Abstract
Paragangliomas (PGLs) are frequently associated with germline mutations in genes involved in energy metabolism. The purpose of the present study was to assess whether the tumor metabolomic profile of patients with hereditary and apparently sporadic PGLs enables the distinction of different subtypes of tumors. Twenty-eight unrelated patients with a histological diagnosis of PGLs were included in the present study. Twelve had germline mutations in SDHx genes (5 SDHB, 7 SDHD), 6 VHL, and 10 were apparently sporadic. Intact tumor samples from these patients (one per patient) were evaluated with 1H high-resolution magic angle spinning (HRMAS) NMR spectroscopy. SDHx-related tumors were characterized by an increase in succinate levels in comparison to other tumor subtypes (p = 0.0001 vs VHL and p = 0.000003 vs apparently sporadic). Furthermore, we found significantly lower values of glutamate in SDHx-related tumors compared to other subtypes (p = 0.0007 vs VHL and p = 0.003 vs apparently sporadic). Moreover, SDHx-tumors also exhibited lower values of ATP/ADP/AMP (p = 0.01) compared to VHL. VHL tumors were found to have the highest values of glutathione (GSH) compared to other tumors. Based on 4 metabolites (succinate, glutamate, GSH, and ATP/ADP/AMP), tumors were accurately distinguished from the other ones on both 3- and 2-class PLS-DA models. The present study shows that HRMAS NMR spectroscopy is a very promising method for investigating the metabolomic profile of various PGLs. The present data suggest the existence of a specific succinate-glutamate hallmark of SDHx PGLs. The relevance of such a metabolomic hallmark is expected to be very useful in designing novel treatment options as well as improving the diagnosis and follow-up of these tumors, including metastatic ones.
Introduction
Paragangliomas (PGLs) are neural crest-derived neuroendocrine neoplasms arising from chromaffin cells that are located in the adrenal medulla (also called pheochromocytomas, PHEOs) or aligned along the para-aortic sympathetic system or from head and neck or thoracic paraganglia, which often act as chemoreceptors. Although most PGLs arise sporadically, PGL susceptibility genes have been identified in approximately one-third of cases and in more than 90% of patients with multifocal tumors [1], [2].
Hereditary PGLs are frequently associated with germline mutations in one of the succinate dehydrogenase subunits genes (collectively named SDHx-related tumors), often in the absence of positive family history or even with negative biochemistry [3]. This is in contrast to other hereditary PHEOs/PGLs, in which the family history is usually very well known and therefore, PHEOs/PGLs related to these syndromes can be diagnosed early and often properly and successfully treated.
Moreover, SDHx tumors are considered to be more aggressive, particularly those with SDHB mutations. Currently there is no cure, and current therapeutic options are either suboptimal or short-lasting despite a good initial response [4]. Therefore, any new information that could bring new insight into the pathogenesis of these tumors, their metabolic activities, and proper monitoring, including early assessment of their therapeutic responses, is very desirable.
SDH (also called respiratory complex II) is an iron-sulfur cluster-containing protein composed of 4 subunits that participates in the tricarboxylic acid (TCA) cycle and electron transport chain (ETC). SDH mediates the transfer of two electrons to CoQ during the oxidation of succinate to fumarate. Inactivation of SDH leads to the accumulation of succinate. The accumulation of specific metabolites has been illustrated in different tumor models with inherited and acquired alterations in enzymes of the TCA cycle, such as fumarate in cases of fumarate hydroxylase gene mutations [5] and 2-hydroxyglutarate (2HG) in mutations in one of the 2 isocitrate dehydrogenase genes (IDH1/2) [6]. The accumulation of these oncometabolites inhibits 2-oxoglutarate (2OG)-dependent dioxygenases that include EglN (also called PHD) family members, methylcytosine dioxygenases and JmjC domain-containing histone demethylases, which induces both pseudohypoxic and hypermethylated phenotypes [7]–[10]. These findings have important implications for our understanding of tumorigenesis via pseudohypoxia, alteration of epigenetic homeostasis and alteration of the cellular redox state.
Simultaneous detection of several metabolites (also called metabolomics) is a fast-growing field, promising to improve our understanding of cell biology and relating the genetics and epigenetics to tumor phenotypes. The metabolites (molecular weight <1 kDa) can be detected in vitro by gas chromatography-mass spectrometry (GCMS), liquid chromatography-mass spectrometry (LCMS), and nuclear magnetic resonance (NMR) spectroscopy [11]. Magnetic resonance spectroscopy also enables in vivo assessment of metabolites (such as 2HG) in the setting of brain tumors [12]. NMR-based methods are less sensitive but more reproducible than LCMS for the quantitative assessment of metabolites.
In recent years, 1H high-resolution magic angle spinning (HRMAS) NMR spectroscopy has been introduced for ex vivo characterization of small intact tissue samples (solid-state NMR). This technique also offers several advantages, such as the simplicity of sample preparation, the intra- and inter-laboratory reproducibility, the relatively low cost to perform the technique, and the availability of major metabolite databases [13]. HRMAS NMR spectroscopy has been suggested as a promising tool in the diagnosis and characterization of cancer.
