (linkage disequilibrium) in our population and provide the same genetic information for the associations, and so as the haplotype. This study has 84.96% statistical power for the effect size 0.08 to detect the association of investigated polymorphisms at p<0.05 in vitiligo patients and control population.
Table 2. Association studies for TNFB gene +252 A/G polymorphism in vitiligo patients and controls from Gujarat.
| SNP | Genotype or allele | Vitiligo Patients(Freq.) | Controls(Freq.) | p for Association | p for HWE | Odds ratio(95% CI) |
| rs909253(+252 A/G) | GenotypeAAAGGG | n = 524285 0.54)193 (0.37)46 (0.09) | n = 592375 (0.63)188 (0.32)29 (0.05) | 0.002a | 0.109(Patients) | |
| AlleleAG | 763 (0.73)285 (0.27) | 938 (0.79)246 (0.21) | 0.001b | 0.389(Controls) | 1.424 (1.171–1.732) |
‘n’ represents number of patients/controls, HWE refers to Hardy-Weinberg Equilibrium, CI refers to confidence interval,
vitiligo patients vs. controls using chi-square test with 3×2 contingency table,
vitiligo patients vs. controls using chi-square test with 2×2 contingency table, values are significant at p≤0.025 due to Bonferroni's correction.
Table 3. Association studies for TNFB gene +252 A/G polymorphism in different clinical types of vitiligo patients and controls from Gujarat.
| SNP | Genotype or allele | Generalized Vitiligo Patients(Freq.) | LocalizedVitiligo Patients(Freq.) | Controls(Freq.) | p for Association | p for HWE | Odds ratio(95% CI) |
| rs909253(+252 A/G) | GenotypeAAAGGGAlleleAG | n = 360184 (0.51)137 (0.38)39 (0.11)505 (0.70)215 (0.30) | n = 164103 (0.63)51 (0.31)10 (0.06)257 (0.78)71 (0.22) | n = 592375(0.63)188(0.32)29 (0.05)938(0.79)246(0.21) | 0.030a<0.0001b0.826c0.006a<0.0001b0.759c | 0.083(GV)0.286(LV)0.389(Controls) | 1.541a (1.133–2.096)1.623b (1.312–2.008)1.053c (0.782–1.419) |
| rs909253(+252 A/G) | GenotypeAAAGGGAlleleAG | ActiveVitiligo(Freq.)n = 393201 (0.51)152 (0.39)40 (0.10)554 (0.70)232 (0.30) | StableVitiligo(Freq.)n = 13185 (0.65)40 (0.30)6 (0.05)210 (0.80)52 (0.20) | Controls(Freq.)n = 592375 (0.63)188 (0.32)29 (0.05)938(0.79)246(0.21) | 0.012d<0.0001e0.945f0.002d<0.0001e0.800f | 0.163(AV)0.645(SV)0.389(Controls) | 1.691d (1.204–2.376)1.597e (1.297–1.966)0.944f (0.676–1.319) |
‘n’ represents number of patients/controls, HWE refers to Hardy-Weinberg Equilibrium, CI refers to confidence interval, GV refers to generalized vitiligo, LV refers to localized vitiligo, AV refers to active vitiligo, SV refers to stable vitiligo,
generalized vitiligo vs. localized vitiligo,
generalized vitiligo vs. controls,
localized vitiligo vs. controls,
active vitiligo vs. stable vitiligo,
active vitiligo vs. controls,
stable vitiligo vs. controls, values are significant at p≤0.025 due to Bonferroni's correction.
Effect of TNFB +252 A/G genotypes on progression of vitiligo
Analysis based on the stage of progression of vitiligo revealed that the increased frequency of the +252 G allele occurred prevalently in the group of patients with active vitiligo compared to the control group (30.00% versus 21.00%, p<0.0001) (Table 3). However, there was no statistically significant difference in the distribution of the +252 G allele between patients with stable vitiligo and control group (20.00% versus 21.00%, p = 0.800) (Table 3). Interestingly, the +252 G allele was more prevalent in patients with active vitiligo as compared to stable vitiligo (30.00% versus 20.00%, p = 0.002) suggesting the important role of +252 G allele in the progression of the disease (Table 3).
Relative gene expression of TNFB in vitiligo patients and controls
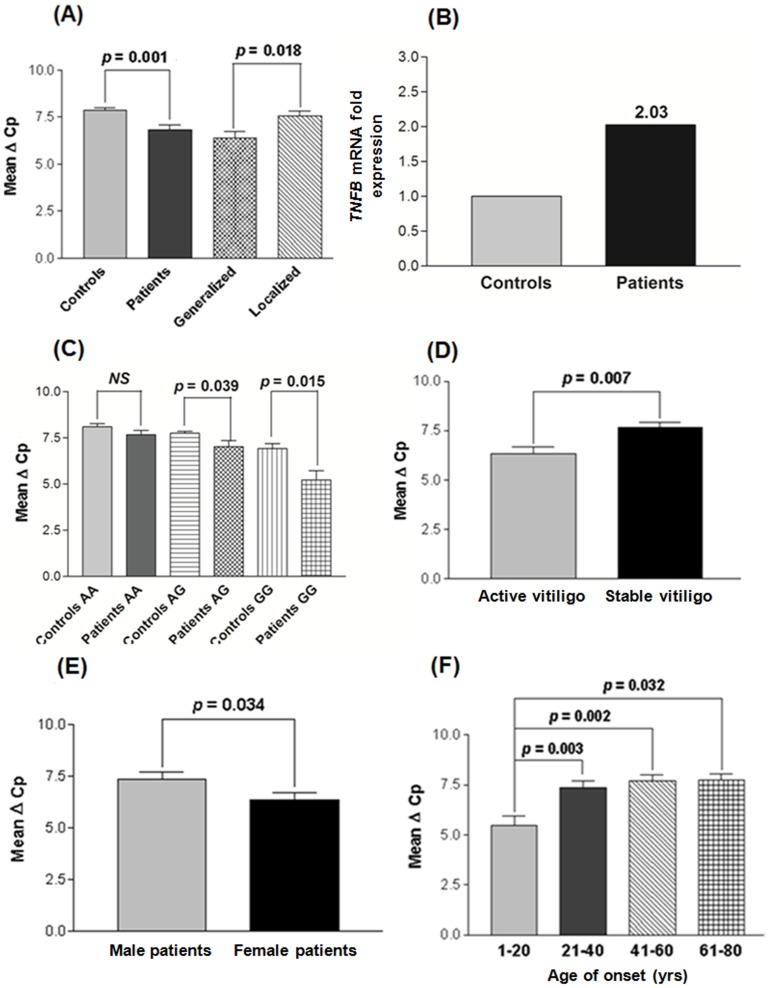
Comparison of the findings showed significant increase in expression of TNFB mRNA in 166 vitiligo patients compared to 175 unaffected controls after normalization with GAPDH expression as suggested by mean ΔCp values (p = 0.001) (Fig. 1A). Moreover, GV patients showed significantly higher expression of TNFB mRNA as compared to LV patients (p = 0.018) (Fig. 1A). The 2−ΔΔCp analysis showed approximately 2.0 fold higher expression of TNFB mRNA in patients as compared to controls (Fig. 1B).
