Abstract
Background
TH2-dependent diseases vary in severity according to genotype, but relevant gene polymorphisms remain largely unknown. The integrin CD11a is a critical determinant of allergic responses, and allelic variants of this gene might influence allergic phenotypes.
Objective
We sought to determine major CD11a allelic variants in mice and human subjects and their importance to allergic disease expression.
Methods
We sequenced mouse CD11a alleles from C57BL/6 and BALB/c strains to identify major polymorphisms; human CD11a single nucleotide polymorphisms were compared with allergic disease phenotypes as part of the international HapMap project. Mice on a BALB/c or C57BL/6 background and congenic for the other strain's CD11a allele were created to determine the importance of mouse CD11a polymorphisms in vivo and in vitro.
Results
Compared with the C57BL/6 allele, the BALB/c CD11a allele contained a nonsynonymous change from asparagine to aspartic acid within the metal ion binding domain. In general, the BALB/c CD11a allele enhanced and the C57BL/6 CD11a allele suppressed TH2 cell–dependent disease caused by the parasite Leishmania major and allergic lung disease caused by the fungus Aspergillus niger. Relative to the C57BL/6 CD11a allele, the BALB/c CD11a allele conferred both greater T-cell adhesion to CD54 in vitro and enhanced TH2 cell homing to lungs in vivo. We further identified a human CD11a polymorphism that significantly associated with atopic disease and relevant allergic indices.
Conclusions
Polymorphisms in CD11a critically influence TH2 cell homing and diverse TH2-dependent immunopathologic states in mice and potentially influence the expression of human allergic disease.
Keywords: Asthma, allergic disease, CD11a, TH2 cell, homing, polymorphism, allele, congenic, biomarker
The mechanisms that control the recruitment, or homing, of TH cells collectively represent an essential immune regulatory checkpoint that crucially influences outcomes of diverse infectious and inflammatory processes.1 The homing of TH2 cells is specifically regulated through the expression and regulation of leukocyte function–associated antigen 1 (LFA-1), representing the heterodimeric association of CD18 and CD11a.2 In the absence of stimulation, LFA-1 is maintained in a low-affinity state that weakly binds the LFA-1 receptor CD54 (intercellular adhesion molecule 1) through the inserted (I) domain of CD11a.3-7 High concentrations of divalent cations stabilize and chemokines activate LFA-1 to a high-affinity state, allowing circulating T cells to tightly adhere to CD54 on endothelial surfaces and extravasate to sites of inflammation and infection.5,8
In part, the importance of LFA-1 for TH2 cell homing is determined by successive rounds of cell division during which LFA-1 expression is maintained or enhanced while expression of alternate homing integrins is reduced.9 Blockade or genetic deletion of LFA-1 does not interfere with the induction of robust TH2 responses but precludes efficient TH2 cell homing to sites of infection and inflammation without interfering with the development and homing of other TH effector subsets.9 Therefore selective blockade of TH2 cell homing through LFA-1 antagonism results in diverse phenotypes in mice, including reduced expression of allergic lung disease and improved control of the intramacrophage parasite Leishmania major in C57BL6 and BALB/c mice.9 The effect of LFA-1 deletion in BALB/c mice is particularly striking, converting the normally progressive and lethal L major infection into a transient illness that more closely resembles the disease course of C57BL/6 mice.9
C57BL/6 mice control L major infection by mounting a dominant TH1 immune response, whereas BALB/c mice do not control the infection because of production of an aberrant TH2-predominant antiparasite response.10,11 Although several genetic loci have been shown to contain genes that promote resistance to L major,12 the specific genetic causes for the dominant TH2 response and susceptibility in BALB/c mice have yet to be clearly defined.13
Airway challenge of C57BL/6 and BALB/c mice with the same allergen also produces variable immune and physiologic responses, the latter assessed as airway hyperreactivity (AHR), although allergic eosinophil- and TH2-predominant inflammation is consistently observed.14 The many genetic factors that underlie the mouse allergic lung inflammatory response15 overlap considerably with those influencing the mouse response to L major infection and include CD11a. BALB/c mice that are deficient in CD11a demonstrate impaired TH2 cell homing to the site of infection and arrested progression of L major infection, which is similar to the healer phenotype observed in the C57BL/6 mouse strain.9 The apparent increase in resistance in the absence of the BALB/c CD11a allele suggests that the BALB/c CD11a allele is an important factor in the pathogenesis of L major in the BALB/c mouse strain. As a gene located within a genomic region identified for carrying L major resistance in C57BL/6 mice, it is possible that polymorphisms in CD11a might in part determine the divergent immune and physiologic outcomes of BALB/c and C57BL/6 mice in models of L major infection and asthma.
To investigate the existence of polymorphisms in CD11a, we sequenced and identified strain-specific polymorphisms in CD11a of BALB/c and C57BL/6 mice and created mice congenic for CD11a on the BALB/c and C57BL/6 backgrounds to test the effect of CD11a alleles in the TH1- and TH2-mediated disease models of L major infection and allergic lung disease.
Methods
Mice
Animal studies were conducted under protocols approved by Baylor College of Medicine's Institutional Animal Care and Use Committee; mice were all housed under specific pathogen-free conditions and were fed standard mouse chow ad libitum. BALB/c, C57BL/6, and recombination-activating gene (Rag) 1−/− mice were purchased from Jackson Laboratories (Bar Harbor, Me). Rag2−/− mice were acquired from Taconic (Hudson, NY). CD11a−/−mice on the BALB/c (10 generations backcrossed) and C57BL/6 (8 generations backcrossed) backgrounds were provided by Dr C. M. Ballantyne.16 CD11a-congenic mice on the BALB/c (BalbC57-CD11a) and C57BL/6 (C57balb-CD11a) backgrounds were created by crossing CD11a−/− mice of one background with the wild-type (WT) strain of the other background. Progeny were then backcrossed to the parent CD11a−/− strain, selecting for CD11a expression on CD4 T cells for 8 generations. Homozygous congenic or litter-mate CD11a−/− mice were recreated by crossing eighth-generation heterozygous mice and selecting for complete loss of the targeting construct by using PCR or CD11a by using flow cytometry, respectively.