The objective of the present study was to find out whether there is a specific metabolomic profile in sympathetic SDHx and other hereditary or apparently sporadic PHEOs/PGLs.
Materials and Methods
Ethics Statement
Data was acquired under regular clinical care conditions, with Ethics Committee approval obtained for the use of these data for scientific purposes at Strasbourg, Marseille, and Nancy University Hospitals. Written informed consent was obtained from all patients included in the present study.
Tissue samples
Twenty-eight specimens of sympathetic PGLs/PHEOs from 28 unrelated patients who fulfilled the following criteria were included:
Histological diagnosis of PGL/PHEO.
Absence of distant metastases within the first year following surgery.
Genetic screening for germline mutations in the SDHB/C/D, SDHAF2 (including large gene rearrangements of all the SDH genes), VHL (including large gene rearrangements), RET, TMEM127, and MAX genes.
Tissue specimens collected just after tumor removal and snap-frozen in liquid nitrogen before storage at −80°C.
The tumors were obtained from 3 different institutions in France (Strasbourg, Marseille, and Nancy University Hospitals) and were distributed as follows: 10 sporadic (9 PHEOs, 1 sympathetic PGL), 5 SDHB (2 PHEOs, 3 sympathetic PGLs), 7 SDHD (1 PHEO, 6 sympathetic PGLs), and 6 VHL (3 PHEOs, 3 sympathetic PGLs).
Tissue sample preparation for HRMAS NMR
The amount of tissue used for HRMAS analysis ranged from 15 to 20 mg. For each sample, the percentage of tumor cells in the total sample of cells and the percentage of necrosis with regard to the total surface were calculated based on frozen sections using a mirror sample stored in the tissue bank. Samples containing at least 30% tumor cells and less than 50% necrosis were used for the study. Each tissue sample was placed in a 30 µL disposable insert. 10 µL of D2O were added to the rotor to provide a lock frequency for the NMR spectrometer. The exact weight of the sample used was determined by weighing the empty insert and the insert containing the tissue sample. The insert was stored at −80°C and placed in a 4-mm ZrO2 rotor just before the HRMAS analysis.
HRMAS NMR data acquisition, spectra processing, and metabolite quantification
1D HRMAS NMR spectra were recorded on a Bruker Avance III 500 spectrometer operating at a proton frequency of 500.13 MHz, installed at the Pathological Department of Strasbourg University Hospitals. A one-dimensional (1D) proton spectrum using a Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence [14] and 1024 transients was acquired for each serum sample. Free induction decays were multiplied by an exponential window function of 0.3 Hz prior to Fourier transformation and were corrected for phase and baseline distortions using TopSpin 2.1 (Bruker GmbH, Germany). The chemical shift was referenced to the peak of the methyl proton of L-lactate at 1.33 ppm. In order to confirm resonance assignments, two-dimensional (2D) homonuclear and heteronuclear experiments were also recorded immediately after the end of 1D spectra acquisition. Because the duration of these experiments is long and significant tissue degradation occurs during NMR acquisition, only a few representative samples were analyzed by 2D experiments. A description of the HRMAS NMR data acquisition and spectra processing has been previously detailed [13].
In the present study, we focused on four selected metabolites with a potential key role in tumoral pathophysiology. Accordingly, succinate, glutamate, glutathione, and energy phosphorylated compounds were precisely identified and quantified. Catecholamines were also measured. The quantification procedure was based on the pulse length-based concentration measurement (PULCON) as previously described [13]. Spectra were normalized according to each sample weight and calibrated using the signal intensity of a 19.3 nmol reference solution of lactate, scanned under the same analytical conditions. The peak integral corresponding to each metabolite's region was normalized to the integral of the entire spectrum within the range of 8.65–1 ppm. Quantification results were expressed as nmol/mg of tissue. Metabolites were assigned using standard metabolite chemical shift tables available in the literature [15], [16].
Succinate concentrations in tissue samples were estimated by integrating the area comprised between 2.39 ppm and 2.43 ppm. The spectral complex previously assigned to ATP/ADP/AMP [17] was identified on our spectra within the range of 6.07–6.11 and thus integrated to quantify the metabolite amount. The glutathione and glutamate amounts were measured from the integral of NMR spectra comprised by 2.93–2.98 ppm and 2.32–2.38 ppm, respectively. To measure epinephrine concentrations in tissue samples, the signal resulting from the N-methyl radical selected at 2.75 ppm was considered. The 3,4-dihydroxybenzene groups of both epinephrine and norepinephrine generate a spectral complex between about 6.85 and 6.98 ppm. The integral of the region corresponding to the 1H in position no. 5 of the aromatic ring (IUPAC nomenclature) was selected to quantify the amount of epinephrine plus norepinephrine in each tissue sample. Finally, the norepinephrine concentration was obtained by subtracting epinephrine from the sum of epinephrine and norepinephrine. The above approach has been previously tested and confirmed (data not shown) by the quantification NMR analysis of epinephrine and norepinephrine standard solutions first separated and subsequently mixed (1/1, v/v). Dopamine could contribute to peaks in the region between 6.85 and 6.98 ppm. However, no triplets at 2.85 ppm and 3.22 ppm, which represent the spectral signature of dopamine, were detected on HRMAS NMR spectra, suggesting an undetectable amount of dopamine in the analyzed tissue.