Figure 1. Relative gene expression of TNFB in controls and vitiligo patients.
(A) Expression of TNFB mRNA in 175 controls, 166 vitiligo patients, 122 generalized vitiligo patients and 44 localized vitiligo patients using Mann-Whitney Wilcoxon test. (B) Fold expression of TNFB mRNA in 166 vitiligo patients and 175 controls, as determined by 2−ΔΔCp method. (C) Expression of TNFB mRNA with respect to TNFB +252 A/G in 166 vitiligo patients and 175 controls using Mann-Whitney Wilcoxon test. (D) Expression of TNFB mRNA with respect to activity of the disease in 121 active vitiligo and 45 stable vitiligo patients using Mann-Whitney Wilcoxon test. (E) Expression of TNFB mRNA with respect to gender differences in 87 male and 79 female vitiligo patients using Mann-Whitney Wilcoxon test. (F) Expression of TNFB mRNA with respect to different age of onset groups in 166 vitiligo patients using Mann-Whitney Wilcoxon test.
Correlation of TNFB transcripts with the TNFB +252 A/G and exon 3 C/A genotypes
Further, the expression levels of TNFB were analyzed with respect to +252 A/G genotypes (Fig. 1C). Interestingly, TNFB expression was significantly increased in patients with susceptible GG genotypes as compared to controls (p = 0.015). Also, patients with genotypes AG showed significant increase in TNFB mRNA as compared to controls (p = 0.039); however, no significant difference was observed in TNFB expression in patients as compared to controls with AA genotypes (p = 0.168) (Fig. 1C). As +252 A/G SNP is strongly linked with the exon 3 C/A SNP, similar results were obtained for TNFB expression with respect to exon 3 C/A genotypes (Fig. S4A in File S2).
Effect of TNFB expression on disease progression
In addition, we also checked the effect of TNFB expression on progression of the disease i.e. active and stable cases of vitiligo (Fig. 1D). Interestingly, active vitiligo patients showed significant increase in expression of TNFB mRNA as compared to the patients with stable vitiligo (p = 0.007) suggesting the involvement of TNFB in disease progression. Moreover, the susceptibility of the disease was also checked based on the gender differences and we found that female patients with vitiligo showed significantly higher TNFB expression as compared to male patients (p = 0.034) (Fig. 1E).
When TNFB expression was monitored in different patient groups of age of onset, patients with the age group 1–20 yrs showed significant increase in expression of TNFB mRNA as compared to the age groups 21–40, 41–60 and 61–80 yrs (p = 0.003, p = 0.002 and p = 0.032 respectively) suggesting the importance of TNFB in early onset of the disease (Fig. 1F).
Relative gene expression of ICAM1 in patients with vitiligo and controls
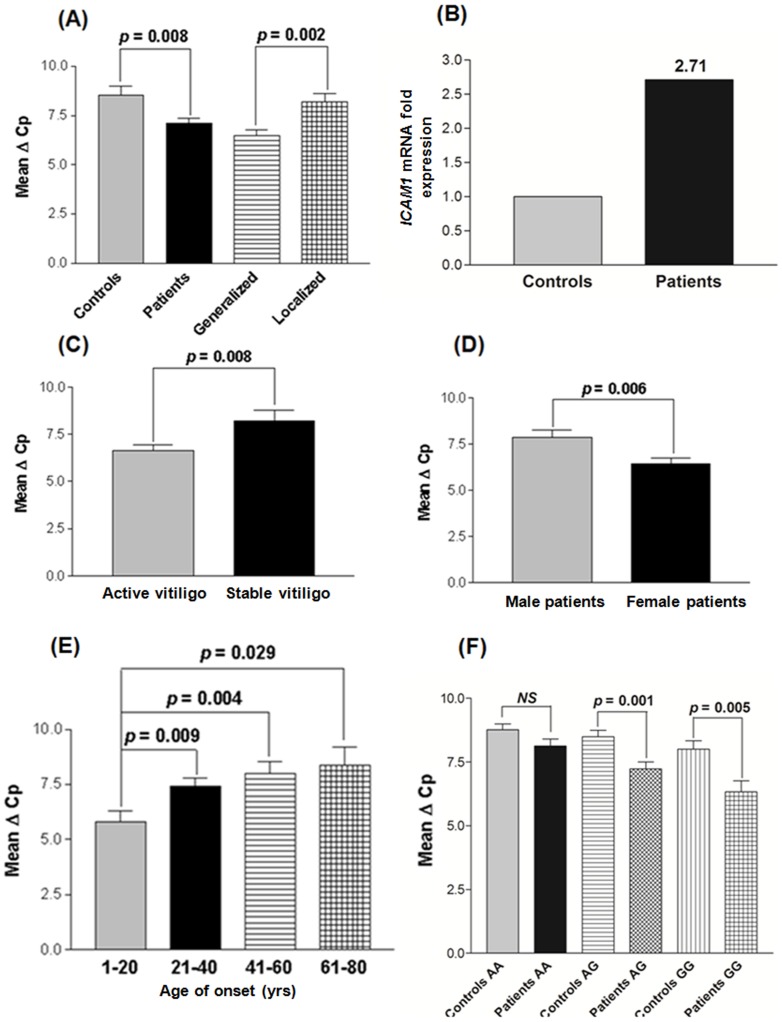
Comparison of the findings showed significant increase in expression of ICAM1 mRNA in 166 vitiligo patients than in 175 unaffected controls after normalization with GAPDH expression as suggested by mean ΔCp values (p = 0.008) (Fig. 2A). GV patients showed significantly higher mRNA levels of ICAM1 as compared to LV patients (p = 0.002) (Fig. 2A). The 2−ΔΔCp analysis showed approximately 2.7 fold higher expression of ICAM1 mRNA in patients as compared to controls (Fig. 2B).
Figure 2. Relative gene expression of ICAM1 in controls and vitiligo patients.
(A) Expression of ICAM1 mRNA in 175 controls, 166 vitiligo patients, 122 generalized vitiligo patients and 44 localized vitiligo patients using Mann-Whitney Wilcoxon test. (B) Fold expression of ICAM1 mRNA in 166 vitiligo patients and 175 controls, as determined by 2−ΔΔCp method. (C) Expression of ICAM1 mRNA with respect to activity of the disease in 121 active vitiligo and 45 stable vitiligo patients using Mann-Whitney Wilcoxon test. (D) Expression of ICAM1 mRNA with respect to gender differences in 87 male and 79 female vitiligo patients using Mann-Whitney Wilcoxon test. (E) Expression of ICAM1 mRNA with respect to different age of onset groups in 166 vitiligo patients using Mann-Whitney Wilcoxon test. (F) Expression of ICAM1 mRNA with respect to TNFB +252 A/G in 166 vitiligo patients and 175 controls using Mann-Whitney Wilcoxon test.
