Abstract
The osmolyte trimethylamine N-oxide (TMAO) stabilizes the tertiary but not the secondary structures of RNA. However, molecular dynamics simulations performed on the PreQl riboswitch showed that TMAO destabilizes the tertiary riboswitch structure, leading us to hypothesize that the presence of RNA could result in enhanced population of the protonated form, TMAOP. Constant pH replica exchange simulations showed that a percentage of TMAO is indeed protonated, thus contributing to the stability of the tertiary but not the secondary structure of PreQl. TMAOP results in an unfavorable dehydration of phosphodiester backbone, which is compensated by electrostatic attraction between TMAOP and the phosphate groups. In addition, TMAOP interacts with specific sites in the tertiary RNA structure, mimicking the behavior of positively charged ions and of the PreQl ligand in stabilizing RNA. Finally, we predict that TMAO-induced stabilization of RNA tertiary structures should be strongly pH dependent.
Keywords: PreQ1 riboswitch, RNA folding, pKa, RNA tertiary structure, constant pH simulations, molecular dynamics simulations, CHARMM
Introduction
Cellular organisms use a variety of osmolytes, which are small molecules, to counteract the effects of osmotic stress, desiccation or high temperatures thereby preserving the molecular stability [1, 2] of proteins and RNA [3, 4]. In all taxa, the concentrations of the osmolytes are regulated in such a way that the functions of biological molecules are uncompromised. Osmolytes, which decrease the stability of the folded states of proteins or RNA, are classified as denaturants whereas those that enhance the native state stability are considered to be protective [1]. Experimental and computational studies on proteins suggest that the greater differential impact of protective osmolytes on the unfolded state relative to the native state leads to an enhancement of the stability of the native state [3]. It has been recently proposed that the action of the protective osmolyte trimethylamine N-oxide (TMAO) is similar to that of nano-crowders whose interactions with proteins leads to their depletion from the protein surface leading to an entropy loss in the osmolyte. Since the surface area is greater in the unfolded than in the folded state protective osmolytes entropically favor the folded state. It is in this sense that osmolytes are like nano-crowding agents. The entropic stabilization mechanism readily explains the impact of osmolytes on proteins and peptides [2–7]. In contrast the mechanism by which osmolytes stabilize RNA tertiary structures is, to a large extent, unexplained.
A few studies have examined the effects of osmolytes on RNA stability [8–10]. Ultraviolet thermal melting experiments have shown that the effect of osmolytes in general and TMAO in particular on the secondary structures of RNA is small [9], which is in contrast with the influence of TMAO on secondary structures of proteins, which are stabilized by TMAO. All-atom MD and coarse-grained (CG) simulations showed that TMAO interacts with the base of guanosine-5’-monophosphate through the formation of a single hydrogen bond with the N2, an interaction that could stabilize the unfolded state, thereby leading to modest destabilization of RNA secondary structures [10]. Additional CG simulations of the RNA hairpin P5GA, using different models of osmolyte-RNA interactions, indicated that TMAO, as well as the majority of osmolytes, will not significantly impact RNA secondary structure stability. The marginal effect of TMAO on RNA secondary structures is confirmed both in experiments and computer simulations [8, 10, 11].
In contrast, the tertiary structures of RNA are greatly affected in the presence of protective osmolytes and denaturants. The denaturant urea destabilized RNA tertiary structures whereas TMAO renders them stable [9]. In a more recent study it was found, using small angle X-ray scattering and hydroxyl radical footprinting, that in the presence of TMAO the radius of gyration (Rg) decreases for two RNA molecules with tertiary interactions [11]. In addition, the structures assessed by distance distribution function and Rg for A-riboswitch in high TMAO concentration are identical to that found in the presence of low concentrations of Mg2+ ions. These observations and the demonstration that TMAO is depleted from the vicinity of dimethyl phosphate (an ion that is a surrogate for RNA backbone) were used to propose that TMAO destabilizes both the unfolded (U) and the folded (F) states by dehydration of the phosphate groups. Because the extent of exposure of phosphate groups is considerably greater in the U than in the N state, it was argued that TMAO destabilizes the U state more than the N state, thus resulting in the net stabilization of the folded state. This proposal is similar to the mechanism suggested for protein stabilization by TMAO although the molecular details are different.