L major infection
L major (strain MRHO/SU/59/P/LV39) was cultured and mouse infection was established, as previously described.17
Macrophage phenotype
Popliteal lymph nodes were harvested from 3-week L major–infected mice and processed for total RNA by using Trizol (Invitrogen, Carlsbad, Calif), according to the manufacturer's methods. Primer sets (Applied Biosystems, Foster City, Calif) for inducible nitric oxide synthase (iNOS; Mm00440502_m;), Arg1 (Mm00475988_m1), found in inflammatory zone 1 (Fizz1; Mm00445109_m1), and 18s RNA (4319413E-0710034) were used to quantify mRNA with the TaqMan One-Step RT-PCR Master Mix (4309169, Applied Biosystems) using the 7500 Real-Time PCR System (Applied Biosystems). Signals were used to calculate relative expression levels in accordance with standard practice and expressed as the relative expression ratios of select genes.
Dynamic adhesion assay
Adhesion of CD4+ cells was quantitated by means of video microscopy under continuous flow conditions in vitro.18,19 For further information, see the Methods section in this article's Online Repository at www.jacionline.org.
Flow cytometry
Lymphocytes were stained with CD4-Pacific Blue, CD3–allophycocyanin, and CD11a–phycoerythrin (PE), CD11b-PE, or CD11c-PE (all from BD PharMingen, San Jose, Calif) by using standard staining methods.
Adoptive transfer of ovalbumin-specific CD4+ T cells
Mice were challenged with ovalbumin (OVA)–alum weekly for at least 3 weeks, spleens were harvested, and CD4+ cells were collected, selected under TH2 responses, differentially labeled fluorescently, and injected into strainspecific Rag-deficient mice for comparison of TH2 cell homing efficiency. For further information, see the Methods section in this article's Online Repository.
Allergic airway disease
Mice were challenged with a clinical isolate of 4 × 105 Aspergillus niger conidia every 2 days for 8 challenges and assessed for allergic airway disease, as previously described.20
Human single nucleotide polymorphism analysis and correlation
Human studies were conducted under Institutional Review Board–approved protocols at Harvard Medical School and Brigham and Women's Hospital.
Genotyping and data management
Of the 1041 participating parent-child trios in the Childhood Asthma Management Program (CAMP) trial,21 92.9% are included in this analysis. Using data from European Americans (CEU) in the International HapMap project,22 we applied a linkage disequilibrium (LD)–tagging algorithm (minor allele frequency, ≥10%; r2 > 0.8) to capture common variation in CD11a and its 10-kb flanks.23 For this study, additional single nucleotide polymorphisms (SNPs) were genotyped to evaluate reported functional variation (rs) within the genomic region of CD11a. All SNPs were genotyped with the SEQUENOM iPLEX platform (Sequenom, San Diego, Calif).24 The 8 polymorphic SNPs successfully genotyped capture of greater than 84% of the HapMap SNPs with a minor allele frequency of 10% or greater in CD11a and its 10-kb flanks in CEU trios at an r2 value of greater than 0.8.
Statistical analysis
Mouse statistics were performed, as described previously.25 For human data, the family-based association analyses in CAMP were performed with the family-based association test statistic implemented in GoldenHelix PBAT version 3.6.26 In family-based samples Hardy-Weinberg equilibrium was tested in parental data.
Results
Identification of polymorphisms in CD11a
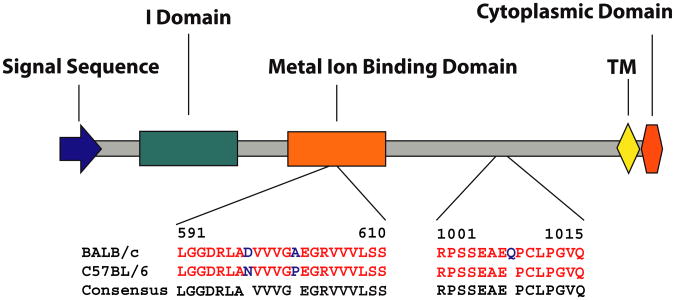
To determine whether CD11a polymorphisms distinguish the divergent responses of BALB/c and C57BL/6 mice to L major infection, we first sequenced the full CD11a mRNA from C57BL/6 and BALB/c mice (GenBank no. JN986841.1, Fig 1). Of 3 variances from the canonical C57BL/6 sequence, 2 of the polymorphisms were found in the metal ion binding domain (MIBD) of CD11a, with the C57BL/6 allele encoding asparagine at position 598 and proline at position 603 and the BALB/c allele encoding aspartic acid and alanine at the same positions, respectively. The BALB/c allele further contained an additional glutamine at position 1007 (Fig 1).
Fig 1.
Mouse CD11a exists in distinct allelic forms. The diagram schematically depicts major functional domains of the extracellular domain of CD11a and the amino acid differences between the BALB/c and C57BL/6 mouse strains. TM, Trans-membrane domain.
Of particular interest were the position 598 and 603 mutations. The change from asparagine (C57BL/6) to aspartic acid (BALB/c) occurs at a site within the MIBD that is predicted to directly bind divalent cations, as required for optimal CD11a function (UniProt). The MIBD is known specifically to regulate the stability and affinity state of LFA-1 and the homing efficiency of T cells.5 The polymorphisms occurring in the BALB/c CD11a allele effectively increase the net negative charge within the MIBD, as well as potentially reducing steric hindrance to cation binding. Together, we hypothesized that this combination of polymorphisms would enhance the binding of divalent cations by the BALB/c CD11a allele, leading to an enhanced affinity for CD54 and potentially augmented TH2 cell homing.
