Abstract
Crohn disease (CD) is a chronic and debilitating inflammatory condition of the gastrointestinal tract.1 Prevalence in western populations is 100–150/100,000 and somewhat higher in Ashkenazi Jews. Peak incidence is in early adult life, although any age can be affected and a majority of affected individuals progress to relapsing and chronic disease. Medical treatments rely significantly on empirical corticosteroid therapy and immunosuppression, and intestinal resectional surgery is frequently required. Thus, 80% of patients with CD come to surgery for refractory disease or complications. It is hoped that an improved understanding of pathogenic mechanisms, for example by studying the genetic basis of CD and other forms of inflammatory bowel diseases (IBD), will lead to improved therapies and possibly preventative strategies in individuals identified as being at risk.
Key words: Atg16L1, dendritic cells, gastrointestinal, genome-wide association, gut microbiota, inflammatory bowel diseases, IRGM, Paneth cell, ulcerative colitis
Introduction
Epidemiological evidence for a genetic contribution to CD is clear-cut; 15% of patients with CD have an affected family member with IBD giving a relative risk to siblings of (λ)17x population risk. In addition, twin studies for CD have shown approximately 50% concordance in monozygotic twins compared to less than 10% dizygotics.2 The most widely accepted model is one in which a genetically susceptible host encounters an environmental bacterial or viral agent, triggering a breach of the intestinal epithelial barrier which separates the lumenal contents and microbial flora from the mucosal immune system. This primary event may lead to inflammatory flares in the gut mucosa. Subsequently, failures to properly downregulate the immunological response and establish epithelial restitution and repair leave the gut primed for future flare-ups, perhaps triggered by progressively less significant environmental insults. Crohn disease is characterized by discontinuous areas of inflammation in the distal small intestine and colon. The distinguishing features of inflammation in CD are that it is transmural and includes frequent lymphoid aggregates that are present even in the outer muscle layer and mesothelial lining as well as non-caseating granulomas composed of fused monocyte-derived cells. The transmural inflammation in CD is significant as it is associated with fistula formation with adjacent tissues. Features that distinguish ulcerative colitis (UC) include diffuse inflammation mostly confined to the inner layer (the mucosa) of the colon. The inflammation tends to be confluent and extends proximally from the rectum to a varying degree throughout the colon and is characterized by a complex mixture of inflammatory cells. In the last decade substantial advances in the understanding of the molecular pathogenesis of CD has been made as a result of multiple related lines of investigation. Importantly, CD has been one of the most tractable multigenic disorders for discovery of susceptibility genes. The genetic associations identified in CD suggest that multiple cell types and pathways are involved in generating the disease phenotype. These new discoveries have underscored the importance of epithelial barrier function, cell specific microbial responses (autophagy, ER stress, phagocytosis), innate (NOD proteins, inflammasome associated NALP3) and adaptive immunity (IL23R/Th17 cells) in disease pathogenesis.
Early work in the study of IBD inflammation focused upon alterations in the activation states of T cells (Th1, Th2 or more recently Th17) and the resultant cytokine profiles that occur in various forms of IBD. However, recent advances have focused on the role of the innate immune system in initiating, modulating and perhaps driving the abnormal inflammatory response that occurs in IBD. Murine models of IBD have proved an important tool to uncover the key pathways and innate cell types that play a role in the pathogenesis/mucosal immunopathology of IBD. In particular, efforts directed towards the identification of environmental factors implicate gut resident commensal microbiota rather than pathogens as drivers of dysregulated immunity and susceptibility to IBD. Commensal enteric bacteria are crucial for development of the host immune system and intestinal epithelial cell repair and homeostasis,3-5 but they are also a critical factor in determining the induction and the course of IBD as indicated by the absence of colitis in several mouse IBD models when they are derived as germ-free.6 In IBD patients, diversion of the fecal stream is associated with improvement in CD7 and some IBD patients improve upon intestinal washes or antibiotic treatment.8-10 Alternatively, administration of probiotic bacteria may contribute to the suppression of colitis.8,9 One theory of IBD is that it involves dysregulation of the normal co-evolved homeostatic relationships between the gut microbiota and the host immune system. The identification of such perturbations and how to effect their restoration is key to understanding the pathogenesis of IBD and routes to therapy.
The intestinal microbiota is comprised of more than 1,000 species of bacteria, which in the colon reach densities of more than 10,12 organisms per milliliter of lumenal content.11,12 Indeed, the lumenal contents of the colon account for 99.9% of the known human indigenous microbiota.13 Indigenous bacteria colonize mucosal surfaces and the gastrointestinal tract at birth in a pattern that resembles ecological succession,14 which reaches climax early in childhood.13 The adult microbiota for each individual is unique and stable over time, as shown by culture-independent small subunit ribosomal RNA gene-based assays.15,16
IBD patients have an altered lumenal- and mucosal-associated microbiome, although the nature of the dysbiosis, and how this relates to patient genotype, remains to be clearly defined. Of interest, a number of groups have recently demonstrated increased numbers of mucosal adherent-invasive E. coli (AIEC) in patients with IBD. Strains of AIEC isolated from CD patients with ileal disease induce epithelial injury and subsequent inflammation. In addition, elegant studies from Garrett et al. have demonstrated that enteric microflora grown in a strain of immunodeficient mice (Tbet and Rag1 null) may transmit colitis to wild-type mice through as-yet undefined microbes.17
Studies have implicated diminished levels of Faecalibacterium prausnitzii as being associated with a higher risk of postoperative recurrence of ileal CD. Collectively, these studies underscore the relevance of the host-microbial interaction in IBD and the importance of achieving better definitions of these processes to reach and understanding of the pathophysiology of these disorders. Despite the abundance of sensing and effector mechanisms that are available to trigger inflammatory immune responses to microbial intruders, the usual response to indigenous gut bacteria is the induction of local and systemic tolerance, often characterized as oral tolerance.18 From this vantage point, one potential model of IBD disease pathogenesis is that a dysregulated innate immune response to intestinal microbes occurs in genetically susceptible individuals. Abnormal cytokine responses are features of both CD and UC, although the model of CD as a T helper, Th1 driven illness and UC as a Th2 bias response now appears overly simplistic with an emerging role imbalance between the Th17 lineage producing IL-23 and IL-17 and regulatory T/B cells in both diseases.
In this review, experts in genetics, mucosal immunity and autophagy provide an up-to-date perspective on Crohn disease.
Genome-Wide Association Studies Reveal an Unexpected Role of Autophagy in Crohn Disease
The last three years have seen dramatic progress in understanding the genetic basis of CD and UC. This is due, in large part, to comprehensive genetic studies of DNA samples from large cohorts of patients and matched controls for over 300,000 genetic variants spread across the human genome. These studies, known as genome-wide association (GWA) studies, enable the examination of the majority of common genetic variation for roles in disease susceptibility. Given that the genetic variants tested in GWA studies are spread throughout the genome, rather than selected to test a specific set of genes, these studies have proven invaluable in identifying biological pathways not previously suspected of playing a role in a disease’s pathogenesis. In the following sections, we will recount how GWA studies have revealed a key role for autophagy in CD and place these discoveries in the broader context of IBD genetics and pathophysiology.
For many years, investigators used affected sibling pair techniques of linkage analysis to try to localize IBD susceptibility genes. Although limited by the intrinsically poor resolution of this method for pinpointing individual genes, some success was achieved using association-based methodologies to fine-map within linkage intervals. The major success in CD, and indeed one of the few successes across the spectrum of complex disease, was the strong association identified with relatively rare variants of the NOD2 gene. This flagged for the first time the importance of defects in innate immunity as a key pathogenic mechanism in IBD.
Nod2 is expressed intracellularly, particularly in monocytes and Paneth cells, and encodes an intracellular receptor for muramyl dipeptide (MDP), a motif present in bacterial cell walls. Although its exact function has yet to be determined, Nod2 appears to play an important role in innate immunity in modulating signaling through Toll-like receptor pathways and activating NFκB. Nod2 may also act as a sensor for intracellular bacteria by triggering autophagy as they enter the cell.19,20 One conundrum is that the NOD2 variants associated with CD result in loss of function of the Nod2 protein, begging the question as to how reduced activity of a key component of an innate immune pathway can lead to an increase in inflammatory activity. Perhaps the most plausible theories suggest that adaptive immune mechanisms may be recruited due to the defective innate immune pathways and that these might drive a physiologically “inappropriate” inflammatory response, but this has yet to be proven.
Recent years have seen a revolution in the ability of investigators to identify the genetic determinants of IBD and indeed all complex diseases. Progress has been made possible by endeavors such as the human genome project and advances in genotyping technologies that make hypothesis-free methods of testing the majority of the human genome for association with disease a tractable proposition. Such genome-wide association (GWA) studies test hundreds of thousands of markers, known as single nucleotide polymorphisms (SNPs) in thousands of individuals, seeking markers at which there are statistically significant differences in allele frequency between cases and controls. GWA studies are being used across the spectrum of complex disease, but nowhere has their application been more successful than in the field of CD genetics.
Since late 2006, three high-density GWA studies and one non-synonymous SNP scan have reported a total of 11 CD susceptibility loci, adding to NOD2 and OCTN/IBD5, previously identified by fine mapping within linkage intervals. A subsequent meta-analysis of GWA studies reported in 2008 brought to >30 the number of confirmed susceptibility loci for CD. These loci have now been studied in large UC cohorts, which identified several loci that are shared between UC and CD, but also a number of loci that are disease-specific.
Results from GWA Studies in IBD
The Th17 pathway is associated with CD and UC
In the first reported GWA study of CD, variants in the interleukin 23 receptor gene (IL-23R) on chromosome 1p31 were found to be unequivocally associated with CD susceptibility.21 The finding was rapidly replicated and extended to confirming association with UC.21,22 Perhaps surprisingly, a rare variant (Arg381Gln) in the IL-23R gene was shown to confer protection against developing IBD, with multiple other IL-23R variants showing independent association. As with NOD2, association of IL-23R was not seen in a well-powered CD association study from Japan.23
The importance of the IL-23 pathway has been further highlighted by the finding of association between variants in IL-12B, which encodes the p40 subunit shared between IL-12 and IL-23, and both CD and UC susceptibility.24-26 Most recently, the CD meta-analysis highlighted a number of other components of the Th17 pathway which are also associated with CD, including the signal transducers STAT2 and JAK3, the chemokine receptor CCR6 and co-stimulatory molecule ICOS-L27 (Table 1). STAT2 and JAK3 are also associated with UC.28
Table 1. Selected other IBD-associated loci.
| Locus | Proposed gene | Pathway | Other evidence |
|---|---|---|---|
| Cluster of SNPs 5.5 kb upstream of PTPN2 |
PTPN2; T cell protein tyrosine phosphatase (TCPTP) |
Negative regulator of inflammatory JAK-STAT signaling |
Mouse knockouts show elevated IFNγ, TNF〈 and IL-12243 |
| ICOSLG |
Inducible T-cell co-stimulator ligand |
Co-stimulatory molecule on antigen presenting cells, key role in Th17 cells from naïve CD4 lymphocytes27 |
|
| STAT327 |
Signal transducer and activator of transcription 3 |
Signal transduction in many cytokine pathways including IL23 and IL6 and their stimulation of Th17 cells244 |
|
| JAK228 |
Janus kinase 2 |
One of the crucial kinases in the STAT3 pathway |
|
| CCR627 |
Chemokine (C-C motif) receptor 6 |
G protein-coupled receptor expressed in immature DCs and memory T cells, mediates tissue-specific migration during epithelial inflammation245 |
Recently shown to be expressed by Th17 cells in CD118 |
| TNFSF1527,246 |
Tumor necrosis factor superfamily, member 15 |
Induces NF|B activation, potentiates IL-2 signaling and secretion of IFNγ by T lymphocytes. |
Strongly associated with CD in Japan, more modestly so in European populations; known to be upregulated in CD247 |
| NKX2-324-26 |
NK2 transcription factor related, locus 3 |
Poorly characterized transcription factor expressed in the intestine |
Variants associated with CD and UC, the former in Japanese as well as Europeans |
| Gene desert on chromosome 5p13 |
PTGER424,26,248 |
SNPs at 5p13 correlate with PTPGER4 expression |
Encodes prostaglandin receptor EP4, strongly implicated in mouse model of IBD249 |
| MST125,26,250 |
Macrophage stimulating 1 |
MST1 induces phagocytosis by resident peritoneal macrophages |
Variants associated with both UC and CD |
| ITLN127 |
Intelectin 1 |
ITLN1 recognizes galactofuranosyl residues found in cell walls of various microorganisms but not in mammals |
Expressed in intestine, including in Paneth cells, suggesting a role in innate immunity251 |
| ECM125 |
Extracellular matrix protein 1 |
Associated with UC in UK-based study, likely due to altered intestinal permeability25 |
|
| IL-10252,253 | Interleukin 10 | Variants associated with UC | IL-10 knock-out mice widely used as models of IBD, colitis depends upon gut flora |
The IL-23/IL-12 pathway has become the subject of intensive study in the field of immunology as it plays a key role in determining differentiation of naïve T cells into effector Th1 cells (driven by IL-12) or Th17 cells (driven by IL-23). Begum et al.29 reported that specific bacterial components such as peptidoglycan can differentially induce antigen presenting cells to produce IL-23 rather than IL-12, with this early regulatory mechanism potentially leading to distinct patterns of inflammatory response. A recent paper has highlighted the role of IL23 in mediating robust early immune control of intracellular bacterial pathogens.30 Riol-Blanco and co-workers demonstrated that the IL-23 receptor is vital for the maintenance of CD4−, CD8− T cells as well as γδ T cell populations, which provide early adaptive immune responses to infection. These IL-17 producing effector T cells are crucial for protection of mice against both L. monocytogenes and Francisella tularensis challenge. This extends the role of IL-23 to protection against intracellular pathogens, as well as differentiating inflammatory, auto-immune predisposing CD4+ major histocompatibility complex (MHC) class II-restricted Th17 cells, from their protective γδ or double-negative T cell counterparts. This, along with the role of Th17 cells in organ-specific inflammation, raises the hope that therapeutic disruption of the IL-23 pathway will control auto-immune inflammation without impairing systemic immunity.