Statistical analysis
As widely suggested [18], [19], a combination of principal component analysis (PCA) and partial least square discriminant analysis (PLS-DA) was adopted. PCA and PLS-DA analysis were done including only succinate, glutamate, glutathione, and energy phosphorylated compounds.
A PCA was performed to evaluate the quality of the data quickly and to identify possible outliers [20]. Then a PLS-DA was employed to optimize the separation between groups and to classify the samples in each of the following 3- or 2-group models: 1. Sporadic vs. SDHx vs. VHL, 2. Sporadic vs. SDHx, 3. Sporadic vs. VHL, 4. SDHx vs. VHL. Cross-validation was used in each PLS-DA model to determine the number of components and to avoid overfitting the data because of the small number of samples. An extensive cross-validation embedded in a Monte-Carlo resampling approach was used during the construction of the model in order to build a confusion matrix that allowed a direct visualization of the performances of the model in term of classification power (sensitivity (Se), specificity (Sp), positive predictive value (PPV), negative predictive value (NPV)) [21], [22]. The non-parametric Mann-Whitney U-test (MWU-test) was utilized for the comparison of metabolite amounts between groups. The results of tissular metabolite concentrations are expressed as medians and ranges. The relationship among the selected metabolites was assessed by the Spearman nonparametric regression coefficient (R).
SIMCA P (version 11.0, Umetrics AB, Umeå, Sweden) and STATISTICA 7 (STATSOFT; www.statsoft.com) packages were used for statistical data analysis. A p value less than 0.05 was considered statistically significant.
Results
General findings
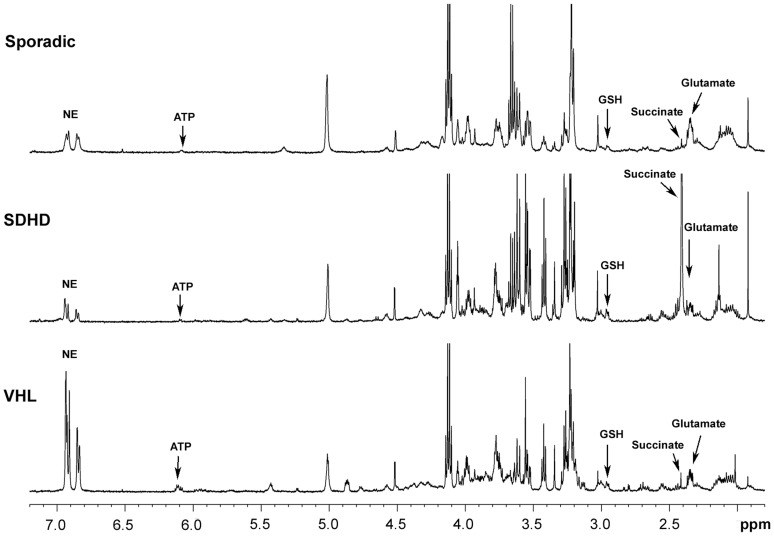
Representative 1D HRMAS CPMG spectra of sporadic, SDHD-, and VHL-related PGLs are presented in Figure 1. Metabolites such as succinate, glutamate, glutathione (GSH), and energy phosphorylated compounds (ATP/ADP/AMP) were identified in 1D spectra, with variable signal intensity according to the genetic background.
Figure 1. Representative 1D 1H-HRMAS spectra of sporadic, SDHx-, and VHL-related PGLs.
The partial metabolite assignment is indicated. The metabolic contents of each spectra are directly comparable, since the intensity of each spectrum has been normalized with respect to the weight of the analyzed sample.