BALB/c CD11a congenic mice are resistant to L major infection
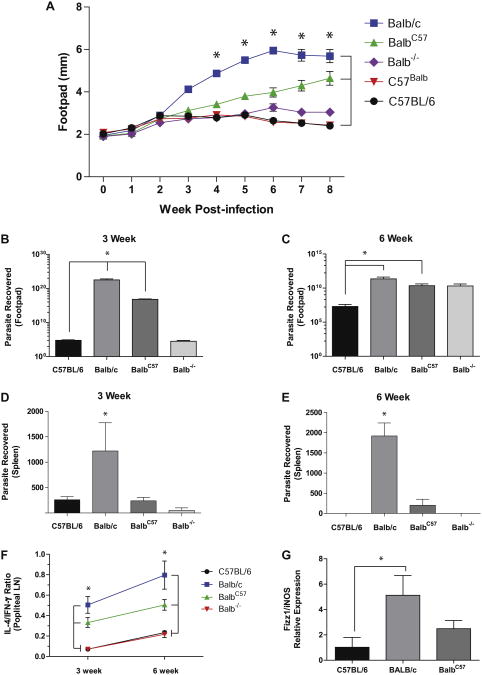
TH cells from BALB/c mice express more robust TH2 cell development under neutral and biased in vitro activation conditions, a finding that parallels the enhanced TH2 cell–dependent susceptibility to L major infection of BALB/c mice.13,27,28 To determine whether such enhanced TH2 responses ascribed to the BALB/c mouse are in part attributable to CD11a polymorphisms, we created mice congenic for CD11a on the BALB/c and C57BL/6 backgrounds, termed BalbC57-CD11a and C57Balb-CD11a, respectively, and assessed their ability to control infection with L major. As expected,9,13,29 wild type C57BL/6 mice challenged with L major controlled the infection, as indicated by an increase followed by a reduction in footpad swelling, whereas BALB/c mice were unable to control the infection, as assessed by progressive footpad swelling (Fig 2, A). C57balb-CD11a mice also controlled infection as efficiently as WT C57BL/6 mice,13,30 as did BALB/c CD11a−/− mice, as previously reported.9 In contrast, relative to WT BALB/c mice, congenic BalbC57-CD11a mice showed a marked attenuation in disease progression (Fig 2, A).
Fig 2.
Effect of CD11a polymorphisms on L major infection. A, Footpad swelling of WT compared with congenic mice infected with L major promastigotes. *P < .001 (n = 5 per group). B-E, Parasite burdens from footpads and spleens at 3 (Fig 2, B and D) and 6 (Fig 2, C and E) weeks. *P < .05. F, Ratio of IL-4– and IFN-γ–producing cells from popliteal lymph nodes at 3 and 6 weeks. *P < .05. G, Ratio of found in inflammatory zone1(Fizz1)/inducible nitric oxide synthase (iNOS), as assessed by using real-time quantitative PCR.*P<.05.
BALB/c mice further showed significantly higher parasite burdens in the footpads than C57BL/6 mice at both 3 and 6 weeks after L major infection (Fig 2, B and C). In contrast, both BalbC57-CD11a and BALB/c-CD11a−/−mice had lower footpad parasite burdens compared with WT BALB/c mice at the 3-week time point. Dissemination of infection from the footpad to the spleen was observed at 3 weeks in all mice. However, WT BALB/c mice had significantly higher parasite burdens in the spleen than all groups at both time points (Fig 2, D and E). Moreover, BALB/c-CD11a−/−and WT C57BL/6 mice resolved splenic infection by 6 weeks, and although BalbC57-CD11a mice retained spleen parasites at 6 weeks, the burdens were approximately 10 times lower relative to WT BALB/c mice (Fig 2, E). For all other parameters, C57balb-CD11a mice did not differ from WT C57BL/6 mice.
The intermediate phenotype of BalbC57-CD11a mice, as assessed based on footpad swelling caused by L major (Fig 2, A), was reflected in 2 additional immune parameters. First, relative to WT BALB/c mice, the relative abundance of IL-4–secreting cells, expressed as the ratio of IL-4–secreting to IFN-γ–secreting cells from popliteal lymph nodes, was lower in BalbC57-CD11a mice, reflecting more predominant IFN-γ secretion. Second, lymph node cells of BalbC57-CD11a mice showed significantly lower expression of a marker of alternatively (ie, TH2 cell) activated macrophages (found in inflammatory zone 1 [Fizz1]) relative to the classically (ie, TH1 cell) activated macrophage marker inducible nitric oxide synthase relative to WT BALB/c mice (Fig 2, G). Together, these findings demonstrate that possessing the C57BL/6 variant of CD11a in BALB/c mice can influence the course of L major infection, favoring weaker TH2 responses and improved disease control.
CD11a polymorphisms influence allergic lung disease
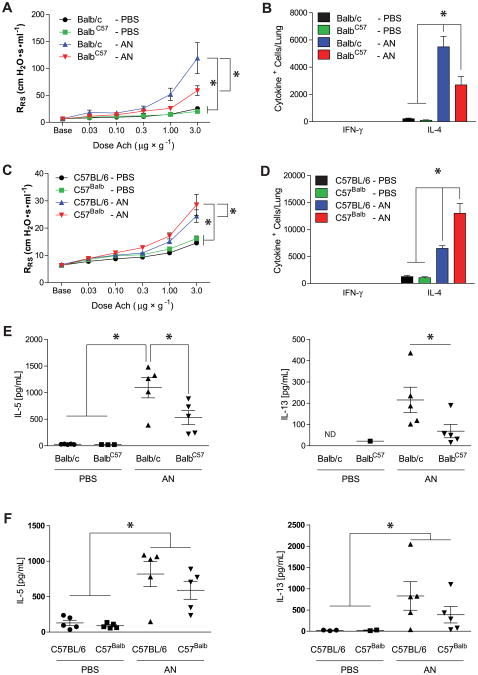
To confirm these findings in a distinct disease model, we next evaluated the effect of CD11a polymorphisms on the development of infectious allergic lung disease, a disease model in which CD11a-dependent TH2 cells exert a more dominant role compared with the L major model.9 WT BALB/c and C57BL/6 mice were challenged intranasally with the conidia of the fungus A niger and assessed for the development of AHR, an index of airway obstruction, in response to acetylcholine challenge, airway eosinophilia, and lung and airway cytokine secretion (Fig 3).20 Based on a much greater increase in respiratory system resistance induced by acetylcholine relative to naive mice, both BALB/c and C57BL/6 mice had AHR, although as expected, this IL-13–dependent response was more pronounced in BALB/c mice (Fig 3, A and B).31,32 However, whereas C57Balb-CD11a mice showed significantly enhanced AHR, BALB/cC57-CD11a mice showed significantly attenuated AHR relative to that seen in genotypematched WT control animals. Similarly, conidia-challenged BalbC57-CD11a mice showed a 2-fold lower recruitment and C57Balb-CD11a mice showed a 2-fold higher recruitment of IL-4– secreting cells to the lung when compared with genotypematched WT control animals (Fig 3, B and D). There were no significant differences in either bronchoalveolar lavage fluid cellularity, especially eosinophils, or airway glycoprotein secretion between the 2 congenic strains (see Figs E1 and E2 in this article's Online Repository at www.jacionline.org). Differences in cellular proliferation could not account for the differences noted between WT and congenic T cells because TH2 cells from both genotypes proliferated identically in vitro (see Fig E3 in this article's Online Repository at www.jacionline.org).