The drastically different impact of TMAO on RNA secondary versus tertiary structures suggests that the molecular mechanism of TMAO on RNA stability may be associated, in part, with interactions with the folded states of RNA. In order to investigate this possibility, namely that TMAO stabilization of the tertiary structure of RNA involves direct interactions with RNA, we examine, at the atomic level, the effects of TMAO on the folded state of PreQ1 riboswitch using atomistic Molecular Dynamics (MD) simulations. Analysis was also performed to understand the impact of TMAO on the secondary structure of that molecule. The riboswitch, which is involved in selecting the intermediate PreQ1 (7-aminomethyl-7-deazaguanine) in queuosine biogenesis [12], was selected for study due to its relatively small size yet complex structure. The PreQ1 riboswitch structure consists of tertiary, non-WC regions including a pseudoknot and adenine- and uracil- rich loop regions, as well as regions of canonical, WC secondary structure (Figure 1). Our studies show that in the presence of TMAO the F state is destabilized by dehydration of the phosphate groups, in accord with the proposal based on thoughtful experiments [11]. We hypothesize that because of possible changes in the pKa of TMAO in the presence of RNA a small fraction of the co-solute can be protonated to produce TMAOP, a positively charged monovalent ion (Figure 1). An implication is that TMAOP stabilizes tertiary RNA structures in the same way as ions (Na+, K+, Mg2+) do. We establish our hypothesis, which differs qualitatively from the mechanism proposed in reference [11], using MD simulations including replica-exchange-constant-pH molecular dynamics (REX-CpHMD) [13], which show a decrease in Rg and an increase in the number of stable tertiary contacts in PreQ1 in aqueous TMAO/TMAOP solutions.
Figure 1.
PreQ1 riboswitch and TMAO/P structures. (A) Secondary structure with nucleotides missing from the crystallographic structure in the red-boxed region. (B) Tertiary structure taken from the crystal structure, (C) TMAO, and (D) protonated form of TMAOP (CPK, atom colored representation).
Methods
Model
We initiated our simulations using the crystal structure of the PreQ1 riboswitch consisting of 34 nucleotides with the queuosine precursor ligand, 7-aminomethyl-7-deazaguanine, and the Ca2+ ion removed (PDB ID: 3FU2) (Figure 1) [12]. Nucleotides 13 to 14, which are not present in the crystal structure, were excluded in the simulations. The RNA system was constructed within the CHARMM program [14] using the CHARMM36 all-atom additive nucleic acid force field [15]. After adding the missing hydrogen atoms to the crystal structure, the RNA molecule was immersed in a pre-equilibrated cubic water box that extended approximately 10 Å beyond all dimensions defined by the RNA non-hydrogen atoms. Overlapping water molecules with their oxygen atoms within 1.8 Å of non-hydrogen RNA atoms were deleted. Potassium ions were randomly placed in the water box to render the system electrically neutral followed by additional KCl to yield an additional ∼ 0.15 M ion concentration.
Simulation details
We performed simulations using periodic boundary conditions. Particle mesh Ewald [16, 17] was used for the calculation of electrostatic interactions with a 12 Å real space cutoff, an approximately 1 Å grid and a 4-order spline interpolation. Lennard-Jones (LJ) interactions were truncated at 12 Å, with a force switch smoothing function from 8 to 12 Å. The Langevin piston algorithm [18] was employed to maintain a pressure of 1 atm and Hoover thermostats [19, 20] were used to maintain the temperature at 298 K. The SHAKE algorithm [21] was used to constrain all the covalent bonds involving hydrogen atoms.
Initially the systems were subjected to 300-step steepest descent and 500-step adopted basis Newton-Raphson minimizations followed by a 500 ps MD simulation in the NPT ensemble with mass weighted restraints with a harmonic force constant of 5 kcal/mol/Å2 on all RNA non-hydrogen atoms. This enabled the equilibration of the solvent molecules and ions around the RNA, filling any voids created by deleting the water molecules. The resultant structures were used as the initial structures for the production MD simulations.