Autophagy genes ATG16L1 and IRGM are specifically associated with CD
Association between the ATG16L1 (autophagy-related 16-like 1) gene on 2q37.1 and CD was by identified by a GWA study, as well as by an independent study of ~7,000 nonsynonymous SNPs.31 Both of these studies identified one amino acid changing polymorphism (T300A, Ala197Thr, rs2241880) within the ATG16L1 gene that was highly associated with CD, a finding replicated in the index report and subsequently widely elsewhere.26,32 Based on haplotype and regression analysis, this variant was found to explain all the signals observed at the ATG16L1 locus. It is not, however, associated with UC.25
Although the detailed functional impact of the Ala197Thr variant remains unclear, it affects a highly conserved motif immediately adjacent to the part of the gene encoding the WD repeat domain. Atg16L1 itself is known to play a key role in mammalian autophagy,32 a process recognized as playing a role in both innate and adaptive immunity. The Atg16L1 protein is widely expressed not only in intestinal epithelial cells, but also in lymphocytes and macrophages. Atg16L1 appears to play multiple roles related to innate immunity, particularly in relationship to Nod proteins. Both Nod1 and Nod2 were recently shown to direct Atg16L1 to bacterial entry sites on the plasma membrane, thus initiating autophagy to remove invading pathogens.20 Initially, the authors demonstrated that Nod agonists induce autophagy in human and mouse cells. In mice, exposure to Nod1 or Nod2 ligands via intraperitoneal injection following thioglycholate priming shows Nod-dependent induction of autophagy within isolated macrophages. However, rapamycin or lipopolysaccharide (LPS) induction of autophagy is normal in cells isolated from either Nod1- or Nod2-deficient animals. Using Shigella flexneri and Listeria monocytogenes infection of mouse embryonic fibroblasts (wild type or Nod1-deficient) Travassos and co-workers demonstrated that autophagic control of both Shigella and Listeria infection is dependent upon Nod1, but independent of RIP2 and NFκB.
The Nod proteins both immunoprecipitated with Atg16L1, and a fraction of these proteins also co-located at the plasma membrane when overexpressed, in a RIP2-independent manner. Both endogenous and overexpressed Nod2 and AtgG16L1 (and overexpressed Nod1) are observed to co-locate preferentially at sites of bacterial entry following short periods of infection with S. flexneri. However, the Nod2 association with Atg16L1 is inhibited by the known CD-associated NOD2 frameshift mutation, resulting in impaired autophagy of invading S. flexneri and reduced cytokine secretion in bone-marrow-derived macrophages from transgenic mice expressing the CD-associated truncation. In addition, the authors show that human cells homozygous for the CD-associated ATG16L1 polymorphism (Ala197Thr) show an impaired autophagic induction upon treatment with peptidoglycan or MDP (the Nod1 and 2 ligands, respectively), suggesting that Nod2 and Atg16L1 localization to the plasma membrane is required for optimal innate immune signaling.
A related study sheds further light upon autophagy and the role of ATG16L1 variants in stimulating innate immunity, and thus driving adaptive immune responses to pathogens. Cooney et al. demonstrated that both functional Nod2 and Atg16L1 are required in dendritic cells (DCs) for the induction of autophagy in response to Nod2 stimulation and proper induction of the antigen-presentation and bacterial handling pathways.19 Unlike in the epithelial and lymphoblastoid cells used by Travassos et al. these effects in DCs are RIP2 dependent. However, the DC system shows similar defects when CD-associated Nod2 and Atg16L1 variants are studied, notably failure to induce autophagy in response to MDP in the presence of either CD-predisposing gene variant. In DCs this led to a failure to translocate bacteria to lysosomes, resulting in reduced bacterial killing. DCs in which Atg5, 7 or 16L1 have been ablated via siRNA also failed to relocate MHC class II to the cell surface following MDP stimulation. This is associated with loss of the localization of MHC class II to autophagic compartments. In DCs from CD patients with either CD-associated Nod2 or ATG16L1 variants, a similar loss of MHC class II localization with the autophagic compartment is observed. Functionally, this results in curtailed antigen presentation and an inability to generate CD4+ T cell responses to bacterially-derived antigens following infection. These studies demonstrate direct links between NOD2 frameshift variants and autophagic bacterial control, as well as a novel Nod/autophagic signaling axis. It will be of interest to see if, as well as colocalization of Atg16L1 and Nod proteins at the plasma membrane, fully-formed autophagsomes provide a platform for Nod and other pathogen-associated molecular pattern (PAMP)-mediated signaling, via the delivery of recognition/ATG complexes to sites of autophagocytosed bacteria or by engulfing pathogen-derived ligands and thus enhancing or turning off signaling.
Further emphasizing the importance of autophagy in CD pathogenesis, the Wellcome Trust Case Control Consortium (WTCCC) identified association between variants in another key autophagy gene, immunity-related guanosine triphosphatase (IRGM) and CD susceptibility. A panel of 2,000 CD cases and 3,000 controls was used in the index GWA scan, with replication in an independent panel.24,33 Resequencing did not identify any coding variants in IRGM, but subsequent work has shown the associated variants to be in tight linkage disequilibrium with a copy number variant 5' of IRGM and has demonstrated impact on both gene expression and adverse impact on handling of intracellular bacteria.34
IRGM
In mice the interferon-induced Irg family contains 20 proteins, many of which are necessary for IFNγ mediated resistance to intracellular pathogens.35-38 Two IRG orthologues are found in humans, the widely expressed IRGM gene and the testis-restricted IRGC gene. IRGM has a role in the autophagy-mediated destruction of Mycobacterium bovis BCG and Salmonella typhimurium, in an expression dependent manner.34,36-38 In the M. bovis BCG model of tuberculosis, IRGM-dependent clearance of bacteria in macrophages is associated with IRGM localization to bacteria-containing compartments. This feature is common to both human IRGM and mouse IRGM1.36,37 Autophagy targeted BCG vacuoles acquire lysosomal markers and the vacuole is acidified, suggesting fusion with the lysosome effects anti-bacterial control.
Single nucleotide polymorphisms (SNPs) close to IRGM have been strongly correlated with Crohn disease (CD) susceptibility in genome wide association studies (GWAS).24,32 A large 20 Kb deletion of potential regulatory sequence is in perfect linkage with the functional polymorphisms associated with CD.34 The regulation of IRGM in humans is complex. Different tissues and cell lines heterozygous for the two haplotypes (CD risk and CD protective) express varying levels from each allele. In a cell culture model of S. typhimurium infection decreased IRGM expression significantly inhibits the efficiency of anti-bacterial autophagy.34 It is possible that IRGM is regulated in a cell-type specific manner and that altered expression induced by the CD risk deletion may result in cell specific phenotypes.
The evolution of the IRG gene family also appears to have been complex. A number of non-human primates, through duplication and diversification events, have evolved up to 21 interferon-regulated genes. Sequenced prosimian lemur species (Lemur catta and Microcebus murinus) have three IRG genes clustered in a phylogenetically similar manner to mice. Through study of Old World and New World monkeys, it appears that ~40 million years ago, IRGM became non-functional in a common ancestor of these species and was resurrected ~20 million years later in the common ancestor of humans and African great apes.39 The original loss of the gene is attributed to the integration of an Alu repeat disrupting the ORF. The restoration of the ORF coincides with the integration of an ERV9 element. Said element serves as a functional promoter for the human IRGM gene, suggesting that alterations of the gene expression are retroposon induced. Interestingly, orangutans and gibbons possess both functional and non-functional IRGM alleles. It seems that the locus is continuing to evolve in humans, possibly reflecting our ever-changing relationship with intracellular pathogens.
The emerging evidence would suggest that dysregulation of autophagy genes may contribute to the pathogenesis of inflammatory bowel disease via several routes: through Atg5 mediated removal of autoreactive CD4+ T cells; through Atg16L1-mediated endotoxin-induced inflammatory signaling in macrophages and Paneth cells, and potentially via autophagy related lymphocyte homeostasis. Each of these will be further explored in subsequent sections of this review. Much remains to be done to elucidate the precise mechanisms of ATG16L1 and IRGM polymorphisms in the pathogenesis of Crohn disease, but autophagy appears to be paramount to the mucosal immune system.
GWA studies have thus yielded many important insights into IBD pathogenic mechanisms, particularly relating to the roles of the Th17 pathway and innate immunity (particularly autophagy), but a number of surprises and challenges have also revealed themselves. It is evident that nearly all of the susceptibility loci identified individually confers only a modest increase risk and that, despite having identified >30 CD susceptibility genes, only ~20% of the genetic variance has yet been accounted for. Characterizing the remainder is one of the major future goals for IBD geneticists.
In addition, for the majority of loci the associated SNPs are simply markers and the true causal variant(s) have yet to be identified. The methodologies for such fine-mapping have yet to be fully worked out. Further hindering functional studies is that the cellular targets for a given IBD-predisposing variant are still unclear for most genes. A further confounding factor is that many causal variants appear to be non-coding,27 most likely predisposing to IBD as a consequence of their impact on the regulation of gene transcription. Indeed, nearly a third of the confirmed loci lie in so-called “gene-deserts”. Determining exactly how each of these affect transcriptional regulation will challenge biologists beyond the IBD community, and may have far-reaching implications for our understanding of genetics in a broader sense.
Autophagy and the Role of Atg16L1—From Yeast to Mammals
Autophagy was first identified as a starvation response in yeast, and the yeast system remains the best studied and characterized, despite considerable recent efforts in other models. It has been assumed that yeast represents the ancestral system, with mammalian autophagy exhibiting increased complexity and regulation. Molecular and genetic evidence suggest that the role of autophagy in multicellular organisms extends beyond starvation responses and has an impact upon other stress-induced pathways, as well as playing tissue-specific roles in the immune response. However, the core functions and apparatus remain conserved from yeast, forming a bulk degradation system, which maintains cellular homeostasis. In concert with the proteasome, autophagy degrades and recycles cellular components, reducing cellular stress and maintaining an amino acid pool essential for survival during periods of starvation.3-5 Autophagy operates through the encapsulation of organelles and cytoplasm within a membrane-bound organelle, termed the autophagosome. The apparatus used to create this unique structure is outlined below.
Yeast genetic analyses have thus far identified 34 autophagy-related (ATG) genes.40,41 Among these genes, ATG1-10, 12–14, 16–18, 29 and 31 are required for autophagosome formation.41,42 These gene products are classified into five functional groups: (1) the Atg1 protein kinase complex, (2) the Atg14-phosphatidylinositol 3-kinase complex, (3) the Atg2-Atg18-Atg9 complex, (4) the Atg12 conjugation system and (5) the Atg8 conjugation system.41
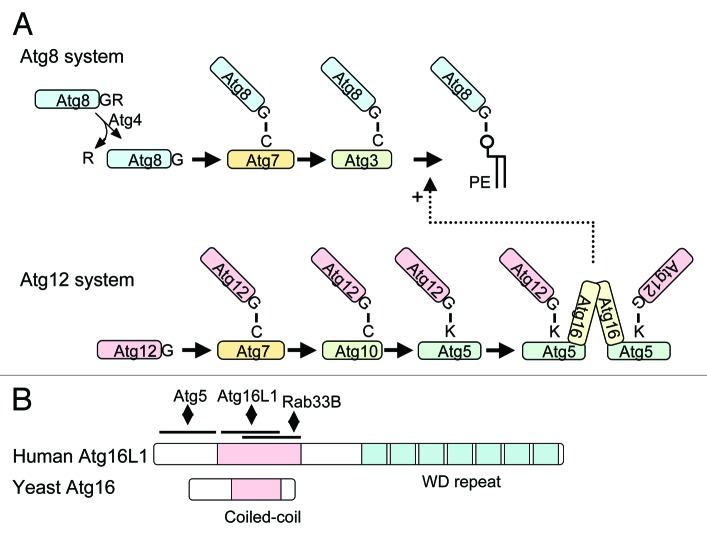
Both the Atg12 and Atg8 conjugation systems are ubiquitin-like systems, which are thought to be involved in the elongation step of the phagophore membrane (Fig. 1A). Atg12, a ubiquitin-like protein, is conjugated to Lys149 of Atg5 through an isopeptide bond.43 This conjugation reaction is mediated by Atg7 (an E1-like enzyme) and Atg10 (an E2-like enzyme). The resulting Atg12–Atg5 conjugate (en dash is used to denote a covalent linkage) is multimerized by Atg16.44 Atg16 is a 150-amino acid protein that contains an Atg5-interacting domain and a coiled-coil domain at its N and C terminus, respectively (Fig. 1B). Atg16 forms a dimer through the coiled-coil domain, thereby cross-linking the Atg12–Atg5 conjugates. Another ubiquitin-like protein, Atg8, is conjugated to the head group of phosphatidylethanolamine (PE) through an amide bond, which is mediated by Atg4 (a C-terminal processing enzyme), Atg7 and Atg3 (E2-like enzyme).45,46
Figure 1. Biochemical features of Atg16L1 and its related proteins. (A) The Atg8/LC3 and Atg12 conjugation systems. In the Atg8/LC3 conjugation system, the C-terminal arginine (this C-terminal extension is much longer in human and mouse) is first cleaved off by Atg4 to expose the glycine residue. Atg8/LC3 is then activated by Atg7, transferred to Atg3 and finally conjugated with PE. Atg8/LC3–PE is deconjugated again by Atg4. In the Atg12 system, Atg12 is activated by Atg7, transferred to Atg10 and then conjugated to Atg5. Atg12–Atg5 is multimerized by Atg16(L). (B) Yeast Atg16 and human Atg16L1 structures. The coiled-coil domain and C-terminal WD repeat domains are shown. Binding sites of interacting partners are also indicated.