Fig 3.
CD11a polymorphisms influence allergic airway disease. A, Respiratory system resistance (RRS) values comparing BALB/c and BALB/cC57 congenic mice challenged with PBS or A niger conidia (AN). *P < .001 (n = 3 for BALB/c mice; n = 5 for all other groups). B, Total lung IFN-γ– and IL-4–producing cells. *P < .001. C, Respiratory system resistance values comparing C57BL/6 and C57Balb congenic mice. *P < .05, (n = 5). D, Total lung IFN-γ– and IL-4–producing cells. *P < .001. E and F, IL-5 and IL-13 levels from lung homogenates. *P < .05 (n = 5). Ach, Acetylcholine; ND, not detectable.
Analysis of culture supernatants of lung homogenates from fungus-challenged mice revealed that BalbC57-CD11a mice secreted less IL-5 and IL-13 relative to WT BALB/c mice, whereas no significant difference was observed between congenic and WT mice on the C57BL/6 background (Fig 3, E and F). Concentrations of other inflammatory chemokines and cytokines (CCL11 [eotaxin], IL-17A, and CCL17 [thymus and activation-regulated chemokine]) did not differ from lung supernatants of the same mice (see Fig E4 in this article's Online Repository at www.jacionline.org). Thus the BALB/c allele confers increased susceptibility to the development of allergic lung disease and more efficient recruitment of IL-4–producing cells to the lungs regardless of the mouse's genetic background. Conversely, having the C57BL/6 CD11a allele correlated with improved resistance to allergic lung disease in either strain.
CD11a polymorphisms influence CD54 adhesion and TH2 cell homing in vivo
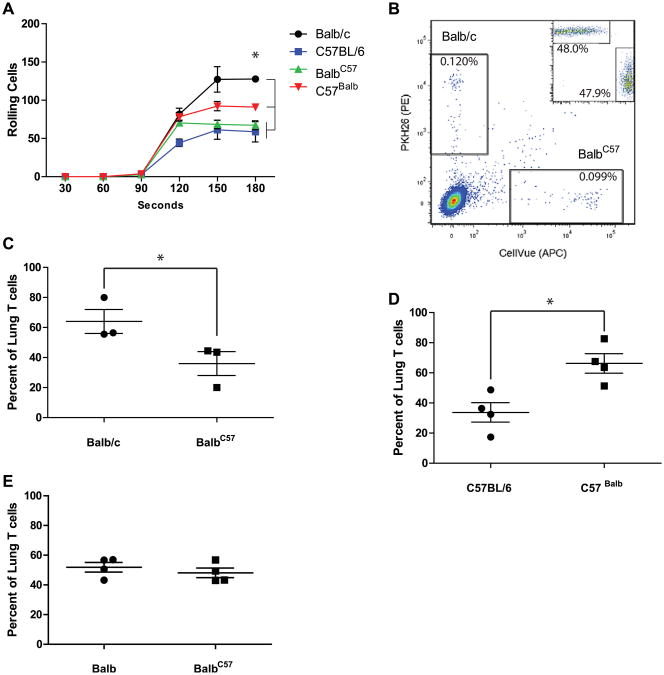
The previous results are potentially explained by several functions ascribed to LFA-1, including modulation of T-cell signaling and development, in addition to homing.33-36 Our prior analysis of CD18 and CD11a function did not reveal an essential role for LFA-1 for either T-cell proliferation or differentiation but did demonstrate its critical importance for TH2 cell homing.2,9 To determine whether allelic variants of CD11a similarly affect T-cell homing, we next sought to demonstrate the effect of polymorphisms of CD11a on adherence to the LFA-1 receptor CD54 under dynamic flow conditions.9 TH cells derived from WT BALB/c mice showed the greatest time-dependent adherence to CD54 while rolling when compared with WT C56BL/6 T cells (Fig 4, A). In contrast, BalbC57-CD11a T cells showed a marked reduction in the number of rolling cells when compared with WT BALB/c T cells and were not significantly different from WT C57BL/6 T cells. Conversely, C57Balb-CD11a T cells showed significantly increased adhesion when compared with both WT C57BL/6 and BalbC57-CD11a T cells. We also assessed the average velocity of rolling T cells but observed no strain-specific differences (see Fig E5 in this article's Online Repository at www.jacionline.org). Differences in CD11a expression or additional integrins known to bind CD54 (CD11b and CD11c) could explain these results but were not detected (see Fig E6 in this article's Online Repository at www.jacionline.org). Thus similar to the observed in vivo phenotypes, allelic variants of CD11a substantially influence the ability of LFA-1 to bind CD54, with the BALB/c CD11a allele conferring increased adherence and the C57BL/6 CD11a allele conferring reduced adherence under dynamic conditions.
Fig 4.