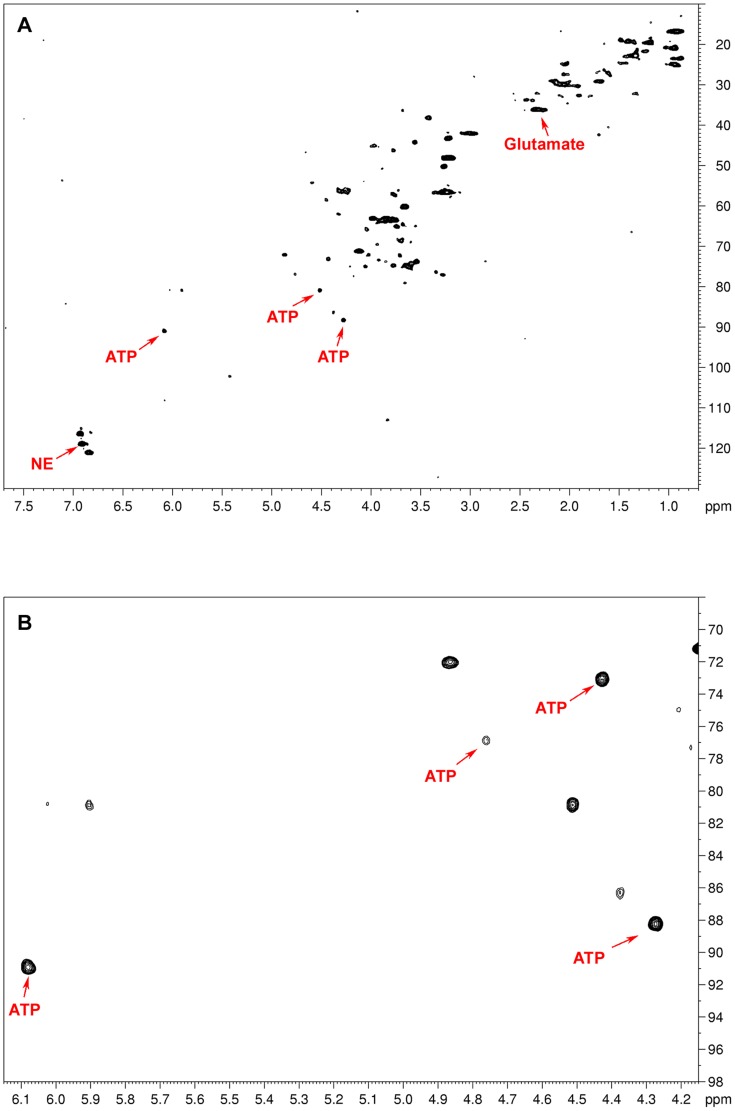
In all the examined tumor samples, no significant spectra overlap that prevented quantification was found. The quality of the examined samples was indirectly proven by the lack of signals arising from both lactate and fatty acids, which are usually considered hallmarks of tissular necrosis. 2D NMR acquisitions were used to better characterize the metabolic substrate appearing on our 1D spectra within the range of 6.07–6.11 ppm and previously assigned to ATP/ADP/AMP (Fig. 2). In particular, the results of 2D HRMAS acquisition from one VHL-related PGL were compared to the widely available NMR spectroscopy database [16], [23]. Our 2D HSQC spectrum was almost superimposable to the known reference of ATP. Nevertheless, a slight chemical shift difference of 0.06 ppm related to the carbon atom linking the ribosyl ring and the aminopurinine may generate confusion with inosine triphosphate (ITP). This difference could be explained by the intratissular pH variability in our samples.
Figure 2. Representative 2D 1H-13C HSQC HRMAS NMR spectra (A), zoomed in on the ATP region (B), obtained from a sample of a VHL-related abdominal PGL.
The 2D NMR experiment confirms the presence of ATP in tumoral tissue as suggested by the 1D spectra of the same patient (Figure 1C). Glutamate and norepinephrine assignments are also indicated. Because of their low concentration in the tissular sample, both succinate and glutathione are not clearly visible in the whole 2D spectrum (7.7-0.7 ppm in 1H dimension) without focusing on their characteristic regions.
All NMR spectra from the SDHx-related tumors showed very high levels of succinate compared to sporadic and VHL-related tumors. On the other hand, apparently sporadic tumors were characterized by high values of glutamate compared to SDHx-related PGLs/PHEOs. Finally, VHL-related tumors exhibited more pronounced ATP/ADP/AMP signals. GSH was predominant in SDHx- and VHL-related tumors.
Three-class model
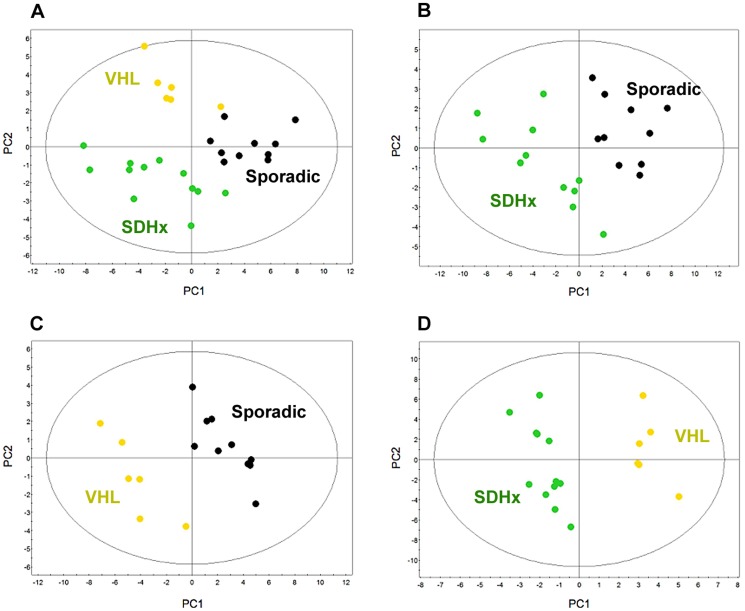
A multivariate two-component PLS-DA based on succinate, glutamate, GSH, and energy phosphorylated compounds (ATP/ADP/AMP) was first generated including patients with apparently sporadic, SDHx-, and VHL-related PGLs/PHEOs. The score plot of the PLS-DA model (Fig. 3A) showed a clear distinction between these three classes of tumors. According to this model, all 12 patients with SDHx-related PGLs/PHEOs were correctly identified. Moreover, eight of ten (80%) and five of six (83%) patients with respectively apparently sporadic and VHL-related PGLs/PHEOs were also accurately classified. To detect patients with germline mutations (SDHx and VHL), the Se and Sp of the established model were respectively 94% and 80%. PPV, NPV, and global accuracy were all 89%.