These two conjugation systems are highly conserved in eukaryotes.47,48 In mammals, Atg8 homologs consist of three distinct families: the LC3, GABARAP and GATE-16 families. All of them can be conjugated to PE and localized on autophagosome membranes.49,50 The Atg12 system is also conserved. Atg12 is conjugated to Lys130 of Atg5 in mammals.51 The mammalian Atg12–Atg5 conjugate is multimerized by Atg16L1 (a mammalian Atg16 homolog) to form a large complex, which is estimated to be 800 kDa based on gel filtration experiments.47 It should be noted that the structures of yeast Atg16 and Atg16L1 are quite different. Atg16L1 of various higher eukaryotes including mammals has a large C-terminal WD repeat domain in addition to the N-terminal Atg16-like region (Fig. 1B). Since the WD repeat domain is involved in protein-protein interactions, there may be an unidentified factor(s) interacting with Atg16L1 through this domain. This WD domain may be important for regulation or specific subtypes of autophagy, perhaps in response to pathogen infection.52 Mice deficient in Atg3, 5, 7 and 16L die within one day after birth,53-56 apparently unable to transition from placental to intermittent oral nutrition. Autophagosome formation is defective in cells derived from these knockout mice, suggesting that these two conjugation systems are essential for autophagy in mammals.
An in vitro reconstitution experiment revealed that the yeast Atg12–Atg5 conjugate interacts with Atg3 and facilitates Atg8–PE conjugation.57 Atg8–PE and LC3–PE conjugation are also severely affected in vivo both in yeast and mammalian cells defective in the Atg12 conjugation system.55,58,59 Thus, the Atg12–Atg5 conjugate behaves as if it is an E3-like enzyme in the Atg8/LC3 conjugation system. In contrast, Atg16 is not essential for Atg8–PE formation, at least in yeast,59 and therefore, the Atg12–Atg5-Atg16 complex must have some function other than simple catalysis of the Atg8–PE conjugation. One possible function is recruitment of Atg8 to the phagophore to form Atg8–PE. If Atg12–Atg5-Atg16L1 is artificially expressed on the plasma membrane in mammalian cells, LC3 can be lipidated on this membrane.60 Another interesting observation is that Atg16L1 can interact with Rab33B in a GTP-dependent manner and this interaction is important to promote autophagy.61 Rab33B is a Golgi-resident protein, but is partially redistributed to the phagophore under starvation conditions. Thus, Rab33B may regulate autophagy through interaction with Atg16L1.
The physiological role of Atg8/LC3 also remains incompletely understood. It has been demonstrated that Atg8–PE can be oligomerized and induces membrane hemifusion in vitro.62 Although this function could be physiologically relevant, it is unknown whether autophagosome formation indeed requires membrane hemifusion. Another yeast study demonstrates that the Atg8 expression level regulates the size of autophagosomes.63 Although its mechanism is unknown, it may be related to a recent finding that human Atg3 interacts with FNBP1L, a human ortholog of yeast Bzz1.64 FNBP1L and Bzz1 contain an F-BAR domain, which is involved in generating membrane curvature generation. FNBP1L is dispensable for canonical autophagy, but is essential for autophagy of intracellular Salmonella. Besides the proposed role of Atg8/LC3 in membrane elongation, it may also be important for completion of autophagosome formation. Unclosed autophagosomes accumulate in Atg3-deficient fibroblasts56 and cells expressing a dominant negative Atg4B (Atg4BC74A).65 These phenotypes may be interpreted as the result of inappropriate elongation of the phagophore membrane. Overall, Atg12–Atg5-Atg16L and Atg8/LC3–PE are important for proper elongation and possibly for completion of the phagophore membrane.
APG15 is identical to APG16
The term ATG is a unified name of several autophagy-related gene collections such as APG, AUT, CVT, GSA, PAZ and PDD.66 The initial screen for autophagy defective mutants performed by Y. Ohsumi’s group isolated 15 apg mutants (apg1-apg15).67 Later, apg7 and apg11 were determined to be allelic.68 In 1999, another novel APG gene was identified by a yeast two-hybrid screen using Apg12 as bait.44 At that time, APG2 and APG15 had not yet been cloned. When a yeast strain deficient for this novel gene was mated with either mutant apg2-1 or apg15-1, autophagy deficiency was restored. This complementation test indicated that this novel gene was neither APG2 nor APG15 and it was named APG16.44 However, cloning of the APG15 gene revealed that it was identical to APG16 and apg15-1 was a nonsense (UGA) mutation of the 83rd CGA codon (arginine) of APG16.69 This result was very surprising because a diploid strain generated by mating of apg15-1 and apg16Δ strains showed normal autophagic activity.44 Further detailed analysis revealed that this discrepancy was due to the strain background used in the mating experiment. A [PSI+]-like cytoplasmic omnipotent suppressor was present in the apg16Δ strain, which decreased translational fidelity and caused misreading of the nonsense mutation in the apg15-1 allele.69 As a result, the phenotype of the apg15-1 allele was partially suppressed. Although the precise nature of this cytoplasmic suppressor remains unknown, it seems to be widely distributed in laboratory yeast strains, but not in the one used in the apg mutant screening. Therefore, the original apg15-1 mutant shows a complete autophagy-defective phenotype. Now, ATG11 and ATG15 are the gene names assigned to the previously named CVT9/GSA9/PAZ6/PDD18 and AUT5/CVT17 genes, respectively.
The Impact of Autophagy upon Inflammation
In mammals, systems for protein degradation are critical for tight control of the inflammatory immune response.70,71 Autophagy has been suggested to be involved in the regulation of inflammation,31,32,72-74 yet the mechanisms underlying the suppression of inflammatory responses by autophagy are poorly understood.
Recently, the Akira laboratory generated ATG16L1-deficient mice and demonstrated that Atg16L1 is essential for autophagosome formation, with most knockout mice dying within a day of birth.55 Therefore studies have utilized Atg16L1-deficient murine embryonic fibroblasts and macrophages, as well as chimeras lacking ATG16L1 expression in specific cell lineages to elucidate the role of Atg16L1 and autophagy in innate immune responses.
Enhanced endotoxin-induced IL-1β production by Atg16L1-Deficient macrophages
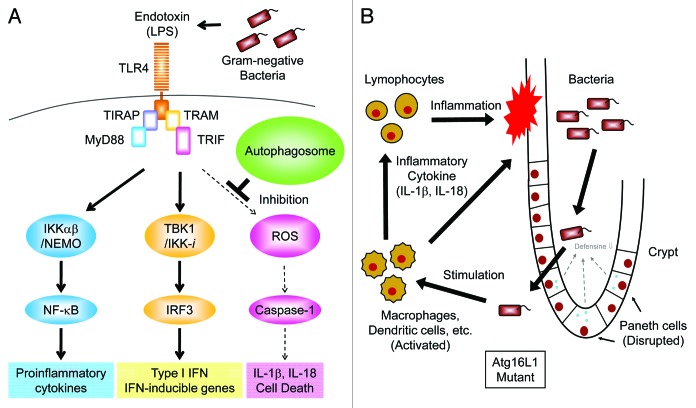
Toll-like receptors (TLRs) are a family of pattern recognition receptors that target microbial components and play a critical role in the innate immune response.75 Aberrant activation of TLR signaling pathways, often induced by disruption of the negative regulatory machinery, results in massive inflammation and may cause septic shock or autoimmune disease.70,75 Therefore, Saitoh et al. attempted to identify the role of Atg16L1 in TLR-mediated innate immune responses, particularly the LPS-induced production of inflammatory cytokines, such as TNFα, IL-6 and IL-1β.55 Although the involvement of autophagy-related proteins in the TLR-mediated antiviral response and in phagocytosis has been reported, it is unclear whether Atg16L1 regulates the production of inflammatory cytokines induced by TLR engagement.76,77 Although the production of TNFα and IL-6 was almost normal in Atg16L1-deficient macrophages, IL-1β production was enhanced compared with that in wild-type macrophages (Fig. 2A).55 Synthetic lipid A, an active component of LPS, also potently induced IL-1β production in Atg16L1-deficient cells. In addition, non-invasive Gram-negative bacteria, such as Enterobacter aerogenes, Escherichia coli and Klebsiella pneumoniae (all found in the commensal flora), potently induced IL-1β production by Atg16L1 deficient macrophages.55
Figure 2. Autophagy suppresses endotoxin-induced inflammatory immune responses. (A) TLR4 triggers both MyD88- and TRIF-dependent signaling pathways in response to LPS. The IKKα-IKKβ-NEMO complex mediates the activation of the transcription factor NFκB which, in turn, induces transcription of pro-inflammatory cytokines, such as TNFα, IL-6 and pro-IL-1β. The TBK1-IKK-i complex mediates the activation of the transcription factor IRF-3, which then induces the transcription of Type I IFN and IFN-inducible genes. In autophagy-deficient cells, high levels of ROS are generated and mediate TRIF-dependent caspase-1 activation resulting in the production of IL-1β. However, in wild-type macrophages, limited amounts of IL-1β are induced by LPS due to a lack of ROS generation. (B) Hypothetical model of intestinal inflammation caused by autophagy deficiency. The intestinal epithelial layer lacking autophagy seems to be susceptible to microbial infection due to the disrupted maturation of Paneth cells. The barrier function of intestinal epithelial cells is also compromised by chemical stress such as DSS exposure resulting in the invasion of commensal bacteria. Macrophages and dendritic cells detect invading bacteria and produce inflammatory cytokines. Autophagy-deficient macrophages produce high levels of IL-1β and IL-18 leading to severe inflammation mediated by lymphocytes.
Basal autophagy, but not stimulation-induced autophagy, regulates IL-1β production
An important question is whether enhanced IL-1β production results from Atg16L1 deficiency per se or autophagy deficiency in general. To address this Saitoh et al. used mutant mice deficient in Atg7, another essential autophagy protein.53 Like Atg16L1-deficient macrophages, Atg7 deficient macrophages show increased production of IL-1β, suggesting that the disruption of autophagy is responsible for the phenotype.
There is increasing evidence that basal autophagy plays an important role under pathophysiological conditions like the immune response, neurodegeneration and hepatic dysfunction.76,78-80 Recently, the potential involvement of TLR signaling in the induction of autophagy has been reported.81,82 In contrast to previous reports, LPS stimulation does not increase the number of endogenous LC3 puncta in mouse primary macrophages.55 However, infection with Salmonella typhimurium, an invasive Gram-negative bacterium, results in Atg16L1-dependent formation of bacteria-containing autophagosomes in macrophages.55 The formation of these autophagosomes does not require engagement of TLRs, since it occurrs in the absence of both MyD88 and TRIF, both essential adaptor molecules for TLR signaling pathways.55,75 The failure of autophagy induction by TLR signaling suggests that inhibition of basal autophagy induces IL-1β overproduction by the macrophages stimulated with endotoxin.
Enhanced TRIF-dependent caspase-1 activation by disruption of Atg16L1
Another question concerns the molecular mechanism underlying enhanced endotoxin-induced IL-1β production in Atg16L1-deficient macrophages. The expression of both IL-1β mRNA and immature pro-IL-1β protein following LPS stimulation of Atg16L1 deficient macrophages was almost comparable with that seen in wild-type cells, indicating an abnormality in post-translational regulation.55 In Atg16L1-deficient macrophages, LPS stimulation induces the cleavage of caspase-1 to an activated form, which mediates the processing of pro-IL-1β to mature IL-1β (Fig. 2A).83,84 IL-18 production, which is mediated by caspase-1, is also enhanced in Atg16L1-deficient macrophages stimulated with LPS. The activation of NFκB and p38 signaling pathways by LPS is similar in wild-type and Atg16L1-deficient macrophages, suggesting that the enhanced IL-1β production in Atg16 deficient cells is not due to disruption of these signaling pathways.55
Among the TLR family members, TLR2, TLR4 and TLR5 detect bacterial components and are required for the antibacterial immune response.75 The Akira group found that the ligand for TLR4, but not the ligands for TLR2 or TLR5, induced IL-1β production by Atg16L1 deficient macrophages.8 These findings prompted them to assess the involvement of TRIF signaling, which is selectively initiated by the engagement of TLR4 on macrophages.75 They generated Atg16L1-TRIF double-deficient mice and found that Atg16L1-TRIF double-deficient macrophages failed to produce IL-1β in response to LPS stimulation because of a lack of caspase-1 activation (Fig. 2A).55 The addition of IFNβ or IFNγ enhances TLR2-mediated IL-1β production by Atg16L1 deficient macrophages.8 These results indicate that disruption of Atg16L1 in macrophages causes enhanced LPS-induced IL-1β production in a TRIF-IFN-dependent manner.
Elevated levels of reactive oxygen species in Atg16L1-deficient macrophages
Recent studies have shown that ROS play an important role in the activation of caspase-1 induced by ATP, uric acid crystals, silicas and asbestos.83,84 Therefore, the potential involvement of ROS in the enhanced IL-1β production by Atg16L1-deficient macrophages was assessed. Strikingly, the enhanced IL-1β production from Atg16L1-deficient macrophages is blocked by the ROS scavengers N-acetyl-L-cysteine and FeTPPS (Fig. 2A).55 Also, the level of ROS in LPS-stimulated Atg16L1-deficient macrophages is higher than that in Atg16L1-TRIF double-deficient or wild-type macrophages. These results suggest that ROS accumulate in Atg16L1-deficient macrophages following LPS stimulation and induce TRIF-dependent production of IL-1β.