CD11a polymorphisms influence TH2 cell homing. A, T-cell adhesion to CD54 under dynamic flow. *P < .05 (n = 3). B-E, Rag-deficient mice were reconstituted with labeled TH2 cells from WT and congenic mice, respectively (Fig 4, B, inset) and intranasally challenged with A niger/OVA to induce T-cell recruitment, after which lung-derived single-cell suspensions were analyzed for donor TH2 (Fig 4, B [n = 1]; C, and D) and TH1 (Fig 4, E) cells. *P < .05. Data from one of 3 comparable experiments analyzing 1 × 106 cells per lung.
We next examined the role of CD11a polymorphisms on the homing of TH2 cells in vivo. OVA-specific TH2 cells from WT and congenic mice were fluorescently labeled, combined in a 1:1 ratio, and adoptively transferred into genotype-appropriate Ragdeficient mice. Recipient mice were then intranasally challenged with OVA and A niger conidia for 3 days, and lungs were assessed for the presence of labeled donor T cells (Fig 4, B-D). Although equivalent donor cells were transferred to each mouse (Fig 4, B, inset), a greater number of WT BALB/c T cells when compared with BalbC57-CD11a T cells were observed in the lungs after A niger and OVA challenge (Fig 4, B). The total numbers of lung T cells (see Fig E7 in this article's Online Repository at www.jacionline.org) and relative percentage of recruited T cells (Fig 4, C) were determined in multiple mice and consistently showed that T cells containing the BALB/c CD11a allele were more efficient at homing to the lungs, regardless of the genetic background of the donor mouse (Fig 4, C and D). In contrast, homing of CD4+ TH1 cells was not affected by CD11a polymorphisms in the in vivo homing model (Fig 4, E). These experiments show that the BALB/c CD11a allele significantly enhances the homing efficiency of TH2 cells when compared with the C57BL/6 CD11a allele.
Human CD11a polymorphism correlates to allergic disease
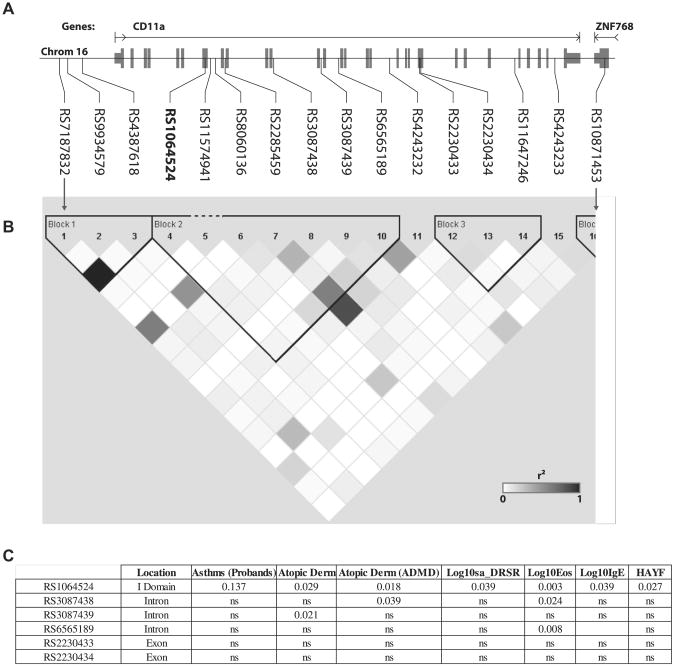
The unexpected importance of allelic variants of CD11a to the expression of mouse allergic disease suggested that human CD11a allelic variants might have genetic importance. To begin to determine this, we first identified 16 SNPs in the region of CD11a on chromosome 16 (Fig 5, A) from participants in the CAMP study.21 Two SNPs (rs1064524 and rs2230433) were found to cause missense mutations and 1 (rs2230434) synonymous mutation in exons of CD11a. LD analysis showed 3 LD blocks across the CD11a SNPs (Fig 5, B).
Fig 5.
Association of human CD11a SNPs to allergic disease. A, Location of SNPs within the human CD11a locus; exons are indicated in gray. B, Pairwise r2 values for correlations between identified SNPs. Black squares, r2 = 1; white squares, r2 = 0; gray squares, 0 < r2 < 1 with intensity proportional to r2. C, Significant correlations between SNPs and the indicated allergy phenotypes. DRSR, Log of dose-response slope to methacholine challenge; ns, not significant; HAYF, hay fever.
Genotyping was performed in 6 SNPs, including the 2 missense mutations. The missense mutation rs1064524 represents a change from a positive polar (Arg) to a hydrophobic (Trp) amino acid in the inserted domain of CD11a. Analysis of the transmission of rs1064524 from parents to asthmatic probands in relation to allergic disease phenotypes was analyzed by using a dominant model of inheritance (Fig 5, C). The frequency of rs1064524 in the CAMP cohort significantly correlated with the occurrence of multiple allergic disease phenotypes and TH2 cell activity, including atopic dermatitis (self-report), peripheral blood eosinophil counts, serum IgE levels, AHR (DRSR), and allergic rhinitis (hay fever; self-report).
Discussion
The divergent but critical roles of CD11a in regulating the mouse immune and physiologic responses in models of L major infection and allergic lung disease suggested a broader role for this adhesion molecule in immune regulation and disease.2,9 We have now identified distinct alleles for CD11a occurring within BALB/c and C57BL/6 mice that explain in part the distinct immune responses of these mouse strains. Functionally, the BALB/c CD11a allele conferred greater adhesion to CD54, more efficient homing of TH2 cells in vivo, and more robust expression of TH2 cell–dependent disease, including more progressive disease caused by L major infection and enhanced allergic lung disease. We further identified a unique allelic variant of CD11a that segregates with multiple indices of human allergic disease. Together, these findings confirm the importance of CD11a to the expression of allergic disease through the regulation of TH2 cell homing and provide the first evidence that polymorphic variants of CD11a are significant disease determinants in human allergic disease.