In addition, the effect of ICAM1 expression on progression of the disease i.e. active and stable cases (Fig. 2C) revealed that active vitiligo patients had significantly increased expression of ICAM1 mRNA as compared to patients with stable vitiligo (p = 0.008) suggesting the involvement ICAM1 in disease progression. Moreover, the susceptibility of the disease was also checked based on the gender differences and we found that female patients with vitiligo showed significantly higher ICAM1 expression as compared to male patients (p = 0.006) (Fig. 2D). When ICAM1 expression was monitored in different age of onset groups of patients, patients with the age of onset group 1–20 yrs showed significantly increased expression of ICAM1 mRNA as compared to the age groups 21–40, 41–60 and 61–80 yrs (p = 0.009, p = 0.004 and p = 0.029 respectively) suggesting the importance of ICAM1 in early onset of the disease (Fig. 2E).
Correlation of ICAM1 transcripts with the TNFB +252 A/G and exon 3 C/A genotypes
Furthermore, the effect of TNFB +252 A/G genotypes was also checked on expression of ICAM1 mRNA (Fig. 2F). Inevitably, ICAM1 expression was significantly increased in patients with susceptible GG genotypes as compared to controls (p = 0.005). Also, patients with genotypes AG showed significant increase in ICAM1 mRNA as compared to controls (p = 0.001); however, no significant difference was observed in the ICAM1 expression in patients as compared to controls with AA genotypes (p = 0.074) (Fig. 2F). As +252 A/G SNP is strongly linked with the exon 3 C/A SNP, similar results were obtained for ICAM1 expression with respect to exon 3 C/A genotypes (Fig. S4B in File S2).
Discussion
TNF plays a central role in the so-called “cytokine storm” characteristic of several autoimmune diseases. As many aspects of the cellular and humoral immune responses are under genetic control and account for individual differences in immune response patterns, there is increasing interest in genetic polymorphisms that affect inflammatory cytokines, which might explain the well-known diversity among clinical findings and outcomes in patients [26]. Therefore, genes encoding cytokines could be considered as the candidate genes for vitiligo susceptibility.
Vitiligo cannot be explained by simple Mendelian genetics, however it is characterized by incomplete penetrance, multiple susceptibility loci and genetic heterogeneity [27]. Autoimmunity has been suggested to play a major role in the pathogenesis of vitiligo since it shows frequent association with other autoimmune disorderse [6], [7]. Furthermore, these autoimmune diseases also occur at elevated frequency among close relatives of vitiligo patients, suggesting that the increased risk of vitiligo and other autoimmune disorders in these families has a genetic basis. Our previous results showed that 21.93% of Gujarat vitiligo patients exhibit positive family history and 13.68% patients have at least one first-degree relative affected [28]. The present study also shows that 12.98% of vitiligo patients have one or more first degree relative affected, suggesting the involvement of genetic factors in vitiligo pathogenesis.
The autoimmune destruction of melanocytes can be explained by the abnormalities in both humoral and cell-mediated immunity [29], [30]. Melanocyte specific circulating autoantibodies [31]–[33], autoreactive CD8+ cytotoxic T-cells and macrophages [34], [35] that recognize pigment cell antigens have been detected in the sera of a significant proportion of vitiligo patients. In particular, active cases of vitiligo were demonstrated to have higher levels of autoantibodies and cytotoxic T-cells [36]. Our recent study has also demonstrated circulating autoantibodies in the sera of 75% of Gujarat vitiligo patients as compared to unaffected individuals (Unpublished data). The autoimmune hypothesis gains further support from immunotherapy studies of melanoma patients [37]. Twenty six percent of melanoma patients responded to IL2 based immunotherapy, developed vitiligo suggesting that antimelanotic T-cells which might be responsible for melanoma regression may also be linked to the destruction of normal melanocytes in vitiligo [38].
The ultimate pathway of destruction of melanocytes in vitiligo is not known. Apoptosis is one of the cell death pathways suggested for melanocyte destruction. Cytokines such as IFNG, TNFA, TNFB and IL1 released by lymphocytes and keratinocytes can initiate apoptosis [22]. Recently, we have shown increased TNFA protein and transcript levels in vitiligo patients, suggesting an early apoptosis of melanocytes [18]. Moreover, imbalance of cytokines in the epidermal microenvironment of lesional vitiligo skin has been demonstrated, which could impair the life span and function of melanocytes [11], [12], [39], [40].
TNFA and TNFB polymorphisms are reported to be associated with the inflammatory and immunomodulatory responses and are involved in the modulation of gene expression, and thus affect the precipitation and progression of the autoimmune diseases [19], [21], [41]–[43]. The +252 A/G polymorphism of TNFB is present in intron 1 and linked to an amino acid substitution at position 26 of the TNFB protein, the substitution being conserved as asparagine in the TNFB +252 G and as threonine in the TNFB +252 A allele [19]. The TNFB +252 G allele has been shown to be associated with increased expression and the TNFB +252 A allele with decreased production of the TNFB protein [19]. The TNFA -308 A and the TNFB +252 G alleles are constituents of one of the extended ancestral haplotypes (AH8.1) located in the chromosomal region 6p21.3–21.1 (MHC). This haplotype is known to be associated with serious disorders of the immune system [44]–[46]. The TNFB +252 G allele was also associated with several autoimmune diseases such as type 1 diabetes mellitus [47], [48] and systemic lupus erythematosus [49], [50]. The present study for the first time reports significant association of TNFB +252 A/G polymorphism with vitiligo susceptibility. In particular, we found that +252 G allele was prevalent in GV patients as compared to LV patients and healthy controls. We also found that +252 G allele was prevalent in active cases of vitiligo as compared to patients with stable vitiligo and controls suggesting the involvement of the allele in the progression of the disease. Moreover, +252 G allele was significantly associated with female patients as compared to male patients suggesting that females are more prone to autoimmune diseases.
The whole TNFB gene is in strong linkage disequilibrium (LD), therefore the +252 G allele naturally coexists with the 804 A allele of the exon 3 C/A polymorphism (Thr26Asn) [51]. We also found that the two SNPs studied are in perfect LD in our population and provide the same genetic information for the associations, and so as the haplotype. Ozaki et al. [21] investigated the functionality of the +252 A/G and 804 C/A SNPs in the TNFB gene. The 804 C/A polymorphism causes an amino-acid change from threonine (T) to asparagine (N) at codon 26. They found that the variant protein 26 N is associated with a two fold increase in the induction of cell adhesion molecules in vascular smooth muscle cells [21].
The TNFB +252 A/G and exon 3 C/A SNPs could affect susceptibility to vitiligo through its influence on the production of TNFA and TNFB. As TNFA and TNFB genes are tandemly arranged within the HLA complex, and it has been shown that the TNFB gene polymorphisms influence the level of production of the TNFA protein [19], [52]. Studies indicate that variations at both TNFA -308 G/A and TNFB +252 A/G can affect TNFA production levels [21], [53]. The exon 3 C/A SNP is in strong LD with +252 A/G SNP of intron 1 and the combination of these allelic forms may lead to different levels of TNFA production in response to various physiological and pathological stimuli [54]. This results in variation in predisposition to vitiligo and is also involved in different clinical aspects of the disease. Japanese and Danish studies showed the polymorphism in the TNFB gene (exon 3 C/A) to be associated with increased susceptibility to type 2 diabetes mellitus (T2DM) [55], [56].