Severe dextran sulphate sodium-induced colitis in chimeric mice lacking Atg16L1 in hematopoietic cells
Elevated expression of IL-1β and IL-18 is involved in the development of intestinal inflammation,85,86 and recent studies have reported that Atg16L1 is a candidate susceptibility gene for Crohn disease.31,32 This led to the question as to whether Atg16L1 is involved in the development of colitis. Atg16L1-deficient chimeric mice do not develop spontaneous colitis and that the large intestines of newborn Atg16L1-deficient mice are not inflamed. The number of bacteria in the feces of wild-type or Atg16L1-deficient chimeric mice is similar with no bacteria detectable in the spleens of these animals.55
A corollary question was whether Atg16L1 deficiency exacerbated inflammation in a dextran sulphate sodium (DSS)-induced experimental model of colitis. Strikingly, chimeric mice lacking Atg16L1 in hematopoietic cells died following severe weight loss after DSS treatment (Fig. 2B).55 Control chimeric mice expressing wild-type Atg16L1 in hematopoietic cells survived. Intraperitoneal administration of 3-methyladenine, an autophagy inhibitor, increases the levels of IL-1β in serum and reduces the survival rate of mice treated with DSS. This suggests that autophagy protects mice from intestinal inflammation. Histological analysis reveals a more severe inflammation in the distal colons of Atg16L1-deficient mice than that seen in control mice with larger areas of ulceration and increased infiltration of lymphocytes. The levels of the pro-inflammatory cytokines IL-1β and IL-18 are significantly elevated in the sera of DSS-treated Atg16L1-deficient chimeric mice relative to controls. Mortality and loss of body weight after DSS exposure in Atg16L1-deficient chimeric mice are reduced by the injection of anti-IL-1β and anti-IL-18 antibodies, thus demonstrating the involvement of excessive production of these cytokines in the development of severe colitis.55
Remaining issues
These studies highlight a novel role for autophagy in the regulation of the inflammatory immune response. Disruption of autophagy causes TRIF-dependent elevation of ROS in LPS-stimulated macrophages.55 Accumulated ROS causes inflammasome activation leading to the production of the inflammatory cytokines IL-1 and IL-18. The source of this ROS has not been identified; however, Tcshopp and colleagues have reported that NADPH oxidase-induced ROS generation is required for the activation of caspase-1 as well as for the production of IL-1β by stimuli such as ATP, uric acid crystals, silica and asbestos.84 However, NADPH oxidase seems not to be involved in the accumulation of ROS in autophagy-deficient macrophages because inhibitors of NADPH oxidase fail to suppress LPS-induced IL-1β production by these cells (Tatsuya Saitoh and Shizuo Akira unpublished data). Other possible sources of ROS include mitochondria. Recent studies clearly demonstrate that autophagy is required for the maintenance of mitochondrial homeostasis and that targeted disruption of the autophagy-related genes results in the accumulation of ROS in immune cells.87,88 S. Akira and colleagues are currently investigating whether mitochondrial ROS are involved in IL-1β production by Atg16L1-deficient macrophages.
The Nod-like receptor (NLR) family members, IPAF and NALP3 have been well studied; they each form a complex called an inflammasome with an adaptor molecule, ASC, which activates caspase-1, resulting in the production of IL-1β.83,84 The IPAF inflammasome, which detects the protein flagellin on S. typhimurium, mediates both IL-1β production and caspase-1-induced cell death and is essential for host defense. The NALP3 inflammasome detects a variety of stimulatory molecules such as ATP, uric acid crystals, silica and asbestos, and plays a major role in the induction of inflammation by regulating caspase-1 activation.83,84 LPS-primed Atg16L1-deficient macrophages produce high levels of IL-1β following stimulation by ATP or uric acid crystals, both stimulators of the NALP3 inflammasome. However, the production of IL-1β induced by S. typhimurium mediated by the IPAF inflammasome shows little elevation in LPS-primed Atg16L1-deficient macrophages.55 Although caspase-1 activation is critical for LPS-induced IL-1β production by autophagy-deficient macrophages, the NLRs responsible for the activation of caspase-1 in autophagy-deficient cells have not been identified. Therefore, current investigations are concentrating on whether activation of the NALP3 inflammasome is required for IL-1β production by Atg16L1-deficient macrophages, and also whether activation of the NALP3 inflammasome is involved in the intestinal inflammation induced by Atg16L1 deficiency. The role of NOD2, another NLR implicated in CD, has also been examined in ATG16L1-deificient cells. Atg16L1 deficient macrophages produced the inflammatory cytokines IL-1β and IL-6 in response to MDP, a ligand for Nod2,55 indicating that Atg16L1 is not required for Nod2-mediated signaling of cytokine production. However, it is still unclear whether the detection of invading microbes by Nod2 or other NLRs induces Atg16L1-dependent autophagosome formation. Further experiments using mutant mice lacking NLRs are needed to address this issue.
In conclusion, Atg16L1 regulates the endotoxin-induced inflammatory response. Given the importance of the elevated expression of IL-1β and IL-18 caused by Atg16L1 deficiency in the pathogenesis of mouse model of colitis, it would be of interest to assess the role of both autophagy and inflammasomes in the development of inflammatory bowel disease in humans.
Autophagy and IBD Immunity— The Adaptive Immune System
While the link between autophagy and innate immunity is being elucidated through recent studies, there remains a substantial gap in our knowledge of the role of autophagy in the adaptive immune system. Key players in linking innate and adaptive immune responses are antigen-presenting cells (APCs), which are capable of presenting both exogenous (e.g., bacteria) and endogenous (self) antigens to T cells for activating adaptive immune responses. Autophagy plays a critical role in the MHC class II-dependent antigen presentation to CD4+ T cells. Indeed, impaired autophagy in a specific APC in the thymus results in the development of colitis.89 In addition, autophagy in T cells per se is implicated in their survival and proliferation.90 Once effector immune responses are generated, regulatory immune cells expand to prevent the responding effector cells from causing harmful effects. Autophagy may directly or indirectly participate in the differentiation of several regulatory (T and B) cell subsets. Therefore, autophagy is required for the entire immune processes involved in the pathogenesis of IBD through controlling the function and development of many different cell subsets in the intestine.91 In this section, we discuss the potential role of autophagy in CD-associated adaptive immune responses.
Antigen sampling by dendritic cells
Bacteria are processed by APCs capable of presenting the antigen to T cells. Dendritic cells (DCs) are professional APCs that can capture enteric bacteria that have invaded the intestinal mucosa by breaching the epithelial barrier. They can also sample the bacteria present in the intestinal lumen by extending dendrites directly into the lumen or by interacting with a specialized epithelial cell termed M cells and through neonatal Fc receptor (FcRn)-dependent transport of IgG/bacterial antigen complexes across the epithelial layer.92,93 Intestinal DCs provide a significant contribution to the direct linkage between innate and adaptive immune responses. There are several DC subsets in the intestine, which play different roles in the pathogenesis of IBD.92 Some DC subsets, particularly those expressing CD103, maintain tolerance towards commensal enteric microorganisms, whereas others are implicated in the generation of effector immune responses against potential pathogens.92 A unique DC subset that shares some phenotypic and functional features with macrophages increases in the inflamed colon of CD patients and in the intestinal granulomas of a mouse CD model.94,95 This unique DC subset is able to produce large amounts of IL-23 in response to bacterial products. In addition, local factors present in the intestine, such as thymic stromal lymphopoietin (TSLP) and retinoic acid, further condition the function of intestinal DCs.92,94,96 Bacteria-derived adenosine 5'-triphosphate (ATP) also activates a unique CD70highCD11low APC to enhance the differentiation of the Th17 subset.97 Involvement of autophagy in DC function has become increasingly apparent. Autophagy is required for the recognition of virus by plasmacytoid DCs76 and it also enhances the antigen presentation by DCs facilitating the efficacy of vaccination.98 However, it is largely unknown whether autophagy plays a role in the intestinal DC function involved in the pathogenesis of IBD. Therefore it is likely that autophagy plays a major role in facilitating and controlling antigen presentation and immune activation in the gut compartment, in both healthy and diseased states.
Effector immune responses
Cellular degradation systems such as proteasomes and lysosomes are used not only for the elimination of intracellular bacteria but also influence adaptive immune responses. Autophagosomes can engulf cytosolic components and thereby deliver cellular and microbial degradation products to MHC class II compartments. This was first suspected during experiments in the early 1990s showing that APCs can process cytoplasmic influenza and measles virus proteins for presentation by MHC class II.99,100 This requires lysosome acidification, but does not require TAP transporters or protein trafficking through the Golgi apparatus, suggesting direct delivery of proteins to lysosomes from the cytosol. This model is consistent with biochemical studies showing that some 35% of peptides bound to HLA-DR molecules are derived from nuclear or cytosolic proteins101 and that the percentage rises following induction of autophagy by starvation. Autophagy also causes downregulation of lysosomal cathepsins and altered patterns of peptide processing, suggesting that autophagy has the potential to change the nature of peptides presented to CD4+ T cells. Recent studies show that autophagosomes labeled with GFP-LC3 fuse directly with MHC class II compartments in both epithelial cells and professional APCs and that LC3 is delivered constitutively to lysosomes over many hours.102,103 Constitutive autophagy is also evident in transgenic mice expressing GFP-LC3 where autophagosomes are seen in tissues in the absence of starvation.104 Autophagy can also deliver model antigens into class II antigen processing pathways if they are targeted to autophagosomes by fusion to LC3. Presentation of fusion proteins to T cells is lost if the lipidation motif in LC3 is mutated to prevent attachment to autophagosomes, or following ATG12 silencing.103 Parallel studies80 have argued that autophagosome processing of proteins for presentation by MHC class II should show a preference for long-lived proteins. This proved correct for lymphoblastic cells infected with Epstein-Barr virus where the long-lived nuclear antigen EBNA1 was presented to CD4+ T cells through a pathway that was blocked by silencing ATG12. In the same study a short-lived EBNA1 lacking the sequences that prevent degradation by the proteasome was presented by MHC class I rather than class II.
The evidence that autophagy delivers cytosolic proteins for processing and presentation to CD4+ T cells by MHC class II is now very strong. Autophagy may be particularly important for intestinal epithelial cells that are not actively endocytic and where class II antigen expression increases in an inflammatory setting in response to INFγ. Interestingly, INFγ does not generate greater numbers of autophagosomes. The increased capacity to present antigens in response to INFγ results from increased MHC class II expression coupled with increased delivery of LC3 positive autophagosomes to MHC class II compartments.103 For an overview of current concepts in the relationship between autophagy and phagocytosis see refs. 105, 106.
In addition to the involvement of autophagy in antigen-presentation that is required for initiating adaptive immune responses, autophagy provides significant contributions to immune regulation through its role in proliferation and survival of T cells after T cell receptor (TCR) ligation. Double-membrane autophagosomes can form in both human and murine T cells and autophagy enhances the survival and proliferation of memory T cells through clearance of superfluous mitochondria.87,89,90,107 Contributions of dysregulated homeostasis of memory T cells have been implicated in the pathogenesis of colitis. Indeed, implantation of Atg5-deficient thymus induces the development of systemic inflammations, including colitis, in the recipient athymic mice, suggesting the requirement of autophagy for the appropriate selection of self antigen-specific CD4+ T cells.89
The antigenic responses of CD4+ T cells are determined by the diversity of the T cell receptor (TCR) repertoire. In both human IBD and experimental models, the TCR repertoire utilized by intestinal CD4+ T cells is sharply skewed, suggesting antigen-specific activation of effector CD4+ T cells.108 Antigens present in high-levels in the gut compartment include bacteria-derived flagellins, which have been demonstrated to serve as IBD-associated antigens capable of stimulating Th1 and Th17 responses.109
In addition to exogenous antigen presentation, a major function of autophagy is the regulation of self-responses.110 Autophagy contributes to the clearance of apoptotic cells that represent the major source of self antigens.111 The role of autophagy in self-responses recalls past findings in IBD models where the involvement of impaired clearance of apoptotic cells has been proposed to exacerbate chronic colitis.112 In addition, commensal bacteria are required for exacerbation, but not development, of ileitis in the SAMP1/YitFc mice, a model of CD.113 These observations raise the possibility that autophagy plays critical roles in the recognition not only of exogenous antigens, but also for self antigens in IBD.
Th1 responses have been implicated in the pathogenesis of CD, whereas UC has a significant contribution from Th2 cytokines such as IL-4, IL-5 and IL-13.114 In addition to the Th1/Th2 theory, recent accumulating studies have unveiled a critical involvement of the IL-23/Th17 pathway in the pathogenesis of IBD.115 IL-12p70 (p40/p35) has generally been believed to be a crucial factor involved in the development of CD because of the beneficial effect of anti-IL-12p40 treatment on CD patients and experimental models.114 However, it is becoming increasingly apparent that IL-23 (a heterodimer of p40 and p19 subunits) rather than IL12 (a heterodimer of p40 and p35 subunits) contributes to the development of CD.115 Indeed, IL-23 receptor polymorphisms are negatively associated with the development of IBD.21 IL-23 serves to maintain a Th17 subset that is characterized by the production of IL-17 (IL17A), IL-17F, IL-21, IL-22 and IL-26, but not IFNγ or IL-4.116 An intestine-specific Th17 subset expressing CD161 has recently been found in humans to be significantly increased in the inflamed colon of CD patients.117 A unique Th17 subset that co-expresses IL-17 and IFNγ has also been identified in the inflamed colon, but not in the peripheral blood, of CD patients118 and a pathogenic role of IL-17 has been demonstrated in some CD models.119,120 These findings highlight the significance of Th17 subsets in CD, although it is unclear whether autophagy plays a common or distinct role in these functionally different T cell subsets.