The unusual susceptibility of BALB/c mice to L major infection has been studied intensively for more than 40 years and is known to be both T cell based and polygenic in nature, involving at least 6 genetic loci.12 Among the many T cell–specific factors linked to resistance to L major infection, our data indicate that the importance of chromosome 7 to L major resistance is in part due to CD11a, the gene for which (Itgal) lies within this chromosome. Replacement of the cognate CD11a allele in BALB/c mice with the C57BL/6 allele improved resistance to L major in mice on the BALB/c background, as assessed based on reduced, although still progressive, footpad swelling; reduced parasite burdens at multiple time points; and reduced alternative macrophage activation. Nonetheless, as expected, the effect was not as pronounced as deletion of CD11a entirely (Fig 2 and Lee et al9). In contrast, congenic C57BL/6 mice were indistinguishable from WT C57BL/6 mice with regard to control of L major infection (Fig 2 and data not shown). Thus CD11a is a major determinant of resistance to L major infection only in BALB/c mice; other factors are more important in determining resistance on the C5BL/6 genetic background. These observations are consistent with the downregulation of LFA-1 expression and CD54-independent homing that has previously been shown for TH1 cells, which predominate in L major–infected C57BL/6 mice.9,37
A more consistent role for CD11a allelic variants was evident in experimental allergic lung disease, a model in which TH2 cells are clearly linked to disease expression across genotypes.14 Regardless of the mouse genetic background, possession of the BALB/c CD11a allele resulted in greater AHR, an important disease marker that is strongly dependent on the recruitment of TH2 cells to the lung.2,9 The BALB/c CD11a allele further enhanced recruitment to the lung of IL-4–secreting cells and the secretion of allergy-related cytokines from the lungs of BALB/c mice (Fig 3). A similar effect of the BALB/c allele was seen in C57BL/6 mice, although enhanced secretion of IL-5 and IL-13 from the lung was not seen. Thus, as with L major infection, although CD11a is important for disease expression in both C57BL/6 and BALB/c mice, additional factors in C57BL/6 mice modify disease expression independently of LFA-1.
Our findings demonstrate that distinct CD11a alleles influence the efficiency of TH2 cell homing. These observations are entirely consistent with the enhanced adhesion of the BALB/c CD11a allele to CD54 in vitro (Fig 4, A) and our prior demonstration of the general importance of CD11a for TH2 cell homing.9 Thus independent of the absolute level of gene expression, mouse CD11a exists in functionally hypomorphic (ie, the C57BL/6 allele) or hypermorphic (ie, the BALB/c allele) variants, as assessed by adhesion to CD54, which critically influence the expression of TH2-dependent diseases, and our preliminary analyses indicate that the same paradigm is also true in human subjects. Although additional study is required, we propose that CD11a might confer enhanced or reduced susceptibility to diverse allergic diseases based on the expressed alleles.
Our findings leave unresolved which amino acid changes or combinations thereof within the BALB/c CD11a allele determine the enhanced adhesion to CD54. We speculate that the greater negative charge within the MIBD of the BALB/c allele promotes more robust interaction with divalent cations, which in turn promotes a more consistently activated conformation that more efficiently engages CD54.4,38 Similarly, we suspect that the Arg to Trp change within the inserted domain of the rs1064524 allele of human CD11a also enhances binding to CD54, although through a mechanism that might not involve cation binding. Obviously, this allele is not present in the mouse, and hence there is no direct polymorphism link between our human genetics and the mouse genetics described here. Therefore much further work is required to resolve the atomic and molecular basis of the distinct CD11a phenotypes. Clarification of these mechanisms and the improved understanding of CD11a function that this will bring might provide new insight into the importance of CD11a as a disease marker and how LFA-1 can be safely manipulated to improve outcomes for a variety of diseases involving TH2 cells.
Supplementary Material
Fig E1. Differential counts of inflammatory cells from bronchoalveolar lavage (BAL) fluid of C57BL/6 (A) and BALB/c (B) background mice challenged with PBS or 4 × 105 A niger conidia (AN; n = 5 mice per group). Eos, Eosinophils; Mac, macrophages; Neut, neutrophils; Lym, lymphocytes.
Fig E2. Quantification of total glycoproteins from bronchoalveolar lavage fluid of C57BL/6 (A) and BALB/c (B) background mice challenged with either PBS or A niger conidia (AN). *P < .05.
Fig E3. Expression histogram depicting dilution of the fluorescent membrane dye PKH26 in OVA-specific TH cells obtained from BALB/c (blue line) and CD11a congenic BalbC57 mice (red line) that were cultured for 5 days in TH2-conditioning medium after labeling.
Fig E4. Concentrations of the indicated cytokines and chemokines from lung homogenate supernatants from mice on the C57BL/6 (A-C) and BALB/c (D-F) genetic backgrounds treated with either PBS or 4.0 × 105 A niger conidia (AN).
Fig E5. Mean velocity of rolling cells from a dynamic adhesion assay performed with CD4+ T cells from the indicated WT and congenic mice (n = 30 per group).
Fig E6. CD11a expression on lymphocytes. A, Expression of CD11a, CD11b, and CD11c on CD4+CD3+ splenocytes from C57BL/6 mice. B, CD11 expression on WT and congenic CD3+CD4+ T cells from the indicated mice. No significant differences were observed (n = 3 per group). MFI, Mean fluorescence intensity.
Fig E7. Effect of CD11a polymorphisms on the recruitment of OVA-specific TH cells to the lung. Rag-deficient mice from the appropriate genetic backgrounds received intraperitoneally a 1:1 mixture of 2 million fluorescently labeled OVA-specific TH2 cells from WT (PKH26 [PE]) and congenic (CellVue [allophycocyanin]) mice of the indicated backgrounds and intranasally challenged (Treated) with A niger/OVA to induce TH2 cell recruitment to lungs. Additional mice received cells but no antigen (Untreated) or no T cells (Treated Rag). T cells were enumerated by using an internal fluorescent bead counting standard. Results for individual mice from the BALB/c (A) and C57BL/6 (B) genetic backgrounds are shown.
Key messages.
Polymorphisms in the integrin CD11a in part explain differences in susceptibility of diverse mouse strains to TH2-dependent disease.