Figure 3. Results of two-component PLS-DA models built on succinate, glutamate, glutathione, and energy phosphorylated compounds (ATP/ADP/AMP) according to the patients' genotypes: A) 3-class model including apparently sporadic (black dots), SDHx- (green dots), and VHL-related tumors (yellow dots); B) 2-class model including apparently sporadic and SDHx-related tumors; C) 2-class model including apparently sporadic and VHL-related tumors; D) 2-class model including SDHx- and VHL-related tumors.
A clear distinction between the different classes of tumors is shown in each model.
In the SDHx-related tumor cohort, PHEOs exhibited higher values of energy-phosphorylated compounds (ATP/ADP/AMP) compared to PGLs (p = 0.04). Succinate, GSH, and glutamate were statistically equivalent regardless of the SDHx tumor location. In contrast, VHL-related PHEOs had higher GSH levels than PGLs (p = 0.04). Considering all tissue samples, a significant negative correlation was shown between succinate and glutamate (R = 0.52), glutamate and energy phosphorylated compounds (ATP/ADP/AMP) (R = 0.54), and succinate and GSH (R = 0.51).
Two-class models
Three multivariate PLS-DA models based on succinate, glutamate, GSH, and energy phosphorylated compounds (ATP/ADP/AMP) were built to compare: (1) apparently sporadic to SDHx- (Fig. 3B), (2) apparently sporadic to VHL- (Fig. 3C), and (3) SDHx- to VHL-related tumors (Fig. 3D). The score plot of each PLS-DA model showed a very clear delineation of the single classes. The results of the MWU-test are detailed in Table 1.
Table 1. Median values and ranges of succinate, glutathione, glutamate, and ATP/ADP/AMP levels measured by 1D 1H-HRMAS spectroscopy in intact tissue samples of 28 apparently sporadic, SDHx, and VHL-related PHEOs/PGLs. Quantification results are expressed as nmol/mg of tissue.
| Sporadic | SDHx | VHL | P Sporadic vs SDHx | P Sporadic vs VHL | P SDHx vs VHL | |
| Succinate | 0.03 (0–0.06) | 3,6 (0.9–9.07) | 0.06 (0.03–0.12) | 0.000003 | 0.02 | 0.0001 |
| Glutathione | 0.04 (0.03–0.22) | 0.15 (0.05–0.39) | 0.32 (0.17–0.43) | 0.01 | 0.003 | 0.08 |
| Glutamate | 1.36 (0.23–2.55) | 0.67 (0.13–1.52) | 1.49 (0.88–2.18) | 0.003 | 1 | 0.0007 |
| ATP/ADP/AMP | 0.10 (0.03–0.18) | 0.05 (0–0.21) | 0.16 (0.06–0.29) | 0.1 | 0.1 | 0.01 |
When comparing apparently sporadic to SDHx-related tumors, the model correctly classified 9 of 10 apparently sporadic cases and 11 of 12 SDHx. The model identified SDHx-related tumors with a Se of 92%, a Sp of 90%, a PPV of 92%, a NPV of 90%, and a global accuracy of 91%. The model did not correctly identify only one SDHD-related PGL. Interestingly, it was easily recognized by visual inspection of the spectra because of a pronounced succinate peak.
In the model comparing apparently sporadic and VHL-related tumors, all cases were correctly classified. The VHL group exhibited a significant increase in both succinate and GSH levels.
Finally, SDHx- and VHL-related PGLs/PHEOs were compared. The model was able to correctly classify all the tumor samples. Succinate remains highly discriminant between the 2 subgroups. No difference in the succinate amount was shown between SDHB and SDHD subtypes, as well as for the remaining selected metabolites. VHL-related tumors showed a more abundant content of energy phosphorylated compounds (ATP/ADP/AMP). Interestingly, despite a greater amount of GSH in VHL vs. SDHx tumors, this parameter did not reach statistical significance (p = 0.08).
In apparently sporadic PGLs/PHEOs, glutamate and energy phosphorylated compounds (ATP/ADP/AMP) (R = 0.71) as well as succinate and GSH (R = 0.68) in SDHx-related tumors were statistically correlated.
Epinephrine was not measurable in any VHL-related tumors and in only one of 12 SDHx samples (1.98 nmol/ml). Only 2 of 10 apparently sporadic tumors showed an epinephrine peak in the NMR spectra (0.93 and 1.04 nmol/ml). On the other hand, norepinephrine was detected in 9 of 10 apparently sporadic tumors (median, 0.90 nmol/ml; range, 0–4.60 nmol/ml), in 9 of 12 SDHx-related PHEOs/PGLs (median, 0.87 nmol/ml; range, 0–2.40 nmol/ml), and in all of the 6 VHL-related PHEOs/PGLs (median, 1.80 nmol/ml; range, 0.26–4.53 nmol/ml). No statistical difference was observed between VHL and SDHx PHEOs/PGLs despite higher values of norepinephrine in VHL tumors.