Using the pre-equilibrated system, a series of five simulations were constructed where the systems differed in the concentration of TMAO (0.4 and 2.0 M) as well as the protonation state of TMAO molecules within the solution. Parameters were taken from the CHARMM General force field (CGenFF) [22] for neutral and protonated TMAO (TMAOP). The osmolyte within each system was placed to reduce van der Waals (vdW) contacts with the riboswitch and upon placement, for each individual osmolyte, approximately three to five water molecules were removed. Subsequently, 120 ns of production simulation was performed on each system using the NPT ensemble and employing the multi-time step integrator within the NAMD program [23]. All the simulations used an integration time step of 2 fs and coordinates were saved every 2.5 ps for analysis. The pressure and temperature were maintained at 1 atm and 298 K. One additional system was constructed using the pre-equilibrated RNA and 20 mM MgCl2 without TMAO/P or potassium to reveal the sites occupied by divalent cations in the folded state.
Constant pH simulations
The replica exchange constant pH molecular dynamics (REX-CpHMD) method, which allows molecules to alternate between protonation states during an MD simulation [13, 24], was used to observe to what extent the presence of RNA leads to the protonation of TMAO as well as the impact of protonated TMAO on RNA structure. The titration of TMAO between the states is based on perturbing the associated non-bonded and bonded parameters that represent the two states. The force-field parameters of protonated TMAO (referred to as TMAOP herein) (Figure 1C, 1D) differ from those of neutral TMAO most significantly in the presence of a proton on the O atom and in the partial charges assigned to the O and N atoms. For neutral TMAO, a titrating (dummy) hydrogen, dHO, is assigned based on the bonded parameters of the OH proton in TMAOP (Table S1, supporting information). This allows for the protonation of TMAO to be defined largely based on the LJ and partial atomic changes, with the TMAO/TMAOP equilibrium associated with a parameter λ (specifying the protonated state) value for each TMAO molecule.
REX-CpHMD is performed by constructing a potential of mean force (PMF) between the protonated (λ = 0) and deprotonated (λ = 1) states of TMAOP that controls the equilibrium between the two states [13]. The PMF may be obtained by thermodynamic integration where the parameters A and B, which define a quadratic function that describes the titration coordinate, are adjusted to yield the correct equilibrium as a function of pH. These terms were empirically optimized such that at a pH of 4.7, corresponding to the pKa of TMAO, the equilibrium between the protonated and deprotonated states was 1, yielding values of −80.6259 and 0.0851797 for A and B, respectively.
Two TMAO-RNA systems were simulated (0.4 and 2.0 M) using REX-CpHMD as implemented in the CHARMM program [24]. A series of 8 replicas were used over a pH range from 6.78 to 7.02, with data from all 8 replicas used for analysis. For each replica, which includes the explicit inclusion of solvent, the GBSW implicit generalized Born (GB) solvent model [25] was applied to calculate the change in free energy as a function of λ as required to titrate TMAO/TMAOP during the REX-CpHMD. In the simulations, a quadratic θ-variable barrier height of 1.5 and a quadratic x-variable barrier height of 2.0 were used for the TMAO protonation PMF. The REX-CpHMD simulations conditions are identical to the standard MD simulations described above. The simulations were run for a total of 10 ns per replica (∼80 ns in total run time) where the frequency of saving the coordinates was 5 ps. The exchange criteria for the replicas was evaluated every 250 ps and the acceptance ratio was 50–60% (Table 1).
Table 1.
The average population of TMAOP (the error analysis is based on random block analysis) and exchange rate for the REX-CpHMD simulations.
| System | % protonated | Exchange rate |
|---|---|---|
| (No RNA) | ||
| 0.4 M | 2 ± 2% | 53% |
| 2.0 M | 1 ± 2% | 57% |
| (RNA) | ||
| 0.4 M | 13 ± 5% | 58% |
| 2.0 M | 11 ± 2% | 54% |
Data Analysis
We analyzed the data using MDAnalysis [26] to reveal the detailed mechanism of TMAO interaction with RNA. The three-dimensional (3D) probability distributions for TMAOP and Mg2+ were based on a grid spacing of 1 Å and were normalized relative to the total number of frames in the trajectory and bulk density of TMAO/P molecules or Mg2+ ions, such that 1 indicates bulk density for either TMAO/P or Mg2+.