A distinct SNP in human CD11a is associated with several allergic disease indices.
CD11a might represent a useful susceptibility biomarker and therapeutic target in allergic disease.
Acknowledgments
Supported by National Institutes of Health grants HL75243, AI057696 and AI070973 (to D.B.C.) and EY018239 (to C.W.S.).
We thank the subjects and their families who made this study possible.
Abbreviations
- AHR
Airway hyperreactivity
- CAMP
Childhood Asthma Management Program
- LD
Linkage disequilibrium
- LFA-1
Leukocyte function–associated antigen 1
- MIBD
Metal ion binding domain, OVA, Ovalbumin
- PE
Phycoerythrin
- Rag
Recombination-activating gene
- SNP
Single nucleotide polymorphism
- WT
Wild-type
Footnotes
Disclosure of potential conflict of interest: L. Roberts, C. W. Smith, and D. B. Corry have received grants from the National Institutes of Health. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Lee SH, Corry DB. Homing alone? CD18 in infectious and allergic disease. Trends Mol Med. 2004;10:258–62. doi: 10.1016/j.molmed.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Lee SH, Prince JE, Rais M, Kheradmand F, Shardonofsky F, Lu H, et al. Differential requirement for CD18 in T-helper effector homing. Nat Med. 2003;9:1281–6. doi: 10.1038/nm932. [DOI] [PubMed] [Google Scholar]
- 3.Leitinger B, Hogg N. Effects of I domain deletion on the function of the beta2 integrin lymphocyte function-associated antigen-1. Mol Biol Cell. 2000;11:677–90. doi: 10.1091/mbc.11.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, et al. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDowall A, Leitinger B, Stanley P, Bates PA, Randi AM, Hogg N. The I domain of integrin leukocyte function-associated antigen-1 is involved in a conformational change leading to high affinity binding to ligand intercellular adhesion molecule 1 (ICAM-1) J Biol Chem. 1998;273:27396–403. doi: 10.1074/jbc.273.42.27396. [DOI] [PubMed] [Google Scholar]
- 6.Rothlein R, Dustin ML, Marlin SD, Springer TA. A human intercellular adhesion molecule (ICAM-1) distinct from LFA-1. J Immunol. 1986;137:1270–4. [PubMed] [Google Scholar]
- 7.Marlin SD, Springer TA. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1) Cell. 1987;51:813–9. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- 8.Rothlein R, Springer TA. The requirement for lymphocyte function-associated antigen 1 in homotypic leukocyte adhesion stimulated by phorbol ester. J Exp Med. 1986;163:1132–49. doi: 10.1084/jem.163.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Prince JE, Rais M, Kheradmand F, Ballantyne CM, Weitz-Schmidt G, et al. Developmental control of integrin expression regulates Th2 effector homing. J Immunol. 2008;180:4656–67. doi: 10.4049/jimmunol.180.7.4656. [DOI] [PubMed] [Google Scholar]
- 10.Heinzel FP, Sadick MD, Holaday BJ, Coffman RL, Locksley RM. Reciprocal expression of interferon gamma or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J Exp Med. 1989;169:59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holaday BJ, Sadick MD, Wang ZE, Reiner SL, Heinzel FP, Parslow TG, et al. Reconstitution of Leishmania immunity in severe combined immunodeficient mice using Th1- and Th2-like cell lines. J Immunol. 1991;147:1653–8. [PubMed] [Google Scholar]
- 12.Beebe AM, Mauze S, Schork NJ, Coffman RL. Serial backcross mapping of multiple loci associated with resistance to Leishmania major in mice. Immunity. 1997;6:551–7. doi: 10.1016/s1074-7613(00)80343-x. [DOI] [PubMed] [Google Scholar]
- 13.Sacks D, Noben-Trauth N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat Rev Immunol. 2002;2:845–58. doi: 10.1038/nri933. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Lamm WJ, Albert RK, Chi EY, Henderson WR, Jr, Lewis DB. Influence of the route of allergen administration and genetic background on the murine allergic pulmonary response. Am J Respir Crit Care Med. 1997;155:661–9. doi: 10.1164/ajrccm.155.2.9032210. [DOI] [PubMed] [Google Scholar]
- 15.Corry DB. Emerging immune targets for the therapy of allergic asthma. Nat Rev Drug Discov. 2002;1:55–64. doi: 10.1038/nrd702. [DOI] [PubMed] [Google Scholar]
- 16.Ding ZM, Babensee JE, Simon SI, Lu H, Perrard JL, Bullard DC, et al. Relative contribution of LFA-1 and Mac-1 to neutrophil adhesion and migration. J Immunol. 1999;163:5029–38. [PubMed] [Google Scholar]
- 17.Corry DB, Reiner SL, Linsley PS, Locksley RM. Differential effects of blockade of CD28-B7 on the development of Th1 or Th2 effector cells in experimental leishmaniasis. J Immunol. 1994;153:4142–8. [PubMed] [Google Scholar]
- 18.Abbassi O, Kishimoto TK, McIntire LV, Anderson DC, Smith CW. E-selectin supports neutrophil rolling in vitro under conditions of flow. J Clin Invest. 1993;92:2719–30. doi: 10.1172/JCI116889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mariscalco MM, Tcharmtchi MH, Smith CW. P-Selectin support of neonatal neutrophil adherence under flow: contribution of L-selectin, LFA-1, and ligand(s) for P-selectin. Blood. 1998;91:4776–85. [PubMed] [Google Scholar]
- 20.Porter P, Susarla SC, Polikepahad S, Qian Y, Hampton J, Kiss A, et al. Link between allergic asthma and airway mucosal infection suggested by proteinase-secreting household fungi. Mucosal Immunol. 2009;2:504–17. doi: 10.1038/mi.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anonymous The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 22.International HapMap C. The international HapMap project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 23.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 24.Little DP, Braun A, O'Donnell MJ, Koster H. Mass spectrometry from miniaturized arrays for full comparative DNA analysis. Nat Med. 1997;3:1413–6. doi: 10.1038/nm1297-1413. [DOI] [PubMed] [Google Scholar]