However, recent evidence has linked autophagy to the production of, and response to, key cytokines, and therefore opened the possibility that some of the immune dysregulation observed in IBD may result from aberrant autophagic processes. There is also evidence that autophagy is affected by the cytokine environment, with Th2 cytokines diminishing the ability of macrophages to mount an effective anti-tuberculosis autophagic response.121
Recent research has highlighted the finding that cytokine secretion is altered in both CD and the tumor microenvironment. Following bacterial exposure, macrophages from CD patients appear to deliver a large proportion of pro-inflammatory cytokines to the lysosomal system, rather than the appropriate secretory pathway.122 This results in reduced bacterial clearance and therefore delays in dealing with infectious insults, potentially leading to increased ongoing inflammatory responses. This failure to secrete adequate levels of cytokines is reminiscent of the Paneth cell phenotype observed in mouse models and human patients with defects in Atg16L1 or other autophagy genes,123,124 where mucin and cryptidin secretion are compromised. It will be crucial to determine whether there are other secretory defects in patients with polymorphisms within autophagy genes.
In the tumor microenvironment, high levels of CCL2 and IL-6 have been observed to drive production of a tumor-promoting macrophage subset, termed M2.125 This polarization occurs as a result of inhibition of apoptosis, via upregulation of autophagy and inhibition of caspase activation, in myeloid cells recruited to the tumor. Thus, autophagy is involved in profound cellular changes in response to cytokines and the impact of defective autophagy upon cytokine signaling in specific cell types is just beginning to be explored.
A major lymphocyte population in the mucosal immune compartment is B cells, with over 80% of activated B cells located within mucosal tissues.126 As compared to T cells, B cells have not typically been considered to be a major source of cytokines. However, it is becoming increasingly apparent that, like T cells, B cells also produce a wide array of cytokines, particularly under inflammatory conditions.127,128 Intestinal inflammatory conditions induce the development of an IL-10-producing regulatory B cell subset termed Bregs, which contribute to improving the ongoing colitis seen in several IBD models.128 This concept of Bregs, which arose from animal models of IBD, is supported by recent case reports showing an exacerbation/development of UC and a reduction of IL-10 production after B cell depletion therapy using humanized mouse anti-human CD20 monoclonal antibody (rituximab).129,130 The intestine contains a unique IgM+ B2 B cell subset that is characterized by very high expression of MHC class II.131 This B cell subset develops in an antigen-independent manner, and significantly expands in the context of inflammation and contributes to host defense against enteric microorganisms by producing IL-12p70. Peritoneal B1 B cells that produce natural IgM against enteric microorganisms have an ability to suppress the development of colitis,132 and their homeostasis is known to be Atg5 dependent.133 Despite the role of autophagy in MHC class II antigen presentation and a known role for Atg5 B cell development and homeostasis, the effect of perturbed autophagy upon B cell repertoires is still largely unknown.
Intestinal Barrier Function and Oral Tolerance
The epithelium of the small intestine and colon create a complex, dynamic chemical and physical barrier between host and microbes, the importance of which is underlined by results from recent human genetic studies as well as data from established model systems. Ideas that have emerged from such studies in model experimental systems are increasingly being translated into human IBD utilizing genotyped patient material. From this growing body of work, functional disease targets of the mucosal barrier have been identified in specific epithelial cell types of the gut. This portion of this review will highlight some of the recent work in this area and the role autophagy plays in maintaining barrier function and oral tolerance.
The chemical barrier I: mucus-producing goblet cells
The first line of defense is a layer of mucus that is produced by specialized cells within the lining of the epithelium called goblet cells. These cells are designed to store mucus for rapid exocytosis. The most abundant mucus components, and the main component of mucus in the intestine, is the Muc family of glycoproteins;134 Muc2 being the most common in the colon,135 although the expression pattern of related gene products varies with location along the proximal to distal intestinal axis.
Mucus is stored in discrete mucin secretory granules (MSGs) that typically undergo merocrine secretion, a process by which discharge of an individual granule occurs without the loss of any part of the apical portion of the cell. This is a rapid process; typically a single granule is secreted within minutes of the application of a given stimulus. A variety of physiological stimuli are known to cause MSG secretion (reviewed in ref. 134 and 136) and include autonomic nerve stimulation (cholinergic), substance P, prostaglandins and ATP/UTP. The latter stimuli bind purinergic receptors on the apical surface of goblet cells. These G-coupled receptors also regulate fluid secretion (e.g., Cl− ions), and PDGF is another known agonist. Neutrophil elastase also has non-receptor-mediated effects on goblet cells to stimulate MSG secretion. Since much of our knowledge is based upon lung derived cells, it will be important to perform studies on mouse and human goblet cells derived from the small intestine and colon to determine the unique properties of goblet cells in this organ. A few human goblet cell lines that are derived from adenocarcinomas have been useful.137 Also, perhaps now the opportunity exists to maintain explants or isolated epithelial cells in culture for studies on non-transformed cells.138,139
The importance of mucin production in the intestine is evidenced by studies of mutant mice that lack the main component of colonic goblet cell mucus, Muc2.140,141 These mice contain decreased lumenal mucus and over time develop spontaneous colitis and epithelial tumors. The enhanced susceptibility to injury of Muc2 deficient mice can be further evoked either by treatment with dextran sodium sulfate or by breeding into an IL-10 knockout background.142,143 Interestingly, the loss of function of other genes such as Mmp9 creates an increased susceptibility to damage by ultimately causing a decrease in Muc2 and, thus, mucus production.144 One proposed mechanism by which loss of mucus creates inflammation is that the indigenous colonic microbes gain direct access to the surface epithelium.145 The Muc2 model, while not a known genetic correlate to human CD, is an instructive model for the importance of the mucus barrier.
Goblet cells also preferentially express other genes that, when knocked-out result in mice with greater susceptibility to gut injury. These include: resistin-like-beta (Relmβ), which has been proposed to augment mucus secretion from goblet cells in an autocrine fashion as well as to stimulate CD4+ T cells to produce cytokines,146-149 and Trefoil factor 3 (TFF3) produced by goblet cells in response to TLR2 signaling and a critical downstream factor of this signaling pathway.150 The lack of production of TFF3 appears to be a feature of the intestinal mucosa in UC patients that carry a polymorphism within TLR2 (R753Q151). Future work in both mouse models and human patient material analysis should further determine if specific IBD susceptibility alleles can affect goblet cell differentiation and/or mucus secretion.
The chemical barrier II: antimicrobial protein-producing Paneth cells
Mucus alone is not a sufficient chemical barrier as many members of the intestinal microbiota contain an abundance of glycosyl hydrolases,152 thus rendering mucus an attractive potential energy source for specific members of the microbiota. However, the mucus layer is an important repository for immunoglobulins (e.g., IgA) and a variety of antimicrobial peptides produced by the intestinal epithelium.
In the small intestine, the primary, but not exclusive, source of antimicrobial proteins and peptides is Paneth cells. Their main job is to produce, package and export a variety of antimicrobial proteins/peptides including multiple α-defensins (e.g., cryptdins), angiogenin-4, lysozyme and secretory phospholipase A2.153,154 These proteins can alter the composition and abundance of individual species of indigenous microbes and appear to play a role in the clearance of pathogens. As an example, mice engineered to overexpress a human cryptdin were less susceptible to death when orally infected with high doses of Salmonella typhimurium.155 These secretory functions may be directly related to IBD since mice with an engineered loss of function for Atg16L1 contain Paneth cells that do not properly package antimicrobial proteins into granules and secrete them into the lumen of the intestine.123,124 This defect appears to be the result of the loss of autophagy since loss of function of Atg5 and Atg7 in the intestinal epithelium recapitulates the Paneth cell phenotype observed in the Atg16L1-deficient mice. In addition, Atg16L1 deficient Paneth cells contain a gain of function whereby they express increased levels of inflammatory cytokines such as adiponectin and leptin. Humans with CD who carry the point mutation in ATG16L1 corresponding to one of the susceptibility alleles for this disease showed similar loss and gain-of-function defects in their Paneth cells as those observed in the mouse model. Mice with loss of function of another CD susceptibility gene, the ER stress response gene Xbp1, contain Paneth cells that appear to undergo premature apoptosis.156 Loss of function of yet another CD susceptibility gene, the MDP receptor Nod2 may play a role in diminished defensin production in Paneth cells, though this result is controversial.157,158 Another link between Paneth cell development and CD susceptibility is the presence of polymorphisms in the Tcf4 promoter.159 Partial loss of function of this gene is related to diminished antimicrobial protein expression.160
Finally, it has been hypothesized that Paneth cells are a source of growth factors, and recent data indicates that this role may be responsive to the bacterial flora. Some intestinal resident microbes such as B. thetaiotaomicron can stimulate Paneth cells to produce mRNAs encoding the antimicrobial protein, angiogenin-4.161 This microbe can also stimulate angiogenesis in the mesenchyme of small intestinal villi indirectly through Paneth cells by an as yet unknown mechanism.162 Paneth cells also produce inflammatory cytokines in both a constitutive (TNFα163) and inducible fashion during injury (e.g., IL-17,164). However, the functional significance of Paneth cell cytokine or growth factor production has still not been tested experimentally.
Clearly, additional studies of Paneth cell function concerning the impact of loss of function (diminished production or delivery of antimicrobial proteins) on the intestinal microbiota and gain of function (increased inflammatory cytokine production) on the mucosal immune system will be critical in the future. Also, understanding the triggers of Paneth cell malfunction (if any) in a genetically susceptible host will be important aspects of IBD pathogenesis.
The cellular barrier: enterocytes
Underlying the mucus layer is a physical barrier that is created primarily by absorptive enterocytes that dominate the epithelium, lining villi in the small intestine and pericryptal cuffs in the colon. The intestinal epithelium is a dynamic barrier in which tightly bound epithelial cells are renewed by accelerated division of crypt cells that migrate upwards from the bottom of intestinal crypts. It is also a highly interactive barrier able to mount rapid responses to pathogens through the production of antimicrobial proteins and mobilization of neutrophils and, through collaborations with antigen presenting cells, induce mucosal adaptive immune responses.165 In the small intestine, four differentiated lineages are produced: absorptive enterocytes, goblet cells, hormone secreting enteroendocrine cells and Paneth cells. All of these lineages except Paneth cells migrate out of crypts onto the surfaces of mucosal evaginations into the lumen called villi, migrating to the villus tip over a period of 3–5 days. Conversely, Paneth cells migrate to the base of crypts where they intermingle with epithelial stem cells and have a much longer life span (at least 30 days). The colon contains the same basic organization of the epithelium with the two notable exceptions that instead of villi, epithelial cells migrate out of each crypt onto a flat surface cuff that surrounds each crypt opening and there are no Paneth cells. The exception to the latter feature is in the proximal portion of the inflamed human colon where the presence of Paneth cells is considered to be metaplastic.166
A critical component of the barrier function of the surface epithelium is the apical junctional complex that consists of tight and adherens junctions; for example overexpression of the myosin light chain kinase in the intestinal epithelium leads to tight junction dysfunction and ultimately broad activation of mucosal immune responses.167 The function of tight junctions in the control of permeability is particularly sensitive to inflammatory cytokines such as tumor necrosis factor. Interestingly, specific pathogens such as enteropathogenic E. coli disrupt tight junctions to increase permeability.168 This pathogen also targets the function of sodium absorption through inhibition of the sodium hydrogen exchanger 3.169 Mice with loss of function of this gene develop spontaneous colitis,170 suggesting an important immune-regulatory function for this ion transporter.
Cytoskeletal alterations can also affect the ability of enterocytes to create an effective barrier. Loss of function of keratins does not appear to cause a defect in handling mechanical stress, but instead affects permeability and subsequent Th2 T cell activation.171 Loss of function of the three Vav GTPases leads to diminished microtubule networks in colonic enterocytes resulting in shortened cells. In combination with immune defects in the Vav knockout mice, this results in a spontaneous breakdown of the colonic intestinal barrier and focal ulceration.
These studies lay the groundwork for potential mechanisms by which enterocytes could be defective in IBD. It will be interesting in the future to see how specific susceptibility genes fit into these pathways. For example, mice with loss of STAT3 function in the intestinal epithelium exhibit worse damage upon dextran sodium sulfate treatment and greater numbers of tumors upon addition of azoxymethane.172 A key question in moving forward is whether these human susceptibility alleles are additive, particularly with regard to the different aspects of barrier function such as chemical, physical, cellular and immune.
Oral tolerance
Oral tolerance has been described in most monogastric mammals, including humans.173 A vast range of antigens can be used to suppress all aspects of systemic immune responses when administered orally, although most of these share the property of being inert and/or soluble thymus-dependent antigens, such as proteins. Generally, T cell-mediated responses and tissue immunopathology are very sensitive to low doses of orally administered antigens. By contrast, antibody responses are less easily inhibited, and the status of local IgA responses in oral tolerance and whether or not they are suppressed as a result of orally administerting antigen are still unclear. Despite its profound effects on systemic immunity and its practical and clinical importance, the mechanisms responsible for oral tolerance remain elusive.