Discussion
In the present study, we analyzed 28 sympathetic PGLs with different genetic backgrounds (10 sporadic, 6 VHL, 12 SDHx). We did not include RET- or NF1-related tumors because these are almost always associated with other syndromic manifestations that enable physicians to easily diagnose them.
In the present study we show that HRMAS NMR-based metabolomics should be considered as a new reliable tool for ex vivo characterization of sympathetic PGLs. The present data suggests the existence of a specific succinate-glutamate hallmark of SDHx sympathetic PGLs. Based on 4 metabolites (succinate, glutamate, GSH, and ATP/ADP/AMP), tumors were accurately distinguished from the other ones on both 3- and 2-class PLS-DA models. This is an extension of the results published by Rao et al., who identified an SDHx signature (increased succinate, decreased ATP/ADP/AMP) but with some overlaps between apparently sporadic and VHL tumors for the other parameters [17].
Until the present, only a few studies employing HRMAS NMR spectroscopy have been published for metabolic profiling of PGLs/PHEOs [17], [24]. An increase in succinate accumulation and a decrease in ATP/ADP/AMP accumulation were observed in SDHx-related PGLs when compared to sporadic PGLs and PGLs of other genotypes [17]. Interestingly, despite methodological differences, we found similar results. Compared to the previous study [17], in which liquid NMR spectroscopy was used to analyze homogenized and centrifuged tissue preparation, our analysis was done on intact tissue samples. This approach may be considered less sensitive compared to liquid-state NMR spectroscopy but also leads to good quality spectra. Furthermore, it has several advantages, such as avoiding technical procedures for tissue preparation prior to the analysis and allowing refreeze and storage of tissue samples for additional analyzes such as pathological analysis in cases of result discrepancies. Small changes (0.1 ppm) in chemical shift in the peak areas assigned to the ATP/ADP/AMP between the 2 studies could be attributed to pH differences (perchloric acid extracts for Rao et al) [17].
We also found lower values of ATP/ADP/AMP in SDHx-related tumors in comparison to VHL tumors, probably because increased glycolysis preferentially occurs in VHL tumors [17], [25].
We have added greater levels of discrimination by using additional metabolites besides succinate and ATP/ADP/AMP peaks. We found that apparently sporadic tumors were characterized by a low level of reduced GSH compared to VHL tumors (p = 0.01) and SDHx-related tumors (p = 0.003). There was a trend towards statistical differences between SDHx- and VHL-related tumors (p = 0.08), but the sample size was too small to achieve statistical significance. GSH is the most prevalent non-protein thiol in animal cells. Its high redox potential renders GSH both a potent antioxidant and a convenient cofactor for enzymatic reactions that require readily available electron pairs. GSH appears to be a sensitive indicator of the ability to resist toxic challenge. It has previously been shown that sporadic tumors may also exhibit reduction of mitochondrial respiration [26]. The low GSH peak in these tumors could be related to several cell events, such as inhibition of the ETC that is needed for GSH regeneration via NADPH synthesis and/or an increase in ROS production.
Interestingly, SDHx-related tumors were also characterized by low glutamate levels in comparison to apparently sporadic (p = 0.003) and VHL tumors (p = 0.0007). This finding is new and provides critical information for accurate classification of tumors. Intracellular glutamate is mainly produced by the oxidative deamination of glutamine. Interestingly, IDH1/2 mutant cells are also characterized by lower levels of glutamate with respect to their wild type counterparts [27]. It is also possible that lower glutamate availability in SDHx-deficient tumors leads to the generation of perooxidant conditions that promote carcinogenesis. There is no clear explanation for the low glutamate level in SDHx-related tumors in comparison to other subtypes. It could be related to increased consumption of glutamate for further biosynthetic pathways or decreased glutamate biosynthesis via modulation of the activity of glutaminase or glutamate dehydrogenase.
In contrast, increased GSH levels in VHL-tumors may be related to increased activity of the pentose phosphate pathway in response to enhanced oxidative stress.
Our 3-class model enabled us to accurately predict the presence of SDHx or VHL germline mutations with the following performances: Se 94%, Sp 80%, and PPV/NPV 89%. The presence of a succinate peak alone easily distinguishes SDHx-related tumors from the other ones. The 2-class model based on succinate/GSH could be used to distinguish VHL from apparently sporadic tumors (global accuracy of 100%). Although our results are promising, they should be validated in a large sample size and independent cohort. This would help build a reliable probability predictive model that may be used in clinics to guide genetic testing and provide functional information about mutations of equivocal pathogenicity (uncertain variants).
Immunohistochemical studies can also be used as a screening method for guiding genetic testing. The absence of SDHB immunoexpression is indicative of a germline mutation in one of the SDHx genes [28], [29]. Additionally, a subset of these tumors may also be immunohistochemically negative for SDHA, which is highly suggestive of the presence of an SDHA mutation [30]. Compared to other methods, metabolomic approaches provide several practical advantages, such as the simplicity of sample preparation, reproducibility, low cost for sample analysis, and simultaneous measurement of several metabolites. Thus, they may enable global metabolomic profiling of tumors with a distinction of different tumor subtypes. Metabotyping various tumors, including PGLs, might indeed identify new oncometabolites for their future diagnostic and therapeutic options.