- 25.Porter PC, Roberts L, Fields A, Knight M, Qian Y, Delclos GL, et al. Necessary and sufficient role for T helper cells to prevent fungal dissemination during mucosal airway infection. Infect Immun. 2011;79:4459–71. doi: 10.1128/IAI.05209-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–9. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh CS, Macatonia SE, O'Garra A, Murphy KM. T cell genetic background determines default T helper phenotype development in vitro. J Exp Med. 1995;181:713–21. doi: 10.1084/jem.181.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Himmelrich H, Launois P, Maillard I, Biedermann T, Tacchini-Cottier F, Locksley RM, et al. In BALB/c mice, IL-4 production during the initial phase of infection with Leishmania major is necessary and sufficient to instruct Th2 cell development resulting in progressive disease. J Immunol. 2000;164:4819–25. doi: 10.4049/jimmunol.164.9.4819. [DOI] [PubMed] [Google Scholar]
- 29.Lee SH, Prince JE, Rais M, Kheradmand F, Shardonofsky F, Lu H, et al. Differential requirement for CD18 in T-helper effector homing. Nat Med. 2003;9:1281–6. doi: 10.1038/nm932. [DOI] [PubMed] [Google Scholar]
- 30.Sakthianandeswaren A, Foote SJ, Handman E. The role of host genetics in leishmaniasis. Trends Parasitol. 2009;25:383–91. doi: 10.1016/j.pt.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewart SL, Kuperman D, Schadt E, Tankersley C, Grupe A, Shubitowski DM, et al. Quantitative trait loci controlling allergen-induced airway hyperresponsiveness in inbred mice. Am J Respir Cell Mol Biol. 2000;23:537–45. doi: 10.1165/ajrcmb.23.4.4199. [DOI] [PubMed] [Google Scholar]
- 33.Van Seventer GA, Bonvini E, Yamada H, Conti A, Stringfellow S, June CH, et al. Costimulation of T cell receptor/CD3-mediated activation of resting human CD4+ T cells by leukocyte function-associated antigen-1 ligand intercellular cell adhesion molecule-1 involves prolonged inositol phospholipid hydrolysis and sustained increase of intracellular Ca2+ levels. J Immunol. 1992;149:3872–80. [PubMed] [Google Scholar]
- 34.Green JM, Zheng XG, Shimizu Y, Thompson CB, Turka LA. T cell receptor stimulation, but not CD28 costimulation, is dependent on LFA-1-mediated events. Eur J Immunol. 1994;24:265–72. doi: 10.1002/eji.1830240141. [DOI] [PubMed] [Google Scholar]
- 35.Salomon B, Bluestone JA. LFA-1 interaction with ICAM-1 and ICAM-2 regulates Th2 cytokine production. J Immunol. 1998;161:5138–42. [PubMed] [Google Scholar]
- 36.Sedwick CE, Morgan MM, Jusino L, Cannon JL, Miller J, Burkhardt JK. TCR, LFA-1, and CD28 play unique and complementary roles in signaling T cell cytoskeletal reorganization. J Immunol. 1999;162:1367–75. [PubMed] [Google Scholar]
- 37.Sasaki K, Tsuji T, Jinushi T, Matsuzaki J, Sato T, Chamoto K, et al. Differential regulation of VLA-2 expression on T(h)1 and T(h)2 cells: a novel marker for the classification of T(h) subsets. Int Immunol. 2003;15:701–10. doi: 10.1093/intimm/dxg066. [DOI] [PubMed] [Google Scholar]
- 38.Qu A, Leahy DJ. The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin. Structure. 1996;4:931–42. doi: 10.1016/s0969-2126(96)00100-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig E1. Differential counts of inflammatory cells from bronchoalveolar lavage (BAL) fluid of C57BL/6 (A) and BALB/c (B) background mice challenged with PBS or 4 × 105 A niger conidia (AN; n = 5 mice per group). Eos, Eosinophils; Mac, macrophages; Neut, neutrophils; Lym, lymphocytes.
Fig E2. Quantification of total glycoproteins from bronchoalveolar lavage fluid of C57BL/6 (A) and BALB/c (B) background mice challenged with either PBS or A niger conidia (AN). *P < .05.
Fig E3. Expression histogram depicting dilution of the fluorescent membrane dye PKH26 in OVA-specific TH cells obtained from BALB/c (blue line) and CD11a congenic BalbC57 mice (red line) that were cultured for 5 days in TH2-conditioning medium after labeling.
Fig E4. Concentrations of the indicated cytokines and chemokines from lung homogenate supernatants from mice on the C57BL/6 (A-C) and BALB/c (D-F) genetic backgrounds treated with either PBS or 4.0 × 105 A niger conidia (AN).
Fig E5. Mean velocity of rolling cells from a dynamic adhesion assay performed with CD4+ T cells from the indicated WT and congenic mice (n = 30 per group).
Fig E6. CD11a expression on lymphocytes. A, Expression of CD11a, CD11b, and CD11c on CD4+CD3+ splenocytes from C57BL/6 mice. B, CD11 expression on WT and congenic CD3+CD4+ T cells from the indicated mice. No significant differences were observed (n = 3 per group). MFI, Mean fluorescence intensity.
Fig E7. Effect of CD11a polymorphisms on the recruitment of OVA-specific TH cells to the lung. Rag-deficient mice from the appropriate genetic backgrounds received intraperitoneally a 1:1 mixture of 2 million fluorescently labeled OVA-specific TH2 cells from WT (PKH26 [PE]) and congenic (CellVue [allophycocyanin]) mice of the indicated backgrounds and intranasally challenged (Treated) with A niger/OVA to induce TH2 cell recruitment to lungs. Additional mice received cells but no antigen (Untreated) or no T cells (Treated Rag). T cells were enumerated by using an internal fluorescent bead counting standard. Results for individual mice from the BALB/c (A) and C57BL/6 (B) genetic backgrounds are shown.