Results
Neutral TMAO destabilizes PreQ1 tertiary structure
The small size of PreQ1 (Figure 1) allows adequate conformational sampling and the presence of secondary and tertiary interactions enables us to explore the effect of TMAO on the two levels of structural organization [9, 11]. The crystal structure of PreQ1 (3FU2) contains the ligand, queuosine, and Ca2+ ions. Two of the ions located within the fold of the RNA and the remaining ions are on the periphery of the tertiary structure. However, we do not include queuosine and Ca2+ in the simulations, which is justified because solution NMR experiments have shown that Ca2+ ions resolved in the crystal structure are not required for folding of PreQ1 [27]. As a result the tertiary structure is modestly destabilized (see below). In the first set of simulations we assumed that TMAO is exclusively present in the zwitterionic neutral form, as would be surmised from the experimental pKa = 4.7 of TMAO [28, 29].
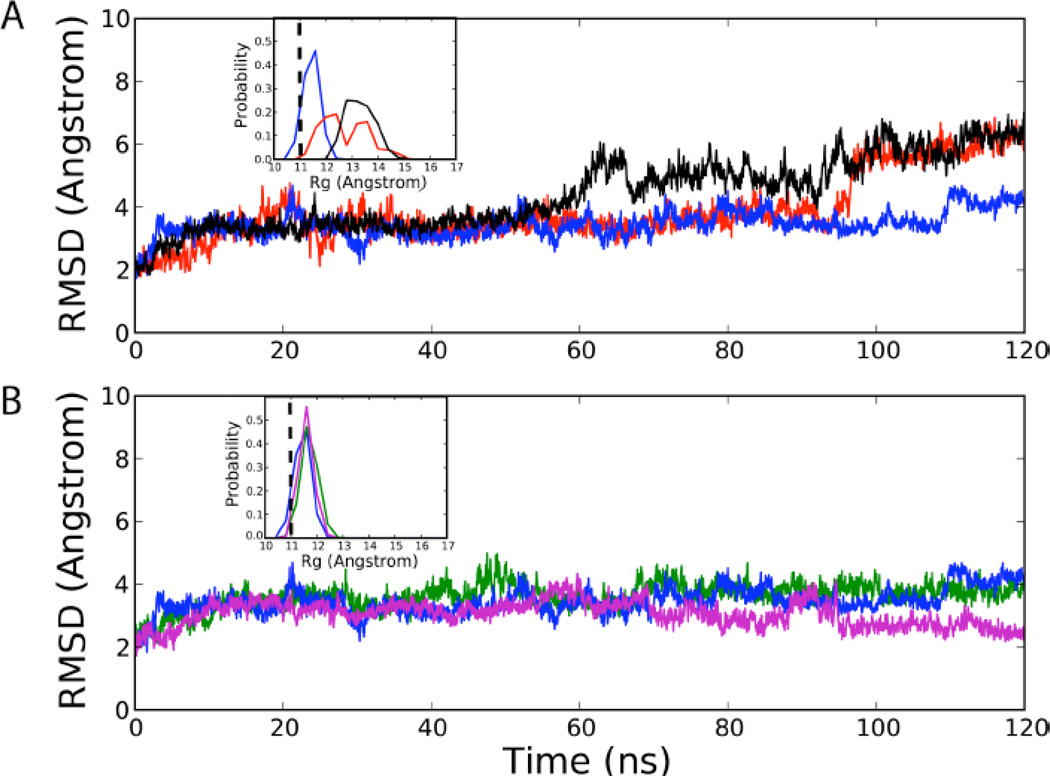
The root mean square differences (RMS) with respect to the crystal structure for all non-hydrogen atoms, excluding terminal nucleotides in 0, 0.4 and 2.0 M TMAO in Figure 2A show that the presence of the osmolyte results in gradual partial unfolding of the RNA. The RMS differences and the radius of gyration (Rg) over the final 60 ns of the simulations (Figure 2A, inset) increase. The enhanced conformational variation in the folded RNA structure, which suggests that TMAO destabilizes the native state, is not inconsistent with experiments. The loss in stability of the folded state may be attributed to dehydration of the phosphate groups promoted by TMAO (see below). Here, we provide an alternate explanation for the increase in stability of tertiary structures of RNA due to TMAO based on our hypothesis that the favorable electrostatic interaction between TMAOP and RNA leads to F state stabilization.
Figure 2.