Pociot et al. [57] reported that TNFA levels were highest in TNFB +252GG homozygotes, lowest in the TNFB +252AA homozygotes, and intermediate in the TNFB +252AG heterozygous individuals. In vitro, direct analysis of skin T-cells from the margins of vitiliginous skin showed that polarized type-1 T cells (CD4+ and particularly CD8+), which predominantly secrete IFNG and TNFA are associated with the destruction of melanocytes during active vitiligo [58]. In vitiligo affected skin, a significantly higher expression of TNFA, IL6, and IFNG was detected compared with healthy controls and perilesional, non-lesional skin [11], [12], [39] indicating that cytokine imbalance plays an important role in the depigmentation process of vitiligo. Since elevated levels of TNFA have been associated with vitiligo, this could explain the association of TNFB +252 G allele with susceptibility to vitiligo in the present study. Homozygosity for the TNFB +252 A allele will be protective because of its association with lower TNFA and TNFB levels. Previously, Hasegawa et al. [59] also suggested the correlation of TNFB +252 A/G polymorphism with elevated levels of TNFA in pulmonary fibrosis in scleroderma. TNFB +252GG homozygosity has also been shown to be associated with susceptibility to systemic lupus erythematosus in some populations from Germany, Korea, and China [60]–[63].
There are no reports available for TNFB gene expression in vitiligo patients till date. Hence, the present study also focuses on the TNFB transcript levels in vitiligo patients. Vitiligo patients showed significant increase in TNFB transcript levels as compared to controls suggesting that melanocyte death in patients could be triggered due to the increased TNFB levels. For the first time we report that GV has significantly higher TNFB transcript levels as compared to LV patients which indicate involvement of autoimmunity in precipitation of GV. Our results also indicate that active vitiligo patients had significantly higher TNFB transcript levels as compared to patients with stable vitiligo, which signifies the role of TNFB in disease progression. Our results also suggest that there are significantly higher transcript levels of TNFB in female patients as compared to male patients suggesting that females have increased susceptibility towards vitiligo as compared to males, implicating gender biasness in the development of autoimmunity [64]–[65]. Furthermore, the genotype-phenotype analysis for +252 A/G polymorphism indicated that patients with GG and AG genotypes had higher expression of TNFB transcripts suggesting the crucial role of +252 G allele in pathogenesis of vitiligo. Also, the genotype-phenotype analysis of TNFB exon 3 C/A showed that patients harboring AA and CA genotypes had higher expression of TNFB suggesting that allele ‘A’ plays an important role in increased expression of TNFB. Hence, genetic association studies with TNFB polymorphisms in different ethnic populations need to be explored.
TNFA and TNFB also have the capacity to inhibit melanogenesis through an inhibitory effect on tyrosinase and tyrosinase related protein [66]. In vitro direct analysis of vitiliginous skin margins showed the presence of polarized CD4+ and CD8+ T-cells, which predominantly secrete IFNG, TNFA and TNFB, might be associated with the destruction of melanocytes during active disease [67]. Our recent study has also shown dramatic increase in CD8+ T-cell number and significant decrease in Tregs number in circulation of active vitiligo patients [68].
Furthermore, IFNG, TNFA and TNFB stimulate the expression of ICAM1 on the cell-surface of melanocytes [23], which is important for activating T-cells and recruiting leukocytes [69], [70]. ICAM1 protein levels are upregulated in vitiligo skin and in melanocytes from perilesional vitiligo skin [71], [24]. The present study also showed increased expression of ICAM1 in vitiligo patients suggesting that increased TNFB levels might be responsible for increased ICAM1 expression in vitiligo patients and our results are in concordance with the previous reports [24], [70], [72], [73]. In addition, we found that ICAM1 expression was increased with the susceptible TNFB +252GG and exon 3 AA genotypes in patients suggesting the crucial role of TNFB and its SNPs in altered ICAM1 expression. It has been reported that increased expression of ICAM1 on the melanocytes enhances T cell -melanocyte attachment in the skin and may lead to the destruction of melanocytes in vitiligo [24], [71]. Moreover, the ICAM1 expression was increased in active cases of vitiligo as compared to stable vitiligo suggesting its role in the progression of the disease. The ICAM1 expression was increased with early age of onset of the disease further implicating the important role of ICAM1 in the early phase of the disease. Also, female patients showed an increased expression of ICAM1 as compared to male patients suggesting that females have more susceptibility towards vitiligo.
Recently, we have shown positive association of HLA-A*33:01, HLA-B*44:03, and HLA-DRB1*07:01 with vitiligo patients from North India and Gujarat suggesting an autoimmune link of vitiligo in these cohorts [74]. We also showed that the three most significant class II region SNPs: rs3096691 (just upstream of NOTCH4), rs3129859 (just upstream of HLA-DRA), and rs482044 (between HLA-DRB1 and HLA-DQA1) are associated with GV [75]. The genotype-phenotype correlation of TNFA, CTLA4, IL4, MYG1, IFNG and NALP1 gene polymorphisms supported the autoimmune pathogenesis of vitiligo in Gujarat population [18], [76]–[80]; whereas our earlier studies on CAT, MBL2, ACE, PTPN22 polymorphisms did not show significant association [81]–[84]. Recently, we have shown that SOD2 and SOD3 polymorphisms may be genetic risk factors for susceptibility and progression of vitiligo and contribute to increased transcript levels and activity of SOD2 and SOD3 in patients [85]. It is worth mentioning that TNFB is one of the major transcriptional activator of SOD2 and SOD3 genes in different tissues and cell types [86], [87]. Hence, increased TNFB production might be responsible for the increased SOD2 and SOD3 transcripts [85] which may lead to accumulation of H2O2 in mitochondrial and extracellular compartments resulting in oxidative damage to the melanocytes.
In conclusion, our findings suggest that the increased TNFB levels in vitiligo patients could result, at least in part, from variations at the genetic level. Moreover, the increased TNFB levels in patients may lead to increased ICAM1 expression which is probably an important link between cytokines and T cells involved in vitiligo pathogenesis. For the first time, we show that the +252 A/G and exon 3 C/A polymorphisms of the TNFB gene are associated with vitiligo and influence the TNFB expression levels. The study also emphasizes the influence of TNFB on the disease progression, onset of the disease and gender biasness for developing vitiligo. More detailed studies regarding the role of TNFB in precipitation of vitiligo and the development of effective anti-TNF agents may prove to be useful as preventive/ameliorative therapies.
Supporting Information
Supplementary Tables. Table S1. Primers and restriction enzymes used for TNFB +252G/A SNP genotyping and gene expression analyses. Table S2. Association studies for TNFB gene +252A/G polymorphism in male and female vitiligo patients from Gujarat. Table S3. Association studies for TNFB gene exon 3 C/A polymorphism in vitiligo patients and controls from Gujarat. Table S4. Association studies for TNFB gene exon 3 C/A polymorphism in different clinical types of vitiligo patients and controls from Gujarat.
