Abstract
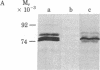
Caulobacter crescentus carries a flagellum and is motile only during a limited time in its cell cycle. We have asked if the biochemical machinery that mediates chemotaxis exists coincident with the cell's structural ability to respond to a chemotactic signal. We first demonstrated that one function of the chemotaxis machinery, the ability to methylate the carboxyl side chains of a specific set of membrane proteins (methyl-accepting chemotaxis proteins, MCPs), is present in C. crescentus. This conclusion is based on the observations that (i) methionine auxotrophs starved of methionine can swim only in the forward direction (comparable to smooth swimming in the enteric bacteria), (ii) a specific set of membrane proteins was found to be methylated in vivo and the incorporated [3H]methyl groups were alkali sensitive, (iii) this same set of membrane proteins incorporated methyl groups from S-adenosylmethionine in vitro, and (iv) out of a total of eight generally nonchemotactic mutants, two were found to swim only in a forward direction and one of these lacked methyltransferase activity. Analysis of in vivo and in vitro methylation in synchronized cultures showed that the methylation reaction is lost when the flagellated swarmer cell differentiates into a stalked cell. In vivo methylation reappeared coincident with the biogenesis of the flagellum just prior to cell division. In vitro reconstitution experiments with heterologous cell fractions from different cell types showed that swarmer cells contain methyltransferase and their membranes can be methylated. However, newly differentiated stalked cells lack methyltransferase activity and membranes from these cells cannot accept methyl groups. These results demonstrate that MCP methylation is confined to that portion of the cell cycle when flagella are present.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aswad D., Koshland D. E., Jr Role of methionine in bacterial chemotaxis. J Bacteriol. 1974 May;118(2):640–645. doi: 10.1128/jb.118.2.640-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. T., Croft R. H., Ferber D. M., Gerardot C. J., Schoenlein P. V., Ely B. Genetic mapping with Tn5-derived auxotrophs of Caulobacter crescentus. J Bacteriol. 1982 Aug;151(2):888–898. doi: 10.1128/jb.151.2.888-898.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Simon M. Bacterial chemotaxis. Annu Rev Physiol. 1982;44:501–517. doi: 10.1146/annurev.ph.44.030182.002441. [DOI] [PubMed] [Google Scholar]
- Contreras I., Shapiro L., Henry S. Membrane phospholipid composition of Caulobacter crescentus. J Bacteriol. 1978 Sep;135(3):1130–1136. doi: 10.1128/jb.135.3.1130-1136.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):384–395. doi: 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Purification of intact flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):376–383. doi: 10.1128/jb.105.1.376-383.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmitt K., Simon M. Purification and thermal stability of intact Bacillus subtilis flagella. J Bacteriol. 1971 Jan;105(1):369–375. doi: 10.1128/jb.105.1.369-375.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P. R., Agabian N. A comparative structural analysis of the flagellin monomers of Caulobacter crescentus indicates that these proteins are encoded by two genes. J Bacteriol. 1982 May;150(2):925–933. doi: 10.1128/jb.150.2.925-933.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Walsh M. P., Ely B., Shapiro L. Flagellar hook and basal complex of Caulobacter crescentus. J Bacteriol. 1979 Jun;138(3):984–989. doi: 10.1128/jb.138.3.984-989.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagenaur C., Agabian N. Physical characterization of Caulobacter crescentus flagella. J Bacteriol. 1976 Oct;128(1):435–444. doi: 10.1128/jb.128.1.435-444.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur C., DeMartini M., Agabian N. Isolation and characterization of Caulobacter crescentus flagellar hooks. J Bacteriol. 1978 Nov;136(2):795–798. doi: 10.1128/jb.136.2.795-798.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhausen M., Gill P. R., Parker G., Agabian N. Cloning of developmentally regulated flagellin genes from Caulobacter crescentus via immunoprecipitation of polyribosomes. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6847–6851. doi: 10.1073/pnas.79.22.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima S., Ohba M., Hayashi H. An in vitro study of the methylation of methyl-accepting chemotaxis protein of Escherichia coli. Construction of the system and effect of mutant proteins on the system. J Biochem. 1981 Feb;89(2):411–420. doi: 10.1093/oxfordjournals.jbchem.a133216. [DOI] [PubMed] [Google Scholar]
- O'Dea R. F., Viveros O. H., Axelrod J., Aswanikaumar S., Schiffmann E., Corcoran B. A. Raipid stimulation of protein carboxymethylation in leukocytes by a chemotatic peptide. Nature. 1978 Mar 30;272(5652):462–464. doi: 10.1038/272462a0. [DOI] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Newton A. Isolation and expression of cloned hook protein gene from Caulobacter crescentus. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4863–4867. doi: 10.1073/pnas.79.16.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike M. C., Kredich N. M., Snyderman R. Requirement of S-adenosyl-L-methionine-mediated methylation for human monocyte chemotaxis. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3928–3932. doi: 10.1073/pnas.75.8.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purucker M., Bryan R., Amemiya K., Ely B., Shapiro L. Isolation of a Caulobacter gene cluster specifying flagellum production by using nonmotile Tn5 insertion mutants. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6797–6801. doi: 10.1073/pnas.79.22.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway H. G., Silverman M., Simon M. I. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J Bacteriol. 1977 Nov;132(2):657–665. doi: 10.1128/jb.132.2.657-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Mansour J., Shaw P., Henry S. Synthesis of specific membrane proteins is a function of DNA replication an phospholipid synthesis in Caulobacter crescentus. J Mol Biol. 1982 Aug 5;159(2):303–322. doi: 10.1016/0022-2836(82)90497-1. [DOI] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Reconstitution and purification of flagellar filaments from Caulobacter crescentus. J Bacteriol. 1977 Dec;132(3):1027–1030. doi: 10.1128/jb.132.3.1027-1030.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T., DeRosier D. J., Aizawa S., Macnab R. M. Flagellar hook structures of Caulobacter and Salmonella and their relationship to filament structure. J Mol Biol. 1982 Nov 25;162(1):69–87. doi: 10.1016/0022-2836(82)90162-0. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T., DeRosier D., Shapiro L., Weissborn A. Three-dimensional reconstruction of the flagellar hook from Caulobacter crescentus. J Mol Biol. 1981 Sep 25;151(3):439–465. doi: 10.1016/0022-2836(81)90005-x. [DOI] [PubMed] [Google Scholar]
- Weissborn A., Steinmann H. M., Shapiro L. Characterization of the proteins of the Caulobacter crescentus flagellar filament. Peptide analysis and filament organization. J Biol Chem. 1982 Feb 25;257(4):2066–2074. [PubMed] [Google Scholar]