RMS difference of the riboswitch non-hydrogen atoms with respect to the crystal structure excluding the terminal residues for the (A) 0, 0.4 M, and 2.0 M neutral TMAO simulations and the (B) 0, 0.4 M, and 2.0 M solution containing 10%TMAOP. The insets show the probability distributions of the radius of gyration of PreQ1 over the final 60 ns of the MD simulations. The vertical dashed line in the inset represents the Rg of the crystal structure. [Blue: 0 M TMAO, Red: TMAO 0.4 M, Black: TMAO 2.0 M, Green: TMAO 0.4 M (10% protonated), Magenta: TMAO 2.0 M (10% protonated)]
PreQ1 RNA increases the population of the protonated state of TMAO
The pKa of TMAO of 4.7 is approximately 2 pH units below the conditions where the experimental studies were performed (pH = 6.8 to 7.0). Thus, a naïve estimate shows that approximately 99% of TMAO is uncharged at neutral pH if no physical interactions were present to perturb the solution pKa of TMAO. We hypothesize that in the presence of RNA there is a shift in the pKa of TMAO such that a small but a significant fraction of molecules in the vicinity of the RNA is protonated to produce TMAOP, a monovalent cation. Consequently, at high enough concentration of TMAO favorable interactions between the positively charged TMAOP and the negatively charged phosphate groups of RNA can occur, which may result in the stabilization of both the U and F states.
The rationale for our hypothesis is based on two observations. First, equilibrium experiments have shown that the stabilizing effect of TMAO on proteins is pH dependent [30]. It seems that only at neutral pH, TMAO stabilizes the folded states, and at lower pH values there is a decrease in stability of the F state. Indeed, at pH values below the pKa of TMAO the native states are destabilized, though the extent of the effect depends on the protein. It is known that proteins typically denature under acidic conditions, and hence it might be assumed that the observed decrease in stability is purely due to acidic pH with TMAO playing no role. However, the data show that not only is TMAO incapable of increasing the stability at pH < pKa but it contributes to additional instability as demonstrated by a decrease in the melting temperatures in the presence of TMAO at low pH [29]. Second, the polyanionic nature of RNA could perturb the pKa of TMAO leading to an increase in the population of TMAOP near neutral pH, thus potentially altering the effect of TMAO on RNA. The expected shift in pKa could arise due to favorable electrostatic interactions between the anionic phosphates and the protonated TMAOP, thereby effectively shifting the pKa of TMAO to higher values. This would increase the probability that a specific TMAO molecule may be in the protonated form at pH 6.8–7.0 in the presence of RNA.
In order to test our hypothesis, we first estimated the population of TMAOP in the presence of the RNA, by performing replica exchange constant pH molecular dynamic (REX-CpHMD) simulations. REX-CpHMD is a free energy perturbation-based approach initially designed to allow the protonation state of ionizable residues on proteins to vary as a function of pH and/or environment [13]. In the present study, we use this approach to allow the protonation state of the osmolyte, TMAO, to vary in the presence of the RNA. The fraction of TMAOP increases from 1 % in the absence of RNA at 0.15 M KCl to 13 and 11 % in the presence of RNA at both 0.4 and 2.0 M TMAO, respectively (Table 1). Thus, in the presence of RNA there is sufficient increase in the concentration of the positively charged TMAOP, which can serve as a surrogate cation to stabilize the folded state of RNA.
TMAOP stabilizes PreQ1 RNA
Having established that there is an increase in the population of TMAOP in the presence of RNA, which could contribute to the stabilization of RNA, we performed simulations by explicitly including TMAOP. Based on the REX-CpHMD we assumed that the pKa of TMAO shifts by approximately 1 pH unit, so that ∼10% of the molecules are in the protonated state. We performed standard MD simulations at both 0.4 and 2.0 M TMAO, with 10 % of TMAO in the TMAOP state. In contrast to the results in Fig. 2A, the RMS difference of PreQ1 is similar in the absence of TMAO and in the 0.4M solution containing TMAOP. In addition, at a higher concentration of 2.0 M TMAO/TMAOP solution the RMS decreases with respect to the crystal structure (Figure 2B). Consistent with these results, the distribution of Rg calculated over the final 60 ns of the simulations in both TMAO/TMAOP mixtures remains narrow with a peak at Rg that coincides with the value in the crystal structure (Figure 2B, inset). These results strongly indicate that the stabilization of the tertiary structures of RNA is due to the presence of TMAOP.