(DOC)
Supplementary Figures. Figure S1. PCR-RFLP analysis of TNFB +252A/G polymorphism on 2.0 % agarose gel electrophoresis: lanes: 1, 3, 4, 5 & 7 show homozygous (GG) genotypes; lane: 2 shows homozygous (AA) genotype; lane: 6 shows heterozygous (AG) genotype. Figure S2. TaqMan end point fluoroscence analysis for TNFB C/A; (Thr26Asn) polymorphism using dual color hydrolysis probes (FAM and VIC) by LightCycler®480Real-Time PCR protocol. The three genotypes identified as: AA, AC and CC, based on fluorescence with Channel 465-510 (FAM for ‘A’ allele) and Channel 536-580 (VIC for ‘C’ allele). A no-template control (NTC) was used with each SNP genotyping assay (shown as grey spot). Figure S3. Melt curve analysis of TNFB, ICAM1 and GAPDH showing specific amplification. Figure S4. Relative gene expression of TNFB and ICAM1 with respect to TNFB exon 3 C/A in controls and vitiligo patients: (A) Expression of TNFB mRNA with respect to TNFB exon 3 C/A in 166 vitiligo patients and 175 controls using Mann-Whitney Wilcoxon test [MeanΔCp ± SEM: Controls CC vs Patients CC: 8.102 ± 0.1747 vs 7.678 ± 0.2379 (p = 0.168); Controls CA vs Patients CA; 7.761 ± 0.0952 vs 7.028 ± 0.3376 (p = 0.039); Controls AA vs Patients AA: 6.918 ± 0.2808 vs 5.217 ± 0.5087 (p = 0.015) (NS = non-significant)]. (B) Expression of ICAM1 mRNA with respect to TNFB exon 3 C/A in 166 vitiligo patients and 175 controls using Mann-Whitney Wilcoxon test [MeanΔCp ± SEM; Controls CC vs Patients CC: 8.767 ± 0.2267 vs 8.133 ± 0.2658 (p = 0.074); Controls CA vs Patients CA: 8.494 ± 0.2434 vs 7.225 ± 0.2729 (p = 0.001); Controls AA vs Patients AA: 8.007 ± 0.3297 vs 6.327 ± 0.4397 (p = 0.005).
(DOCX)
Acknowledgments
We sincerely thank all the vitiligo patients and healthy unaffected controls for their participation in this study. NCL thanks CSIR, New Delhi for SRF and MSM thanks UGC, New Delhi for JRF.
Funding Statement
This work was supported by grants to RB (BMS/Adhoc/122/11-2012) ICMR, New Delhi, India; (GSBTM/MD/PROJECTS/SSA/453/2010-2011), GSBTM, Gandhinagar, Gujarat, India and (BT/PR9024/MED/12/332/2007) DBT, New Delhi, India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ortonne JP, Bose SK (1993) Vitiligo: Where do we stand? Pigment Cell Res 8: 61–72. [DOI] [PubMed] [Google Scholar]
- 2. Majumder PP, Nordlund JJ, Nath SK (1993) Pattern of familial aggregation of vitiligo. Arch Dermatol 129: 994–998. [PubMed] [Google Scholar]
- 3. Nath SK, Majumder PP, Nordlund JJ (1994) Genetic epidemiology of vitiligo: multilocus recessivity cross-validated. Am J Hum Genet 55: 981–990. [PMC free article] [PubMed] [Google Scholar]
- 4. Sun X, Xu A, Wei X, Ouyang J, Lu L, et al. (2006) Genetic epidemiology of vitiligo: a study of 815 probands and their families from south China. Int J Dermatol 45: 1176–1181. [DOI] [PubMed] [Google Scholar]
- 5. Taieb A, Picardo M (2007) VETF Members (2007) The definition and assessment of vitiligo: a consensus report of the Vitiligo European Task Force. Pigment Cell Res 20: 27–35. [DOI] [PubMed] [Google Scholar]
- 6. Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA (2003) Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16: 208–214. [DOI] [PubMed] [Google Scholar]
- 7. Laberge G, Mailloux CM, Gowan K, Holland P, Bennett DC, et al. (2005) Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res 18: 300–305. [DOI] [PubMed] [Google Scholar]
- 8. Spritz RA (2007) The genetics of generalized vitiligo and associated autoimmune diseases. Pigment Cell Res 20: 271–278. [DOI] [PubMed] [Google Scholar]
- 9. Spritz RA (2008) The genetics of generalized vitiligo. Curr Dir Autoimmunity 10: 244–257. [DOI] [PubMed] [Google Scholar]
- 10. Feldmann M, Brennan FM, Maini R (1998) Cytokines in autoimmune disorders. Int Rev Immunol 17: 217–228. [DOI] [PubMed] [Google Scholar]
- 11. Moretti S, Spallanzani A, Amato L, Hautmann G, Gallerani I, et al. (2002) New insights into the pathogenesis of vitiligo: imbalance of epidermal cytokines at sites of lesions. Pigment Cell Res 15: 87–92. [DOI] [PubMed] [Google Scholar]
- 12. Moretti S, Spallanzani A, Amato L, Hautmann G, Gallerani I, et al. (2002) Vitiligo and epidermal microenvironment: possible involvement of keratinocyte-derived cytokines. Arch. Dermatol 138: 273–274. [DOI] [PubMed] [Google Scholar]
- 13. Smith CA (1994) The TNF receptor super family of cellular and viral proteins-activation, co stimulation and death. Cell 76: 959–962. [DOI] [PubMed] [Google Scholar]
- 14. Vassalli P (1992) The pathophysiology of tumor necrosis factors. Annu Rev Immunol 10: 411–452. [DOI] [PubMed] [Google Scholar]
- 15. Funahashi I, Kawatsu M, Kajikawa T, Takeo K, Asahi T, et al. (1991) Usefulness of glycosylated and recombinant human lymphotoxin for growth inhibition of human and murine solid tumors and experimental metastasis in mice. J Immunother 10: 28–38. [DOI] [PubMed] [Google Scholar]
- 16. Lucas R, Lou J, Morel DR, Ricou B, Suter PM, et al. (1997) TNF receptors in the microvascular pathology of acute respiratory distress syndrome and cerebral malaria. J Leukocyte Biol 61: 551–558. [DOI] [PubMed] [Google Scholar]
- 17. Beutler B, Bazzoni F (1998) TNF, apoptosis and autoimmunity: a common thread? Blood Cells Mol Dis 24: 216–230. [DOI] [PubMed] [Google Scholar]
- 18. Laddha NC, Dwivedi M, Begum R (2012) Increased Tumor Necrosis Factor (TNF)-α and its promoter polymorphisms correlate with disease progression and higher susceptibility towards vitiligo. PLoS ONE 7: e52298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Messer G, Spengler U, Jung MC, Honold G, Blomer K, et al. (1991) Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-b gene correlates with a variant amino acid in position 26 and a reduced level of TNF-b production. J Exp Med 173: 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whichelow CE, Hitman GA, Raafat I, Bottazzo GF, Sachs JA (1996) The effect of TNF*B gene polymorphism on TNF-alpha and –beta secretion level in patients with insulin dependent diabetes mellitus and healthy controls. Eur J Immunogenet 23: 425–435. [DOI] [PubMed] [Google Scholar]