Mechanisms of oral tolerance
Using very different in vitro or in vivo approaches, immune tolerance can occur through active suppression, anergy or apoptosis of (self) antigen-reactive lymphocytes. However, no clear consensus has been reached about the relative importance or contribution of each of these. Indeed it is possible that there is a wide range of different regulatory mechanisms that may either be important under different circumstances or that collectively are needed to ensure systemic tolerance.174 The question of whether tolerance to the antigens of the microbiota exists is unanswered, although some data suggests that this is the case. For example, the absence or diminishment of antibody responses in animals systemically administered indigenous gut bacteria,175,176 differences in immunogenicity of different members of the microbiota177 and the finding that normal individuals are tolerant to their own microbiota, whereas they respond to bacteria of others with a breakdown of tolerance to antigens of an individuals own microbiota, resulting in IBD.178
Oral tolerance and intestinal epithelial cells
Intestinal epithelial cells (IECs) possess many of the attributes of professional APCs including their ability to sample soluble proteins via non-selective (fluid-phase pinocytosis) and selective receptor-mediated routes179,180 and express MHC class I and II molecules.181,182 However, as IECs normally lack expression of the co-stimulatory molecules required for full T cell activation,183 they are good candidates for tolerogenic antigen presenting cells in vivo. This may not, however, be the only mechanism by which IECs can control and suppress mucosal T cell responses. For example, primary colonic epithelial cells, some of which express co-stimulatory molecules, are still effective at suppressing CD4+ T cell activation by professional APCs.184 IECs are also a source of immunosuppressive cytokines such as TGFβ.185 By a variety of means, therefore, IECs can exert the global hypo-responsiveness that is characteristic of oral tolerance with the dysregulation of any of their pathways of suppression able to contribute to the uncontrolled inflammation seen in IBD.
One way in which IEC ability to induce or preserve mucosal T cell tolerance would be compromised is through perturbations in the processing and presentation of self antigens. Several lines of evidence suggest that autophagy pathways might be involved in intracellular antigen processing for MHC class II loading and considerable levels of autophagy have been observed in thymic epithelial cells and immature dendritic cells involved in central and peripheral T cell tolerance induction, respectively.102,104 Second, the activation of autophagy that invariably follows microbial infection has the potential to increase processing of host, microbial and dietary antigens by MHC class II pathways, and this may alter the spectrum of peptides presented to the immune system.
Activation of autophagy by bacteria
It has become clear that despite different intracellular niches, diverse bacterial pathogens can be engulfed by autophagosomes and delivered to lysosomes. Thus, autophagy acts as a defense against bacteria that are unable to tolerate lysosomes, and for these bacteria activation of autophagy results in microbial degradation, increased class II antigen presentation and decreased microbial yields. Bacteria can, however, survive if they are able to block autophagy or prevent delivery to lysosomes. Most Salmonella remain within modified phagosomes called Salmonella-containing vacuoles (SCV) and are unaffected by autophagy. Damage to the SCVs or release of bacteria into the cytosol, results in engulfment by autophagosomes, delivery to lysosomes and bacterial destruction.186 Shigella flexneri resides within the host cytosol and secretes the VirG protein that is required for actin-based motility and transfer to neighboring cells. VirG binds to Atg5 and activates autophagy, but virulent isolates survive because this interaction is masked by the Shigella IcsB protein.187 Listeria monocytogenes evades autophagosomes via ActA-dependent actin-based motility and by secreting bacterial phospholipases that may compromise the autophagosome membrane.188,189 Other bacteria have evolved to replicate in the lysosome, and in these cases bacteria may even activate autophagy to accelerate delivery to lysosomes, as seen for example with Coxiella burnetii.190 Francisella tularensis disrupt the phagosome membrane soon after cell entry and are engulfed by autophagosomes and delivered to lysosomes. Francisella tularensis is, however, resistant to degradation in lysosomes and activation of autophagy increases microbial replication.191 Thus, autophagy can protect the cell cytosol from infection, but is also subverted by specialized pathogens via specific virulence factors.
Activation of autophagy by viruses
Much of what we know about autophagy and viral infection comes from studies on herpes simplex virus where inhibition of autophagy by the viral ICP34.5 protein leads to neurovirulence192 and from studies of the positive-stranded RNA viruses such as picornaviruses and coronaviruses, which induce double-membrane vesicles for use as platforms for replication.193-195 There are no examples of intestinal infection by human coronaviruses viruses and the role played by autophagy during cornavirus replication has recently become controversial.196,197 Picornaviruses such as poliovirus and coxsackieviruses do, however, replicate in gastrointestinal epithelial cells and induce LC3 processing and translocation of GFP-LC3 from the cytosol to autophagosomes in cell culture models.198,199 Poliovirus replicase proteins co-localize with LC3 suggesting that replication takes place on autophagosomes, and assembly of the replicase complex may be provide the signal that activates autophagy.198 Replicase protein 2BC alone causes conversion of LC3-I to LC3-II and recruitment of LC3 to membranes, but co-expression of poliovirus 3A and 2BC replicase proteins is required to convert these membranes into double-membraned vesicles resembling autophagosomes that stain for LC3 and LAMP-1.200,201 Similarly, infection of IPCs by rotaviruses, which cause viral gastroenteritis, results in recruitment of LC3 to virus-induced membranes,202 again providing a link between viral infection of IPCs and activation of autophagy. Since our knowledge of the human virome is still nascent,203,204 it will be of great interest to observe whether specific intestine-tropic viruses are identified as being associated with gut inflammation and perhaps inducing chronic inflammatory states in susceptible individuals. In a recently published study in a mouse model of Atg16L-deficient mice, investigators found that the persistent infection of a common enteric virus in mouse colonies, murine norovirus, is required to trigger abnormal Paneth cell secretion and responses to DSS toxin injury. Interestingly, autophagy function is not responsible for differences in innate immunity and does not play a role in the clearance of the virus. Instead, Atg16L1 function is required for the proper function of specific epithelial host cell types in response to this persistent infection. Thus, this study is a specific virus-plus-susceptibility gene interaction in the context of CD.
Pattern recognition during phagocytosis by antigen-presenting cells
Autophagy can also be activated from outside the cell following recognition of PAMPS by Toll-like receptors (TLRs). This makes it possible that constant exposure to gut microbiota could deliver sustained activation autophagy of gastrointestinal APCs and other cells with TLR receptors. A screen for PAMPS able to activate autophagy through TLRs of phagocytic cells has shown potent activation following recognition of single stranded RNA by TLR7.81 TLR7 is mostly confined to endocytic compartments and provides a defense against viral infection, as demonstrated by activation of plasmacytoid dendritic cells by TLR7 signaling during infection by vesicular stomatitis virus.76,205 Induction of autophagy by TLR7 ligands requires the MyD88 adaptor protein and activation may be mediated through displacement of Beclin 1 from Bcl-2 following binding of Beclin 1 to MyD88 and TRIF.206 Beclin 1 may also be released from Bcl-2 during TLR signaling following phosphorylation by JNK.207 Activation of autophagy by TLR7 accelerates elimination of Mycobacteria from macrophages81 showing that activation of TLR7 during viral infection can affect the outcome of bacterial infection. Bacterial products such as LPS have also been shown to activate autophagy via TLR4.82 TLR4 induction of autophagy requires class III PtdIns 3-kinases and p38 MAPK. TLR4 signaling also upregulates LRG47 GTPase, and in common with TLR7, increases elimination of Mycobacteria from macrophages.82
Activation of autophagy following TLR signaling from phagosomes has also been studied using TLR ligands immobilized on latex beads.77 Beads bound to synthetic bacterial lipopeptides (Pam3CSK4, TLR1/2), zymosan (TLR2) or LPS (TLR4) stimulate autophagy and caused rapid recruitment of Beclin 1, PtdIns 3-kinase activity and LC3 to the phagosome membrane. Recruitment is dependent on Atg5 and Atg7, and results in increased fusion of LC3 with lysosomes; even so it was not possible to detect double-membraned vesicles associated with phagosomes. This makes it possible that TLR signaling results in direct recruitment of LC3 and PtdIns 3-kinase activity from the cytosol to the phagosome membrane to facilitate fusion with lysosomes to enhance elimination of incoming microorganisms. This may also be the case for Nod2, indicating that autophagy may be a general response to PAMP-derived signals that serves as an anti-infection mechanism.
The Microbial Hypothesis of IBD
CD has several features reminiscent of a microbial or infectious process in the gut114,208,209 and this has led many investigations into finding the potential infectious agent responsible. Mycobacterium avium subspecies paratuberculosis (MAP) was perhaps the bacterium with the longest history as a putative pathogenic role in CD.210,211 MAP had been initially favored as a candidate pathogen because there are clinical similarities between Johne disease, a spontaneous MAP infection in ruminants and CD. In the past few decades, several dozen published studies have provided evidence that both supports and disputes the association of MAP with CD.210,212,213 However there remains no direct evidence that MAP plays a causal role in human CD and despite the establishment of culture-free molecular tools for microbial identification, MAP has not been observed in the majority of CD or IBD cases. Similar cases have been made for other putative IBD-associated pathogens, including Listeria monocytogenes and Yersinia spp., but again evidence has been lacking, despite some circumstantial observations.214-216
What has become evident is that IBD patients exhibit an intestinal dysbiosis (microbial imbalance)—studies of lumenal bacterial composition in patients with IBD using both culture and molecular biological techniques have provided evidence for a decrease in beneficial bacteria such as the Bifidobacteria, Lactobacilli and Firmicutes and an increase in putative pathogenic bacteria such as Bacteroides and Escherichia coli.217-220 This dysbiosis may promote inflammation by inducing a breakdown in the balance between putative protective versus harmful intestinal bacteria. Indeed, a recent study showed that a low proportion of Faecalibacterium prausnitzii, bacteria with high anti-inflammatory properties, is associated with endoscopic recurrence at 6 months on resected ileal Crohn mucosa.221 In addition, various studies in CD patients reported that high concentrations of bacteria forming a biofilm are found on the surface of the gut mucosa217 and that increased numbers of E. coli are associated with the epithelial intestinal layer.219,222-224 Phenotypic characterization of CD-associated E. coli, which are AIEC, shows that they are able to adhere to and invade intestinal epithelial cells, but also to survive and replicate within macrophages, leading to the secretion of high amounts of TNFα.225,226 Such invasive E. coli have been observed by many groups in Europe and the United States.222,227-231 Like other pathogenic E. coli strains, AIEC are potent pro-inflammatory inducers since the interaction between bacteria and host cells activates several pathways which lead to NFκB activation, upregulated IL-8 expression and transmigration of polymorphonuclear leukocytes.228,232 This inflammatory response is thought to result from the interaction of flagellin with Toll-like receptor 5 (TLR5) and IPAF,233 but the roles of the NOD family are yet to be fully elucidated in these models. In contrast to many pathogens that escape from the normal endocytic pathway, CD-associated invasive E. coli are taken up by macrophages within phagosomes, which mature without diverting from the classical endocytic pathway and which share features with phagolysosomes.234 Because of their ability to survive and replicate within epithelial cells, we could hypothesize that these pathogenic bacteria possess mechanisms to circumvent the innate host response and survive under hostile intracellular conditions. So far, such factors have yet to be identified in AIEC, since these organisms lack canonical type III secretion for delivery of effector proteins to host cells. This is in contrast to Shigella spp. which shares a similar lifestyle within the host epithelium and for which virulence factors required for autophagy avoidance have been identified.187 Further investigations are needed to determine whether AIEC can escape from or modulate autophagy in order to survive and/or replicate and whether they could replicate at a higher level in host cells with altered autophagy. Recent data suggest that AIEC are subject to autophagic control within epithelial cells, and that this process is influenced by ATG16L1 genotype.235
Despite these findings, it remains unclear whether such alterations in flora represent a proximal cause of inflammation, or are a consequence of disease. Such inflammation-induced shifts in the gut flora have been observed following infection in mouse models, with both Citrobacter rodentium and Salmonella typhimurium infection promoting growth of Enterobacteriaceae.236,237
E. coli overgrowth could also be due to abnormal expression of host molecules acting as receptors for bacterial adhesion. In CD patients with ileal involvement of the disease, Barnich et al. recently observed an abnormal ileal expression of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) and showed that CEACAM6 acts as a very efficient receptor mediating binding of pathogenic CD-associated E. coli.238 Interestingly, in vitro studies demonstrate that CEACAM6 expression is increased in cultured intestinal epithelial cells not only after IFNγ or TNFα stimulation,239 but also after infection with AIEC bacteria, indicating that AIEC bacteria can promote their own colonization in CD patients.238 In addition, using the transgenic CEABAC10 mice expressing human CEACAM6,240 the Darfeuille-Michaud laboratory showed that CD-associated AIEC bacteria can colonize the intestinal mucosa and induce gut inflammation (Carvalho et al. manuscript submitted). Perhaps some of these questions may be answered in longitudinal metagenomic studies of large cohorts, but such efforts are only just getting underway.
The implication of the Nod2-autophagy-innate immune axis through genetic studies have strengthened the idea that AIEC, but also other as-yet unidentified microbes such as enteric viruses, could be candidates for a role in CD. The basis of these assumptions rests upon the imputed host genetic susceptibility due to impaired function in host defense against intracellular bacteria, induced by polymorphisms in pathogen sensing and innate effector systems, such as Nod2 and autophagy.
Despite years of study, bacterial/CD correlations in humans have not yet approached fulfilling Hill’s criteria for causation;241 however, recent work demonstrating norovirus-autophagy interactions in an Atg16L1-hypomorphic mouse model of IBD shows promise in beginning to define specific pathogenic interactions in susceptible hosts.242 With current high-throughput technologies and large, genotyped patient cohorts it may be possible to identify putative initiating agents for further testing. Studies using new molecular microbiological tools, in subsets of patients with known predisposing Atg16L1, IRGM and Nod2 polymorphisms are needed to determine the extent of correlation between colonization/infection with particular microbial agents and CD. It would be of utility to begin such studies in affected families prior to onset of overt disease in order to examine the longitudinal variation of the flora and attempt to establish a baseline, pre-disease “map” of the gut microbiota for comparison with later putative dysbioses.