The REX-CpHMD provided additional insights into the molecular details of the interaction of TMAO/P with RNA. For example, Figure S1 of the SI shows the Rg distributions for the final 60 ns in the absence of TMAO, a solution containing 10% TMAOP, along with those from the REX-CpHMD simulations. The stabilization of the RNA in the presence of a mixture of TMAO/TMAOP in the context of the REX-CpHMD methodology is similar to that obtained in the standard MD simulations containing 10% TMAOP. These results provide the first indication that TMAOP, with ionic character, is needed to stabilize the F state of PreQ1.
TMAO has negligible influence on the secondary structure of PreQ1 RNA
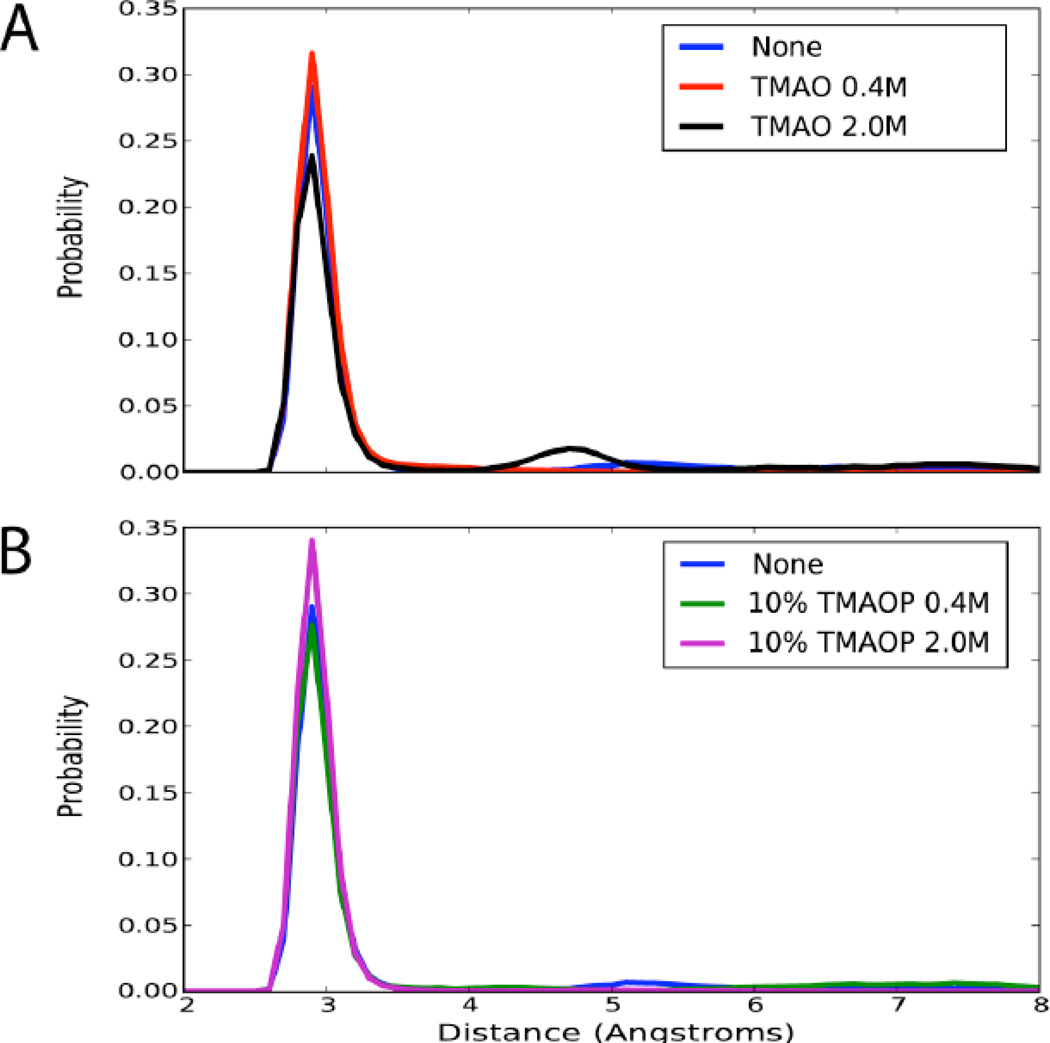
We assessed the impact of TMAO on the secondary structure of PreQ1 by computing the probability distributions of the N1-N3 distances. In the absence of TMAO the N1-N3 distances of the WC pairs are well preserved with a major peak at 2.8 Å, although some disruption is evident as distances beyond 4 Å are sampled (Figure 3A). In the presence of 2.0 M neutral TMAO, modest destabilization of the secondary structure occurs, as indicated by the presence of a minor peak at 4.7 Å, which suggests additional base pair opening (Figure 3A). In simulations containing 10% TMAOP simulations, the distributions are nearly identical to that found at 0 M TMAO. The N1- N3 interactions are well maintained, with some evidence of minor disruption (Figure 3B). Thus, TMAO has only a negligible effect on the secondary structure of PreQ1, which adds support to previous experiments [8,9] on different RNA structures.