- 21. Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, et al. (2002) Functional SNPs in the lymphotoxin alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 32: 650–654. [DOI] [PubMed] [Google Scholar]
- 22. Huang CL, Nordlund JJ, Boissy R (2002) Vitiligo: A manifestation of apoptosis. Am J Clin Dermatol 3: 301–308. [DOI] [PubMed] [Google Scholar]
- 23. Yohn JJ, Critelli M, Lyons MB, Norris DA (1990) Modulation of melanocyte intercellular adhesion molecule-1 by immune cytokines. J Invest Dermatol 90: 233–237. [DOI] [PubMed] [Google Scholar]
- 24. Al Badri AM (1993) Abnormal expression of MHC class ll and ICAM-l by melanocytes in vitiligo. J Pathol 169: 203–206. [DOI] [PubMed] [Google Scholar]
- 25. Faul F, Erdfelder E, Lang AG, Buchner A (2007) G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 39: 175–191. [DOI] [PubMed] [Google Scholar]
- 26. Gottenberg JE, Busson M, Loiseau P, Dourche M, Cohen-Solal J, et al. (2004) Association of transforming growth factor-β1 and tumor necrosis factor alpha polymorphisms with anti-SSB/La antibody secretion in patients with primary Sjogren's syndrome. Arthritis Rheum 50: 570–580. [DOI] [PubMed] [Google Scholar]
- 27. Zhang XJ, Chen JJ, Liu JB (2005) The genetic concept of vitiligo. J Dermatol Sci 39: 137–146. [DOI] [PubMed] [Google Scholar]
- 28. Shajil EM, Agrawal D, Vagadia K, Marfatia YS, Begum R (2006) Vitiligo: Clinical profiles in Vadodara, Gujarat. Ind J Dermatol 51: 100–104. [Google Scholar]
- 29. Shajil EM, Chatterjee S, Agrawal D, Bagchi T, Begum R (2006) Vitiligo: Pathomechanisms and genetic polymorphism of susceptible genes. Ind J Exp Biol 44: 526–539. [PubMed] [Google Scholar]
- 30. Kemp EH, Waterman EA, Weetman AP (2001) Immunological pathomechanisms in vitiligo. Expert Rev Mol Med 23: 1–22. [DOI] [PubMed] [Google Scholar]
- 31. Cui J, Bystryn JC (1995) Melanoma and vitiligo are associated with antibody responses to similar antigens on pigment cells. Arch Dermatol 131: 314–318. [PubMed] [Google Scholar]
- 32. Harning R, Cui J, Bystryn JC (1991) Relation between the incidence and level of pigment cell antibodies and disease activity in vitiligo. J Invest Dermatol 97: 1078–1080. [DOI] [PubMed] [Google Scholar]
- 33. Kemp EH, Gawkrodger DJ, MacNeil S, Watson PF, Weetman AP (1997) Detection of tyrosinase autoantibodies in patients with vitiligo using 35Slabeled recombinant human tyrosinase in a radioimmunoassay. J Invest Dermatol 109: 69–73. [DOI] [PubMed] [Google Scholar]
- 34. Le Gal FA, Avril M, Bosq J, Lefebvre P, Deschemin JC, et al. (2001) Direct evidence to support the role of antigen-specific CD8 (+) T-cells in melanoma-associated vitiligo. J Invest Dermatol 117: 1464–1470. [DOI] [PubMed] [Google Scholar]
- 35. Mandelcorn-Monson RL, Shear NH, Yau E, Sambhara S, Barber BH, et al. (2003) Cytotoxic T lymphocyte reactivity to gp100, MelanA/MART-1, and tyrosinase, in HLA-A2- positive vitiligo patients. J Invest Dermatol 121: 550–556. [DOI] [PubMed] [Google Scholar]
- 36. Le Poole I, Luiten R (2008) Autoimmune etiology of generalized vitiligo. Curr Dir Autoimmun 10: 227–243. [DOI] [PubMed] [Google Scholar]
- 37. Rosenberg SA (1997) Cancer vaccines based on the identification of genes encoding cancer regression antigens. Immunol Today 18: 175–182. [DOI] [PubMed] [Google Scholar]
- 38. Zeff RA, Freitag A, Grin CM, Grant-Kels JM (1997) The immune response in halo nevi. J Am Acad Dermatol 37: 620–624. [DOI] [PubMed] [Google Scholar]
- 39. Grimes PE, Morris R, Avaniss-Aghajani E, Soriano T, Meraz M, et al. (2004) Topical tacrolimus therapy for vitiligo: therapeutic responses and skin messenger RNA expression of proinflammatory cytokines. J Am Acad Dermatol 51: 52–61. [DOI] [PubMed] [Google Scholar]
- 40. Laddha NC, Dwivedi M, Mansuri MS, Gani AR, Ansarullah, et al (2013) Vitiligo: interplay between oxidative stress and immune system. Exp Dermatol 22: 245–250. [DOI] [PubMed] [Google Scholar]
- 41. Kaluza W, Reuss E, Grossmann S, Hug R, Schoopf RE, et al. (2000) Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNFalpha 238A promoter polymorphism. J Invest Dermatol 114: 1180–1183. [DOI] [PubMed] [Google Scholar]
- 42. Wilson A, Symons J, McDowell TL, McDevitt HO, Duff GW (1997) Effects of a polymorphism in the human tumor necrosis factor-a promoter on transcriptional activation. Proc Natl Acad Sci USA 94: 3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gonza' lez S, Rodrigo L, Martı'nez-Borra J, Lo‘pez-Va’ zquez A, Fuentes D, et al. (2003) TNF-a -308A promoter polymorphism is associated with enhanced TNF-a production and inflammatory activity in Crohn's patients with fistulizing disease. Am J Gastroenterol 98: 1101–1106. [DOI] [PubMed] [Google Scholar]
- 44. Dawkins RL, Christiansen FT, Kay PH, Garlepp M, McCluskey J, et al. (1983) Disease associations with complotypes, supratypes and haplotypes. Immunol Rev 70: 1–22. [DOI] [PubMed] [Google Scholar]
- 45. Candore G, Lio D, Colonna Romano G, Caruso C (2002) Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev 1: 29–35. [DOI] [PubMed] [Google Scholar]
- 46. Candore G, Modica MA, Lio D, Colonna-Romano G, Listi F, et al. (2003) Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: a genetically determined defect of C4 influences immunological parameters of healthy carriers of the haplotype. Biomed Pharmacother 57: 274–277. [DOI] [PubMed] [Google Scholar]
- 47. Dawkins RL, Spies T, Strominger JL, Hansen JA (1989) Some disease-associated ancestral haplotype carry a polymorphism of TNF. Hum Immunol 26: 91–94. [DOI] [PubMed] [Google Scholar]