The present study further supports the existence of different secretory phenotypes in PGLs according to their hereditary background. In our series, epinephrine was not measurable in any VHL-related tumors and in only one of the 12 SDHx samples. By contrast, norepinephrine was detected in 9 out of 12 SDHx-related tumors and in all of the 6 examined VHL-related PHEOs/PGLs. The lack of epinephrine secretion in VHL- and SDHx-related PGLs was previously reported [31]–[33] and linked to reduced or absent expression of phenylethanolamine-N-methyltransferase (PNMT). PNMT catalyzes the last step in catecholamine biosynthesis, the conversion of norepinephrine to epinephrine. SDHx-related tumors exhibit an immature catecholamine secretory profile by the absence of epinephrine, possibly explained by hypermethylation of the PMNT promoter [34]. These tumors accumulate succinate, which inhibits 2-oxoglutarate-dependent histone and DNA demethylase enzymes, resulting in epigenetic modifications [8]. These findings further emphasize the interplay between the Krebs cycle, oxidative phosphorylation, and epigenetic events [35], [36].
We acknowledge several limitations to the present study: a) the absence of carriers of mutations in the TMEM127/SDHAF2/MAX/SDHA genes; and b) the absence of screening of tumors for somatic mutations, mainly in apparently sporadic tumors, that could potentially occur and influence the presented tumor metabolomic profiles. We also have to admit that the high proportion of hereditary PGLs in our series is related to a selection bias. Nevertheless, the present study provides new data about specific genotype-metabolomic phenotype of SDHx-related PGLs compared to VHL-related and apparently sporadic counterparts.
The present study advances our knowledge of the pathogenesis of PGLs and also provides important and promising information about the classification of these tumors. It is expected that in the near future, various metabolomic approaches will reveal new diagnostic and therapeutic targets. A large-scale study comparing PGL clinical and imaging phenotypes, genotypes, somatic mutations, gene expression levels, DNA methylation phenotypes, protein spectra, and metabolomic profiles will have a great impact on our understanding of how they function and contribute to the development of various PGLs, resulting in the most optimal early and accurate diagnosis, treatment, and outcome.
Acknowledgments
Victoria Martucci and the personnel of the Tumor Bio-bank (Centre de Ressources Biologiques) of the Strasbourg University Hospitals are gratefully acknowledged for their technical assistance.
Funding Statement
This work is part of the CARMeN project and was supported by grants from Région Alsace, Oséo, Communauté Urbaine de Strasbourg, Conseil Départemental du Bas-Rhin, Bruker BioSpin, University of Strasbourg and Strasbourg University Hospital. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jafri M, Whitworth J, Rattenberry E, Vialard L, Kilby G, et al. (2013) Evaluation of SDHB, SDHD and VHL gene susceptibility testing in the assessment of individuals with non-syndromic phaeochromocytoma, paraganglioma and head and neck paraganglioma. Clinical endocrinology 78: 898–906. [DOI] [PubMed] [Google Scholar]
- 2. Piccini V, Rapizzi E, Bacca A, Di Trapani G, Pulli R, et al. (2012) Head and neck paragangliomas: genetic spectrum and clinical variability in 79 consecutive patients. Endocr Relat Cancer 19: 149–155. [DOI] [PubMed] [Google Scholar]
- 3. Timmers HJ, Gimenez-Roqueplo AP, Mannelli M, Pacak K (2009) Clinical aspects of SDHx-related pheochromocytoma and paraganglioma. Endocr Relat Cancer 16: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang H, Abraham J, Hung E, Averbuch S, Merino M, et al. (2008) Treatment of malignant pheochromocytoma/paraganglioma with cyclophosphamide, vincristine, and dacarbazine: recommendation from a 22-year follow-up of 18 patients. Cancer 113: 2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, et al. (2005) HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8: 143–153. [DOI] [PubMed] [Google Scholar]
- 6. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaelin WG Jr, McKnight SL (2013) Influence of metabolism on epigenetics and disease. Cell 153: 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, et al. (2013) SDH Mutations Establish a Hypermethylator Phenotype in Paraganglioma. Cancer Cell 23: 739–752. [DOI] [PubMed] [Google Scholar]
- 9. Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, et al. (2005) Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet 14: 2231–2239. [DOI] [PubMed] [Google Scholar]
- 10. Xiao M, Yang H, Xu W, Ma S, Lin H, et al. (2012) Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development 26: 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart DS (2013) Exploring the human metabolome by nuclear magnetic resonance spectroscopy and mass spectroscopy; Lutz NW, editor. Cambridge: Cambridge University Press.