Prior to the identification of the role of autophagy in CD, it was known that the bacterial flora played a crucial role in disease pathogenesis, but no single cellular pathway had been identified which might confer the immune regulatory defects seen in patients. Now autophagy integrates several aspects of CD pathogenesis including: known Nod2 associations; innate immune activation; defects in bacterial handling and antigen presentation; and the homeostasis of the epithelial barrier. Thus, autophagy provides a defined pathway with which to study the interaction of host and gut flora and how this is perturbed in disease states.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/13074
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Breslin NP, Todd A, Kilgallen C, O’Morain C. Monozygotic twins with Crohn’s disease and ulcerative colitis: a unique case report. Gut. 1997;41:557–60. doi: 10.1136/gut.41.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 6.Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073–83. doi: 10.1016/j.addr.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet. 1991;338:771–4. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 8.Rutgeerts P, Van Assche G, Vermeire S, D’Haens G, Baert F, Noman M, et al. Ornidazole for prophylaxis of postoperative Crohn’s disease recurrence: a randomized, double-blind, placebo-controlled trial. Gastroenterology. 2005;128:856–61. doi: 10.1053/j.gastro.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics and prebiotics. Gastroenterology. 2004;126:1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 10.Sands BE. Inflammatory bowel disease: past, present and future. J Gastroenterol. 2007;42:16–25. doi: 10.1007/s00535-006-1995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 13.Savage DC. Mucosal microbiota. In: Ogra PL, Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee J, Eds. Mucosal Immunology, 2nd Edition. San Diego: Adademic Press 1999; 19-30 [Google Scholar]
- 14.Copperstock MS, Zedd AJ. Intestinal flora of infants. In: Hentges DA, ed. Human intestinal microflora in health and disease. New York: Academic Press 1983; 79-100 [Google Scholar]
- 15.Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–9. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoetendal EG, Ben-Amor K, Harmsen HJ, Schut F, Akkermans AD, de Vos WM. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl Environ Microbiol. 2002;68:4225–32. doi: 10.1128/AEM.68.9.4225-4232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3:331–41. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 19.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2009;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 20.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immun. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 21.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tremelling M, Cummings F, Fisher SA, Mansfield J, Gwilliam R, Keniry A, et al. IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology. 2007;132:1657–64. doi: 10.1053/j.gastro.2007.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet. 2007;52:575–83. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]
- 24.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40:710–2. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson CA, Massey DC, Barrett JC, Prescott NJ, Tremelling M, Fisher SA, et al. Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523–9. doi: 10.1053/j.gastro.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begum NA, Ishii K, Kurita-Taniguchi M, Tanabe M, Kobayashi M, Moriwaki Y, et al. Mycobacterium bovis BCG cell wall-specific differentially expressed genes identified by differential display and cDNA subtraction in human macrophages. Infect Immun. 2004;72:937–48. doi: 10.1128/IAI.72.2.937-948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riol-Blanco L, Lazarevic V, Awasthi A, Mitsdoerffer M, Wilson BS, Croxford A, et al. IL-23 Receptor Regulates Unconventional IL-17-Producing T Cells That Control Bacterial Infections. J Immunol. 2010;184:1710–20. doi: 10.4049/jimmunol.0902796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 32.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med. 2001;194:181–8. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFNgamma-inducible LRG-47. Science. 2003;302:654–9. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 37.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 38.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J Immunol. 2007;179:6963–72. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 39.Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, et al. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 42.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356:405–10. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–96. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 46.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 48.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 50.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–76. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–92. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 52.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS ONE. 2008;3:3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 55.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 56.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 58.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008;19:2916–25. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 63.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huett A, Ng A, Cao Z, Kuballa P, Komatsu M, Daly MJ, et al. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J Immunol. 2009;182:4917–30. doi: 10.4049/jimmunol.0803050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–9. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 67.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 68.Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–43. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- 69.Okazaki H, Ono B, Ohsumi Y, Ohsumi M. apg15-1, a UGA mutant allele in the Saccharomyces cerevisiae APG16 gene, and its suppression by a cytoplasmic factor. Biosci Biotechnol Biochem. 2004;68:1541–8. doi: 10.1271/bbb.68.1541. [DOI] [PubMed] [Google Scholar]
- 70.Liu YC, Penninger J, Karin M. Immunity by ubiquitylation: a reversible process of modification. Nat Rev Immunol. 2005;5:941–52. doi: 10.1038/nri1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Chen T, Han C, He D, Liu H, An H, et al. Lysosome-associated small Rab GTPase Rab7b negatively regulates TLR4 signaling in macrophages by promoting lysosomal degradation of TLR4. Blood. 2007;110:962–71. doi: 10.1182/blood-2007-01-066027. [DOI] [PubMed] [Google Scholar]
- 72.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–77. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 75.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 76.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 77.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 78.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 79.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 80.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–6. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 81.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–21. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–7. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–65. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 85.Ishikura T, Kanai T, Uraushihara K, Iiyama R, Makita S, Totsuka T, et al. Interleukin-18 overproduction exacerbates the development of colitis with markedly infiltrated macrophages in interleukin-18 transgenic mice. J Gastroenterol Hepatol. 2003;18:960–9. doi: 10.1046/j.1440-1746.2003.03097.x. [DOI] [PubMed] [Google Scholar]
- 86.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, et al. Nod2 mutation in Crohn’s disease potentiates NFκB activity and IL-1β processing. Science. 2005;307:734–8. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 87.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, Cadwell K, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–35. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–5. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 90.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–55. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 91.Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–49. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 92.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–46. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshida M, Kobayashi K, Kuo TT, Bry L, Glickman JN, Claypool SM, et al. Neonatal Fc receptor for IgG regulates mucosal immune responses to luminal bacteria. J Clin Invest. 2006;116:2142–51. doi: 10.1172/JCI27821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, et al. Unique CD14+ intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFNγ axis. J Clin Invest. 2008;118:2269–80. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mizoguchi A, Ogawa A, Takedatsu H, Sugimoto K, Shimomura Y, Shirane K, et al. Dependence of intestinal granuloma formation on unique myeloid DC-like cells. J Clin Invest. 2007;117:605–15. doi: 10.1172/JCI30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mora JR. Homing imprinting and immunomodulation in the gut: role of dendritic cells and retinoids. Inflamm Bowel Dis. 2008;14:275–89. doi: 10.1002/ibd.20280. [DOI] [PubMed] [Google Scholar]
- 97.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 98.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr., Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 99.Jaraquemada D, Marti M, Long EO. An endogenous processing pathway in vaccinia virus-infected cells for presentation of cytoplasmic antigens to class II-restricted T cells. J Exp Med. 1990;172:947–54. doi: 10.1084/jem.172.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Malnati MS, Marti M, LaVaute T, Jaraquemada D, Biddison W, DeMars R, Long EO. Processing pathways for presentation of cytosolic antigen to MHC class II-restricted T cells. Nature. 1992;357:702–4. doi: 10.1038/357702a0. [DOI] [PubMed] [Google Scholar]
- 101.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmid D, Münz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sanjuan MA, Green DR. Eating for good health: linking autophagy and phagocytosis in host defense. Autophagy. 2008;4:607–11. doi: 10.4161/auto.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev. 2009;227:203–20. doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- 107.Kasai M, Tanida I, Ueno T, Kominami E, Seki S, Ikeda T, Mizuochi T. Autophagic compartments gain access to the MHC class II compartments in thymic epithelium. J Immunol. 2009;183:7278–85. doi: 10.4049/jimmunol.0804087. [DOI] [PubMed] [Google Scholar]
- 108.Probert CS, Saubermann LJ, Balk S, Blumberg RS. Repertoire of the alpha beta T-cell receptor in the intestine. Immunol Rev. 2007;215:215–25. doi: 10.1111/j.1600-065X.2006.00480.x. [DOI] [PubMed] [Google Scholar]
- 109.Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yoshimori T. Autophagy: paying Charon’s toll. Cell. 2007;128:833–6. doi: 10.1016/j.cell.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 111.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 112.Mizoguchi A, Mizoguchi E, Smith RN, Preffer FI, Bhan AK. Suppressive role of B cells in chronic colitis of T cell receptor alpha mutant mice. J Exp Med. 1997;186:1749–56. doi: 10.1084/jem.186.10.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bamias G, Okazawa A, Rivera-Nieves J, Arseneau KO, De La Rue SA, Pizarro TT, Cominelli F. Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J Immunol. 2007;178:1809–18. doi: 10.4049/jimmunol.178.3.1809. [DOI] [PubMed] [Google Scholar]
- 114.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–21. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Abraham C, Cho JH. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu Rev Med. 2009;60:97–110. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- 116.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 117.Kleinschek MA, Boniface K, Sadekova S, Grein J, Murphy EE, Turner SP, et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206:525–34. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–61. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Z, Zheng M, Bindas J, Schwarzenberger P, Kolls JK. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm Bowel Dis. 2006;12:382–8. doi: 10.1097/01.MIB.0000218764.06959.91. [DOI] [PubMed] [Google Scholar]
- 121.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–17. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 122.Smith AM, Rahman FZ, Hayee B, Graham SJ, Marks DJ, Sewell GW, et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn’s disease. J Exp Med. 2009;206:1883–97. doi: 10.1084/jem.20091233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cadwell K, Patel KK, Komatsu M. Virgin HWt, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–2. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J Biol Chem. 2009;284:34342–54. doi: 10.1074/jbc.M109.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brandtzaeg P, Johansen FE. Mucosal B cells: phenotypic characteristics, transcriptional regulation and homing properties. Immunol Rev. 2005;206:32–63. doi: 10.1111/j.0105-2896.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 127.Fillatreau S, Gray D, Anderton SM. Not always the bad guys: B cells as regulators of autoimmune pathology. Nat Rev Immunol. 2008;8:391–7. doi: 10.1038/nri2315. [DOI] [PubMed] [Google Scholar]
- 128.Mizoguchi A, Bhan AK. A case for regulatory B cells. J Immunol. 2006;176:705–10. doi: 10.4049/jimmunol.176.2.705. [DOI] [PubMed] [Google Scholar]
- 129.El Fassi D, Nielsen CH, Kjeldsen J, Clemmensen O, Hegedus L. Ulcerative colitis following B lymphocyte depletion with rituximab in a patient with Graves’ disease. Gut. 2008;57:714–5. doi: 10.1136/gut.2007.138305. [DOI] [PubMed] [Google Scholar]
- 130.Goetz M, Atreya R, Ghalibafian M, Galle PR, Neurath MF. Exacerbation of ulcerative colitis after rituximab salvage therapy. Inflamm Bowel Dis. 2007;13:1365–8. doi: 10.1002/ibd.20215. [DOI] [PubMed] [Google Scholar]
- 131.Shimomura Y, Ogawa A, Kawada M, Sugimoto K, Mizoguchi E, Shi HN, et al. A unique B2 B cell subset in the intestine. J Exp Med. 2008;205:1343–55. doi: 10.1084/jem.20071572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shimomura Y, Mizoguchi E, Sugimoto K, Kibe R, Benno Y, Mizoguchi A, Bhan AK. Regulatory role of B-1 B cells in chronic colitis. Int Immunol. 2008;20:729–37. doi: 10.1093/intimm/dxn031. [DOI] [PubMed] [Google Scholar]
- 133.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–14. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 134.Rogers DF. Airway goblet cells: responsive and adaptable front-line defenders. Eur Respir J. 1994;7:1690–706. [PubMed] [Google Scholar]
- 135.Tytgat KM, Buller HA, Opdam FJ, Kim YS, Einerhand AW, Dekker J. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology. 1994;107:1352–63. doi: 10.1016/0016-5085(94)90537-1. [DOI] [PubMed] [Google Scholar]
- 136.Davis CW, Dickey BF. Regulated airway goblet cell mucin secretion. Annu Rev Physiol. 2008;70:487–512. doi: 10.1146/annurev.physiol.70.113006.100638. [DOI] [PubMed] [Google Scholar]
- 137.Roumagnac I, Laboisse C. A mucus-secreting human colonic epithelial cell line responsive to cholinergic stimulation. Biol Cell. 1987;61:65–8. doi: 10.1111/j.1768-322x.1987.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 138.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–6. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 140.Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–9. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 142.van der Sluis M, Bouma J, Vincent A, Velcich A, Carraway KL, Buller HA, et al. Combined defects in epithelial and immunoregulatory factors exacerbate the pathogenesis of inflammation: mucin 2-interleukin 10-deficient mice. Lab Invest. 2008;88:634–42. doi: 10.1038/labinvest.2008.28. [DOI] [PubMed] [Google Scholar]
- 143.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–29. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 144.Garg P, Ravi A, Patel NR, Roman J, Gewirtz AT, Merlin D, Sitaraman SV. Matrix metalloproteinase-9 regulates MUC-2 expression through its effect on goblet cell differentiation. Gastroenterology. 2007;132:1877–89. doi: 10.1053/j.gastro.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 145.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hogan SP, Seidu L, Blanchard C, Groschwitz K, Mishra A, Karow ML, et al. Resistin-like molecule beta regulates innate colonic function: barrier integrity and inflammation susceptibility. J Allergy Clin Immunol. 2006;118:257–68. doi: 10.1016/j.jaci.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krimi RB, Kotelevets L, Dubuquoy L, Plaisancie P, Walker F, Lehy T, et al. Resistin-like molecule beta regulates intestinal mucous secretion and curtails TNBS-induced colitis in mice. Inflamm Bowel Dis. 2008;14:931–41. doi: 10.1002/ibd.20420. [DOI] [PubMed] [Google Scholar]