- 48. Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, et al. (2006) Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci USA 103: 14074–14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smerdel-Ramoya A, Finholt C, Lilleby V, Gilboe IM, Harbo HF, et al. (2005) Systemic lupus erythematosus and the extended major histocompatibility complex- evidence for several predisposing loci. Rheumatology (Oxford) 44: 1368–1373. [DOI] [PubMed] [Google Scholar]
- 50. McHugh NJ, Owen P, Cox B, Dunphy J, Welsh K (2006) MHC Class II, tumour necrosis factor alpha, and lymphotoxin alpha gene haplotype associations with serological subsets of systemic lupus erythematosus. Ann Rheum Dis 65: 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clarke R, Xu P, Bennett D, Lewington S, Zondervan K, et al. (2006) Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet 2: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kobayashi Y, Miyomotu D, Asada M, Obinata M, Osawa T (1986) Cloning and expression of human lymphotoxin mRNA derived from a human T cell hydrydoma. J Biochem 100: 727–730. [DOI] [PubMed] [Google Scholar]
- 53. Abraham LJ, Kroeger KM (1999) Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol 66: 562–566. [DOI] [PubMed] [Google Scholar]
- 54. Steinman L (1995) Escape from ‘horror autotoxicus’: pathogenesis and treatment of autoimmune disease. Cell 80: 7–10. [DOI] [PubMed] [Google Scholar]
- 55. Yamada A, Ichihara S, Murase Y, Kato T, Izawa H, et al. (2004) Lack of association of polymorphisms of the lymphotoxin alpha gene with myocardial infarction in Japanese. J Mol Med 82: 477–483. [DOI] [PubMed] [Google Scholar]
- 56. Hamid YH, Urhammer SA, Glqmer C, Borch-Johnsen K, Jørgensen T, et al. (2005) The common T60N polymorphism of the lymphotoxin-alpha gene is associated with type 2 diabetes and other phenotypes of the metabolic syndrome. Diabetologia 48: 445–451. [DOI] [PubMed] [Google Scholar]
- 57. Pociot F, Briant L, Jongeneel CV, Molvig J, Worsaae H, et al. (1993) Association of tumor necrosis factor (TNF) and class II major histocompatibility complex alleles with the secretion of TNF-a and TNF-b by human mononuclear cells: a possible link to insulin-dependent diabetes mellitus. Eur J Immunol 23: 224–231. [DOI] [PubMed] [Google Scholar]
- 58. Wajkowicz-Kalijska A, van den Wijngaard RM, Tigges BJ, Westerhof W, Ogg GS, et al. (2003) Immunopolarization of CD4+ and CD8+ T cells to Type-1-like is associated with melanocyte loss in human vitiligo. Lab Invest 83: 683–695. [DOI] [PubMed] [Google Scholar]
- 59. Hasegawa M, Fujimoto M, Kikuchi K, Takehara K (1983) Elevated serum tumor necrosis factor-a levels in patients with systemic sclerosis: association with pulmonary fibrosis. J Rheumatol 24: 663–665. [PubMed] [Google Scholar]
- 60. Bettinotti MP, Hartung K, Deicher H, Messer G, Keller E, et al. (1993) Polymorphism of the tumor necrosis factor beta gene in systemic lupus erythematosus: TNFB MHC haplotypes. Immunogenetics 37: 449–454. [DOI] [PubMed] [Google Scholar]
- 61. Kim TG, Kim HY, Lee SH, Cho CS, Park SH, et al. (1996) Systemic lupus erythematosus with nephritis is strongly associated with the TNFB*2 homozygote in the Korean population. Hum Immunol 46: 10–17. [DOI] [PubMed] [Google Scholar]
- 62. Lee SH, Park SH, Min JK, Kim SI, Yoo WH, et al. (1997) Decreased tumour necrosis factor-beta production in TNFB*2 homozygote: an important predisposing factor of lupus nephritis in Koreans. Lupus 6: 603–609. [DOI] [PubMed] [Google Scholar]
- 63. Zhang J, Ai R, Chow F (1997) The polymorphisms of HLA-DR and TNF B loci in northern Chinese Han nationality and susceptibility to systemic lupus erythematosus. Chin Med Sci J 12: 107–110. [PubMed] [Google Scholar]
- 64. Whitacre CC (2001) Sex differences in autoimmune disease. Nat Immunol 2: 777–780. [DOI] [PubMed] [Google Scholar]
- 65. Panchanathan R, Choubey D (2012) Murine BAFF expression is up-regulated by estrogen and interferons: Implications for sex bias in the development of autoimmunity. Mol Immunol 53: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Martinez-Esparza M, Jimenez-Cervantes C, Solano F, Lozano JA, Garcia-Borron JC (1998) Mechanisms of melanogenesis inhibition by tumor necrosis factor-α in B16–F10 mouse melanoma cells. Eur J Biochem 255: 139–146. [DOI] [PubMed] [Google Scholar]
- 67. Wajkowicz-Kalijska A, van den Wijngaard RM, Tigges BJ, Westerhof W, Ogg GS, et al. (2003) Immunopolarization of CD4+ and CD8+ T cells to Type-1-like is associated with melanocyte loss in human vitiligo. Lab Invest 83: 683–695. [DOI] [PubMed] [Google Scholar]
- 68. Dwivedi M, Laddha NC, Arora P, Marfatia YS, Begum R (2013) Decreased regulatory T-Cells and CD4+/CD8+ ratio correlate with disease onset and progression in patients with generalized vitiligo. Pigment Cell Melanoma Res 26: 586–591. [DOI] [PubMed] [Google Scholar]
- 69. Hedley SJ, Metcalfe R, Gawkrodger DJ, Weetman AP, Mac Neil S (1998) Vitiligo melanocytes in long-term culture show normal constitutive and cytokine-induced expression of intercellular adhesion molecule-1 and major histocompatibility complex class I and class II molecules. Br J Dermatol 139: 965–973. [DOI] [PubMed] [Google Scholar]
- 70. Ahn SK, Choi EH, Lee SH, Won JH, Hann SK, et al. (1994) Immunohistochemical studies from vitiligo- comparison between active and inactive lesions. Yonsei Med J 35: 404–410. [DOI] [PubMed] [Google Scholar]
- 71. Morelli JG, Norris DA (1993) Influence of inflammatory mediators and cytokines on human melanocyte function. J Invest Dermatol 100: 191–195. [PubMed] [Google Scholar]
- 72. Reimann E, Kingo K, Karelson M, Reemann P, Loite U, et al. (2012) The mRNA expression profile of cytokines connected to the regulation of melanocyte functioning in vitiligo skin biopsy samples and peripheral blood mononuclear cells. Hum Immunol 73: 393–398. [DOI] [PubMed] [Google Scholar]