- 12. Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, et al. (2012) Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. Journal of neuro-oncology 107: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piotto M, Moussallieh FM, Imperiale A, Benahmed MA, Detour J, et al.. (2013) Reproducible sample preparation and spectrum acquisition techniques for metabolic profiling of human tissues by proton high-resolution magic angle spinning nuclear magnetic resonance; Lutz NW, editor. Cambridge: Cambridge University Press. 627 p. [Google Scholar]
- 14. Meiboom S, Gill D (1958) Modified spin echo method for measuring nuclear relaxation times. The Review of scientific instruments 29: 688–691. [Google Scholar]
- 15. Martinez-Bisbal MC, Marti-Bonmati L, Piquer J, Revert A, Ferrer P, et al. (2004) 1H and 13C HR-MAS spectroscopy of intact biopsy samples ex vivo and in vivo 1H MRS study of human high grade gliomas. NMR in biomedicine 17: 191–205. [DOI] [PubMed] [Google Scholar]
- 16. Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, et al. (2007) HMDB: the Human Metabolome Database. Nucleic acids research 35: D521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao JU, Engelke UF, Rodenburg RJ, Wevers RA, Pacak K, et al. (2013) Genotype-Specific Abnormalities in Mitochondrial Function Associate with Distinct Profiles of Energy Metabolism and Catecholamine Content in Pheochromocytoma and Paraganglioma. Clin Cancer Res 19: 3787–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, et al. (2010) High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nature protocols 5: 1019–1032. [DOI] [PubMed] [Google Scholar]
- 19. Sitter B, Bathen TF, Tessem MB, Gribbestad IS (2009) High-resolution magic angle spinning (HR MAS) MR spectroscopy in metabolic characterization of human cancer. Progress in nuclear magnetic resonance spectroscopy 54: 239–254. [Google Scholar]
- 20. Ebbels TMD, Cavill R (2009) Bioinformatic methods in NMR-based metabolic profiling. Progress in nuclear magnetic resonance spectroscopy 55: 361–374. [Google Scholar]
- 21. Picard RR, Cook RD (1984) Cross-Validation of Regression Models. Journal of the American Statistical Association 79: 575–583. [Google Scholar]
- 22. Xu QS, Liang YZ (2001) Monte Carlo cross validation. Chemometrics and Intelligent Laboratory Systems 56: 1–11. [Google Scholar]
- 23. Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, et al. (2013) HMDB 3.0–The Human Metabolome Database in 2013. Nucleic acids research 41: D801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Imperiale A, Elbayed K, Moussallieh FM, Reix N, Piotto M, et al. (2013) Metabolomic profile of the adrenal gland: from physiology to pathological conditions. Endocr Relat Cancer 20: 705–716. [DOI] [PubMed] [Google Scholar]
- 25. Favier J, Briere JJ, Burnichon N, Riviere J, Vescovo L, et al. (2009) The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One 4: e7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feichtinger RG, Zimmermann FA, Mayr JA, Neureiter D, Ratschek M, et al. (2011) Alterations of respiratory chain complexes in sporadic pheochromocytoma. Frontiers in bioscience 3: 194–200. [DOI] [PubMed] [Google Scholar]
- 27. Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, et al. (2011) Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A 108: 3270–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Nederveen FH, Gaal J, Favier J, Korpershoek E, Oldenburg RA, et al. (2009) An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. The lancet oncology 10: 764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gill AJ, Benn DE, Chou A, Clarkson A, Muljono A, et al. (2010) Immunohistochemistry for SDHB triages genetic testing of SDHB, SDHC, and SDHD in paraganglioma-pheochromocytoma syndromes. Hum Pathol 41: 805–814. [DOI] [PubMed] [Google Scholar]
- 30. Korpershoek E, Favier J, Gaal J, Burnichon N, van Gessel B, et al. (2011) SDHA Immunohistochemistry Detects Germline SDHA Gene Mutations in Apparently Sporadic Paragangliomas and Pheochromocytomas. J Clin Endocrinol Metab 96: E1472–1476. [DOI] [PubMed] [Google Scholar]
- 31. Eisenhofer G, Lenders JW, Linehan WM, Walther MM, Goldstein DS, et al. (1999) Plasma normetanephrine and metanephrine for detecting pheochromocytoma in von Hippel-Lindau disease and multiple endocrine neoplasia type 2. N Engl J Med 340: 1872–1879. [DOI] [PubMed] [Google Scholar]
- 32. Eisenhofer G, Pacak K, Huynh TT, Qin N, Bratslavsky G, et al. (2011) Catecholamine metabolomic and secretory phenotypes in phaeochromocytoma. Endocr Relat Cancer 18: 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eisenhofer G, Walther MM, Huynh TT, Li ST, Bornstein SR, et al. (2001) Pheochromocytomas in von Hippel-Lindau syndrome and multiple endocrine neoplasia type 2 display distinct biochemical and clinical phenotypes. J Clin Endocrinol Metab 86: 1999–2008. [DOI] [PubMed] [Google Scholar]
- 34. Burnichon N, Cascon A, Schiavi F, Morales NP, Comino-Mendez I, et al. (2012) MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clin Cancer Res 18: 2828–2837. [DOI] [PubMed] [Google Scholar]
- 35. Yang M, Pollard PJ (2013) Succinate: a new epigenetic hacker. Cancer Cell 23: 709–711. [DOI] [PubMed] [Google Scholar]
- 36. Burgess DJ (2013) Metabolism: Epigenetic links to the web of Krebs. Nat Rev Cancer 13: 437. [DOI] [PubMed] [Google Scholar]