- 148.McVay LD, Keilbaugh SA, Wong TM, Kierstein S, Shin ME, Lehrke M, et al. Absence of bacterially induced RELMbeta reduces injury in the dextran sodium sulfate model of colitis. J Clin Invest. 2006;116:2914–23. doi: 10.1172/JCI28121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nair MG, Guild KJ, Du Y, Zaph C, Yancopoulos GD, Valenzuela DM, et al. Goblet cell-derived resistin-like molecule β augments CD4+ T cell production of IFNγ and infection-induced intestinal inflammation. J Immunol. 2008;181:4709–15. doi: 10.4049/jimmunol.181.7.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Podolsky DK, Gerken G, Eyking A, Cario E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology. 2009;137:209–20. doi: 10.1053/j.gastro.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang F, Tahara T, Arisawa T, Shibata T, Nakamura M, Fujita H, et al. Genetic polymorphisms of CD14 and Toll-like receptor-2 (TLR2) in patients with ulcerative colitis. J Gastroenterol Hepatol. 2007;22:925–9. doi: 10.1111/j.1440-1746.2007.04909.x. [DOI] [PubMed] [Google Scholar]
- 152.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–6. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 153.Mukherjee S, Vaishnava S, Hooper LV. Multi-layered regulation of intestinal antimicrobial defense. Cell Mol Life Sci. 2008;65:3019–27. doi: 10.1007/s00018-008-8182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ouellette AJ. Paneth cells and innate immunity in the crypt microenvironment. Gastroenterology. 1997;113:1779–84. doi: 10.1053/gast.1997.v113.pm9352884. [DOI] [PubMed] [Google Scholar]
- 155.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–6. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 156.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, Radford-Smith GL. Reduced α-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut. 2008;57:903–10. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 158.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal α-defensin expression. Gut. 2004;53:1658–64. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koslowski MJ, Kubler I, Chamaillard M, Schaeffeler E, Reinisch W, Wang G, et al. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn’s disease. PLoS ONE. 2009;4:4496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wehkamp J, Wang G, Kubler I, Nuding S, Gregorieff A, Schnabel A, Kays RJ, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn’s disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–18. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 161.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–73. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 162.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–5. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tan X, Hsueh W, Gonzalez-Crussi F. Cellular localization of tumor necrosis factor (TNF)alpha transcripts in normal bowel and in necrotizing enterocolitis. TNF gene expression by Paneth cells, intestinal eosinophils and macrophages. Am J Pathol. 1993;142:1858–65. [PMC free article] [PubMed] [Google Scholar]
- 164.Takahashi N, Vanlaere I, de Rycke R, Cauwels A, Joosten LA, Lubberts E, et al. IL-17 produced by Paneth cells drives TNF-induced shock. J Exp Med. 2008;205:1755–61. doi: 10.1084/jem.20080588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–64. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 166.Symonds DA. Paneth cell metaplasia in diseases of the colon and rectum. Arch Pathol. 1974;97:343–7. [PubMed] [Google Scholar]
- 167.Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–63. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shifflett DE, Clayburgh DR, Koutsouris A, Turner JR, Hecht GA. Enteropathogenic E. coli disrupts tight junction barrier function and structure in vivo. Lab Invest. 2005;85:1308–24. doi: 10.1038/labinvest.3700330. [DOI] [PubMed] [Google Scholar]
- 169.Hecht G, Hodges K, Gill RK, Kear F, Tyagi S, Malakooti J, et al. Differential regulation of Na+/H+ exchange isoform activities by enteropathogenic E. coli in human intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:370–8. doi: 10.1152/ajpgi.00432.2003. [DOI] [PubMed] [Google Scholar]
- 170.Laubitz D, Larmonier CB, Bai A, Midura-Kiela MT, Lipko MA, Thurston RD, et al. Colonic gene expression profile in NHE3-deficient mice: evidence for spontaneous distal colitis. Am J Physiol Gastrointest Liver Physiol. 2008;295:63–77. doi: 10.1152/ajpgi.90207.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Toivola DM, Krishnan S, Binder HJ, Singh SK, Omary MB. Keratins modulate colonocyte electrolyte transport via protein mistargeting. J Cell Biol. 2004;164:911–21. doi: 10.1083/jcb.200308103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu JY, Seno H, Miletic AV, Mills JC, Swat W, Stappenbeck TS. Vav proteins are necessary for correct differentiation of mouse cecal and colonic enterocytes. J Cell Sci. 2009;122:324–34. doi: 10.1242/jcs.033720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mowat AM, Weiner HL. Oral tolerance: Physiological basis and clinical application. In: Ogra PL, Mestecky J, MEL, Strober W, Bienenstock J, McGhee J, Eds. Mucosal Immunology 2nd Edition. San Diego: Academic Press 1999; 586-617 [Google Scholar]
- 174.Mowat AM, Parker LA, Beacock-Sharp H, Millington OR, Chirdo F. Oral tolerance: overview and historical perspectives. Ann N Y Acad Sci. 2004;1029:1–8. doi: 10.1196/annals.1309.001. [DOI] [PubMed] [Google Scholar]
- 175.Berg RD, Savage DC. Immune responses of specific pathogen-free and gnotobiotic mice to antigens of indigenous and nonindigenous microorganisms. Infect Immun. 1975;11:320–9. doi: 10.1128/iai.11.2.320-329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Foo MC, Lee A. Immunological response of mice to members of the autochthonous intestinal microflora. Infect Immun. 1972;6:525–32. doi: 10.1128/iai.6.4.525-532.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peach S, Lock MR, Katz D, Todd IP, Tabaqchali S. Mucosal-associated bacterial flora of the intestine in patients with Crohn’s disease and in a control group. Gut. 1978;19:1034–42. doi: 10.1136/gut.19.11.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Duchmann R, Kaiser I, Hermann E, Mayet W, Ewe K, Meyer zum Buschenfelde KH. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD) Clin Exp Immunol. 1995;102:448–55. doi: 10.1111/j.1365-2249.1995.tb03836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mazanec MB, Kaetzel CS, Lamm ME, Fletcher D, Nedrud JG. Intracellular neutralization of virus by immunoglobulin A antibodies. Proc Natl Acad Sci USA. 1992;89:6901–5. doi: 10.1073/pnas.89.15.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Warshaw AL, Walker WA, Cornell R, Isselbacher KJ. Small intestinal permeability to macromolecules. Transmission of horseradish peroxidase into mesenteric lymph and portal blood. Lab Invest. 1971;25:675–84. [PubMed] [Google Scholar]
- 181.Hershberg RM, Cho DH, Youakim A, Bradley MB, Lee JS, Framson PE, Nepom GT. Highly polarized HLA class II antigen processing and presentation by human intestinal epithelial cells. J Clin Invest. 1998;102:792–803. doi: 10.1172/JCI3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mayer L, Eisenhardt D, Salomon P, Bauer W, Plous R, Piccinini L. Expression of class II molecules on intestinal epithelial cells in humans. Differences between normal and inflammatory bowel disease. Gastroenterology. 1991;100:3–12. doi: 10.1016/0016-5085(91)90575-6. [DOI] [PubMed] [Google Scholar]
- 183.Sanderson IR, Ouellette AJ, Carter EA, Walker WA, Harmatz PR. Differential regulation of B7 mRNA in enterocytes and lymphoid cells. Immunology. 1993;79:434–8. [PMC free article] [PubMed] [Google Scholar]
- 184.Cruickshank SM, McVay LD, Baumgart DC, Felsburg PJ, Carding SR. Colonic epithelial cell mediated suppression of CD4 T cell activation. Gut. 2004;53:678–84. doi: 10.1136/gut.2003.029967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4+CD25+ regulatory T cells is mediated by cell surface-bound transforming growth factor β. J Exp Med. 2001;194:629–44. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–83. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 187.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 188.Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–51. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 189.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–25. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 190.Gutierrez MG, Vázquez CL, Munafó DB, Zoppino FC, Berón W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cellular Micro. 2005;7:981–93. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 191.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci USA. 2006;103:14578–83. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 193.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–14. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–82. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–8. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- 196.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jackson WT, Giddings TH, Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–53. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Suhy DA, Giddings TH, Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–65. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–53. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol. 2006;80:6061–71. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Micro. 2008;159:367–73. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 204.Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques. 2005;39:729–36. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- 205.Lee HK, Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20:23–9. doi: 10.1016/j.coi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–82. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 209.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 210.Sartor RB. Does Mycobacterium avium subspecies paratuberculosis cause Crohn’s disease? Gut. 2005;54:896–8. doi: 10.1136/gut.2004.055889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Golde DW. Aetiology of regional enteritis. Lancet. 1968;1:1144–5. doi: 10.1016/s0140-6736(68)90202-x. [DOI] [PubMed] [Google Scholar]
- 212.Selby W, Pavli P, Crotty B, Florin T, Radford-Smith G, Gibson P, et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin and clofazimine for Crohn’s disease. Gastroenterology. 2007;132:2313–9. doi: 10.1053/j.gastro.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 213.Feller M, Huwiler K, Stephan R, Altpeter E, Shang A, Furrer H, et al. Mycobacterium avium subspecies paratuberculosis and Crohn’s disease: a systematic review and meta-analysis. Lancet Infect Dis. 2007;7:607–13. doi: 10.1016/S1473-3099(07)70211-6. [DOI] [PubMed] [Google Scholar]
- 214.Lamps LW, Madhusudhan KT, Havens JM, Greenson JK, Bronner MP, Chiles MC, et al. Pathogenic Yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn’s disease. Am J Surg Pathol. 2003;27:220–7. doi: 10.1097/00000478-200302000-00011. [DOI] [PubMed] [Google Scholar]
- 215.Kallinowski F, Wassmer A, Hofmann MA, Harmsen D, Heesemann J, Karch H, et al. Prevalence of enteropathogenic bacteria in surgically treated chronic inflammatory bowel disease. Hepatogastroenterology. 1998;45:1552–8. [PubMed] [Google Scholar]
- 216.Liu Y, van Kruiningen HJ, West AB, Cartun RW, Cortot A, Colombel JF. Immunocytochemical evidence of Listeria, Escherichia coli and Streptococcus antigens in Crohn’s disease. Gastroenterology. 1995;108:1396–404. doi: 10.1016/0016-5085(95)90687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 218.Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–11. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 219.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–13. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 220.Neut C, Bulois P, Desreumaux P, Membre JM, Lederman E, Gambiez L, et al. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am J Gastroenterol. 2002;97:939–46. doi: 10.1111/j.1572-0241.2002.05613.x. [DOI] [PubMed] [Google Scholar]
- 221.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 223.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2 + D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–75. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, Seganti L, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55:1760–7. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect Immun. 1999;67:4499–509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529–37. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 228.Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Jezek GE, et al. Escherichia coli isolated from a Crohn’s disease patient adheres, invades and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397–409. doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 229.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–18. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 230.Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009;15:872–82. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 231.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042–54. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 232.La Ferla K, Seegert D, Schreiber S. Activation of NFκB in intestinal epithelial cells by E. coli strains isolated from the colonic mucosa of IBD patients. Int J Colorectal Dis. 2004;19:334–42. doi: 10.1007/s00384-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 233.Carvalho FA, Barnich N, Sauvanet P, Darcha C, Gelot A, Darfeuille-Michaud A. Crohn’s disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm Bowel Dis. 2008;14:1051–60. doi: 10.1002/ibd.20423. [DOI] [PubMed] [Google Scholar]
- 234.Bringer MA, Glasser AL, Tung CH, Meresse S, Darfeuille-Michaud A. The Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cellular Micro. 2006;8:471–84. doi: 10.1111/j.1462-5822.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 235.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cellular Micro. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 237.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–74. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Fahlgren A, Baranov V, Frangsmyr L, Zoubir F, Hammarstrom ML, Hammarstrom S. Interferon-gamma tempers the expression of carcinoembryonic antigen family molecules in human colon cells: a possible role in innate mucosal defence. Scand J Immunol. 2003;58:628–41. doi: 10.1111/j.1365-3083.2003.01342.x. [DOI] [PubMed] [Google Scholar]
- 240.Chan CH, Stanners CP. Novel mouse model for carcinoembryonic antigen-based therapy. Mol Ther. 2004;9:775–85. doi: 10.1016/j.ymthe.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 241.Hill AB. The environment and disease: Association or causation? Proc R Soc Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Heinonen KM, Nestel FP, Newell EW, Charette G, Seemayer TA, Tremblay ML, Lapp WS. T-cell protein tyrosine phosphatase deletion results in progressive systemic inflammatory disease. Blood. 2004;103:3457–64. doi: 10.1182/blood-2003-09-3153. [DOI] [PubMed] [Google Scholar]
- 244.Achkar JP, Dassopoulos T, Silverberg MS, Tuvlin JA, Duerr RH, Brant SR, et al. Phenotype-stratified genetic linkage study demonstrates that IBD2 is an extensive ulcerative colitis locus. Am J Gastroenterol. 2006;101:572–80. doi: 10.1111/j.1572-0241.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 245.Vanbervliet B, Homey B, Durand I, Massacrier C, Ait-Yahia S, de Bouteiller O, et al. Sequential involvement of CCR2 and CCR6 ligands for immature dendritic cell recruitment: possible role at inflamed epithelial surfaces. Eur J Immunol. 2002;32:231–42. doi: 10.1002/1521-4141(200201)32:1<231::AID-IMMU231>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 246.Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 247.Bamias G, Mishina M, Nyce M, Ross WG, Kollias G, Rivera-Nieves J, et al. Role of TL1A and its receptor DR3 in two models of chronic murine ileitis. Proc Natl Acad Sci USA. 2006;103:8441–6. doi: 10.1073/pnas.0510903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kabashima K, Nagamachi M, Honda T, Nishigori C, Miyachi Y, Tokura Y, Narumiya S. Prostaglandin E2 is required for ultraviolet B-induced skin inflammation via EP2 and EP4 receptors. Lab Invest. 2007;87:49–55. doi: 10.1038/labinvest.3700491. [DOI] [PubMed] [Google Scholar]
- 250.Goyette P, Lefebvre C, Ng A, Brant SR, Cho JH, Duerr RH, et al. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunol. 2008;1:131–8. doi: 10.1038/mi.2007.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wrackmeyer U, Hansen GH, Seya T, Danielsen EM. Intelectin: a novel lipid raft-associated protein in the enterocyte brush border. Biochemistry. 2006;45:9188–97. doi: 10.1021/bi060570x. [DOI] [PubMed] [Google Scholar]
- 252.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 253.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–20. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]