- 73. Yagi H, Tokura Y, Furukawa F, Takigawa M (1997) Vitiligo with raised inflammatory borders: involvement of T cell immunity and keratinocytes expressing MHC class II and ICAM-1 molecules. Eur J Dermatol Clin 7: 19–22. [Google Scholar]
- 74. Singh A, Sharma P, Kar HK, Sharma VK, Tembhre MK, et al. (2012) HLA alleles and amino acid signatures of the peptide binding pockets of HLA molecules in Vitiligo. J Invest Dermatol 132: 124–134. [DOI] [PubMed] [Google Scholar]
- 75. Birlea SA, Ahmad FJ, Uddin RM, Ahmad S, Pal SS, et al. (2013) Association of Generalized Vitiligo with HLA Class II Loci in Patients from the Indian Subcontinent. J Invest Dermatol 133: 1369–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dwivedi M, Laddha NC, Imran M, Shah BJ, Begum R (2011) Cytotoxic T-lymphocyte associated antigen-4 (CTLA-4) in isolated vitiligo: a genotype-phenotype correlation. Pigment cell Melanoma Res 24: 737–740. [DOI] [PubMed] [Google Scholar]
- 77. Imran M, Laddha NC, Dwivedi M, Mansuri MS, Singh J, et al. (2012) Interleukin-4 genetic variants correlate with its transcript and protein levels in vitiligo patients. Brit J Dermatol 167: 314–323. [DOI] [PubMed] [Google Scholar]
- 78. Dwivedi M, Laddha NC, Begum R (2013) Correlation of increased MYG1 expression and its promoter polymorphism with disease progression and higher susceptibility in vitiligo patients. J Dermatol Sci 71: 195–202. [DOI] [PubMed] [Google Scholar]
- 79.Dwivedi M, Laddha NC, Shah K, Shah BJ, Begum R (2013) Involvement of interferon-gamma genetic variants and intercellular adhesion molecule-1 in disease onset and progression of generalized vitiligo. J Interferon Cytokine Res. doi:10.1089/jir.2012.0171. [DOI] [PMC free article] [PubMed]
- 80.Dwivedi M, Laddha NC, Mansuri MS, Marfatia YS, Begum R (2013) Association of NLRP1 genetic variants and mRNA overexpression with generalized vitiligo and disease activity in a Gujarat population. Brit J Dermatol. doi: 10.1111/bjd.12467. [DOI] [PubMed]
- 81. Shajil EM, Laddha NC, Chatterjee S, Gani AR, Malek RA, et al. (2007) Association of catalase T/C exon 9 and glutathione peroxidase codon 200 polymorphisms in relation to their activities and oxidative stress with vitiligo susceptibility in Gujarat population. Pigment Cell Res 20: 405–407. [DOI] [PubMed] [Google Scholar]
- 82. Dwivedi M, Gupta K, Gulla KC, Laddha NC, Hajela K, et al. (2009) Lack of genetic association of promoter and structural variants of Mannan-binding lectin (MBL2) gene with susceptibility to generalized vitiligo. Brit J Dermatol 161: 63–69. [DOI] [PubMed] [Google Scholar]
- 83. Dwivedi M, Laddha NC, Shajil EM, Shah BJ, Begum R (2008) The ACE gene I/D polymorphism is not associated with generalized Vitiligo Susceptibility in Gujarat population. Pigment Cell Melanoma Res 21: 407–408. [DOI] [PubMed] [Google Scholar]
- 84. Laddha NC, Dwivedi M, Shajil EM, Prajapati H, Marfatia YS, et al. (2008) Association of PTPN22 1858C/T polymorphism with vitiligo susceptibility in Gujarat population. J Dermatol Sci 49: 260–262. [DOI] [PubMed] [Google Scholar]
- 85. Laddha NC, Dwivedi M, Gani AR, Shajil EM, Begum R (2013) Involvement of superoxide dismutase isoenzymes and their genetic variants in progression of and higher susceptibility to vitiligo. Free Radic Biol Med 65: 1110–1125. [DOI] [PubMed] [Google Scholar]
- 86. Wong GH, Goeddel DV (1988) Induction of manganous superoxide dismutase by tumor necrosis factor: possible protective mechanism. Science 242: 941–944. [DOI] [PubMed] [Google Scholar]
- 87. Marklund SL (1992) Regulation by cytokines of extracellular superoxide dismutase and other superoxide dismutase isoenzymes in fibroblasts. J Biol Chem 267: 6696–6701. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables. Table S1. Primers and restriction enzymes used for TNFB +252G/A SNP genotyping and gene expression analyses. Table S2. Association studies for TNFB gene +252A/G polymorphism in male and female vitiligo patients from Gujarat. Table S3. Association studies for TNFB gene exon 3 C/A polymorphism in vitiligo patients and controls from Gujarat. Table S4. Association studies for TNFB gene exon 3 C/A polymorphism in different clinical types of vitiligo patients and controls from Gujarat.
(DOC)
Supplementary Figures. Figure S1. PCR-RFLP analysis of TNFB +252A/G polymorphism on 2.0 % agarose gel electrophoresis: lanes: 1, 3, 4, 5 & 7 show homozygous (GG) genotypes; lane: 2 shows homozygous (AA) genotype; lane: 6 shows heterozygous (AG) genotype. Figure S2. TaqMan end point fluoroscence analysis for TNFB C/A; (Thr26Asn) polymorphism using dual color hydrolysis probes (FAM and VIC) by LightCycler®480Real-Time PCR protocol. The three genotypes identified as: AA, AC and CC, based on fluorescence with Channel 465-510 (FAM for ‘A’ allele) and Channel 536-580 (VIC for ‘C’ allele). A no-template control (NTC) was used with each SNP genotyping assay (shown as grey spot). Figure S3. Melt curve analysis of TNFB, ICAM1 and GAPDH showing specific amplification. Figure S4. Relative gene expression of TNFB and ICAM1 with respect to TNFB exon 3 C/A in controls and vitiligo patients: (A) Expression of TNFB mRNA with respect to TNFB exon 3 C/A in 166 vitiligo patients and 175 controls using Mann-Whitney Wilcoxon test [MeanΔCp ± SEM: Controls CC vs Patients CC: 8.102 ± 0.1747 vs 7.678 ± 0.2379 (p = 0.168); Controls CA vs Patients CA; 7.761 ± 0.0952 vs 7.028 ± 0.3376 (p = 0.039); Controls AA vs Patients AA: 6.918 ± 0.2808 vs 5.217 ± 0.5087 (p = 0.015) (NS = non-significant)]. (B) Expression of ICAM1 mRNA with respect to TNFB exon 3 C/A in 166 vitiligo patients and 175 controls using Mann-Whitney Wilcoxon test [MeanΔCp ± SEM; Controls CC vs Patients CC: 8.767 ± 0.2267 vs 8.133 ± 0.2658 (p = 0.074); Controls CA vs Patients CA: 8.494 ± 0.2434 vs 7.225 ± 0.2729 (p = 0.001); Controls AA vs Patients AA: 8.007 ± 0.3297 vs 6.327 ± 0.4397 (p = 0.005).
(DOCX)