Figure 3.
Probability distributions of the N1-N3 distances for WC base pairs for the (A) 0, 0.4 M, and 2.0 M neutral TMAO simulations and the (B) 0, 0.4 M, and 2.0 M with 10% TMAOP. [Blue: No TMAO, Red: TMAO 0.4 M, Black: TMAO 2.0 M, Green: TMAO 0.4 M (10% protonated), Magenta: TMAO 2.0 M (10% protonated)]
In order to further probe the influence of TMAO on the secondary structure of PreQ1 we computed the distribution of the backbone and glycosidic bond dihedral angles for the canonical regions (Figure S2, supporting information) and the RMS differences of the canonical regions with respect to themselves (Table S2, supporting information). Although the secondary structure is modestly destabilized in neutral TMAO simulations, it is not significantly impacted when TMAOP was included in the simulations. These results are consistent with experiments [8,9] and previous [10] and present simulations, which showed that TMAO has at best modest influence on RNA secondary structures.
TMAO/TMAOP mixture increases the fraction of tertiary contacts
We calculated the dynamics of native contact formation to determine the effects of TMAO/TMAOP on the tertiary structure. We considered only the native contacts associated with the tertiary fold of the PreQ1 RNA as defined in the list of tertiary interactions reported in the crystallography paper (Table S3, supporting information) [12]. The contacts exclude WC basepair interactions and focus on Hoogsteen interactions as well as additional interactions that stabilize the tertiary fold of the riboswitch. Figure 4 shows the time series of the fraction of native contacts in the absence of TMAO, neutral TMAO, and the TMAO/TMAOP simulations. The probability distribution of the fraction of native contacts (QN) over the last 60 ns for each system is in the inset of Figure 4. In the absence of the osmolyte, the tertiary structure is partially disrupted, which is expected due to the absence of Mg2+ or Ca2+ in the simulations as well as the omission of queuosine and selected nucleotides. Interestingly, the presence of neutral TMAO does not compensate for the loss of tertiary contacts and, in fact, leads to a noticeable decrease in QN, consistent with the overall increase in both the RMSD and Rg (Figure 2). In striking contrast, QN is significantly higher in the presence of 10% TMAOP, reaching values in the range 0.6 to 0.7, results that are also consistent with the decrease in RMSD and Rg shown in Figure 2. Taken together our simulations show that in the presence of TMAOP the tertiary structure of PreQ1 is stabilized.
Figure 4.
Fraction of tertiary native contacts for the last 60 ns of the simulations. The contacts are those defined by Klein et. al12 in their supplemental materials based on the native crystallographic structure. The inset is the probability distribution of the fraction of native contacts. [Blue: No TMAO, Red: TMAO 0.4 M, Black: TMAO 2.0 M, Green: TMAO 0.4 M (10% protonated), Magenta: TMAO 2.0 M (10% protonated)]
TMAOP promotes dehydration of the phosphate groups to a greater extent than TMAO
It has been suggested that TMAO-induced stabilization could be due to larger differential dehydration of the phosphodiester moieties between the U and F states, while the possibility of specific interactions of TMAO with the RNA were excluded. To better understand the role of TMAO-induced dehydration, analysis of the distribution of water, TMAO, and TMAOP around the RNA was undertaken. We used hydration number analysis (Table 2) and water radial-distribution functions (RDFs) (Figure S4, supporting information) calculated around different regions of the RNA to dissect the potential dehydration of the folded RNA in the presence of TMAO/P. The extent of hydration was determined for the entire RNA, the non-canonical (non-WC) regions, and the canonical (WC) regions in the 0 M TMAO and 2.0 M TMAO/TMAOP systems. TMAO leads to various levels of dehydration of the RNA with the most affected region being the oxygen atoms of the anionic phosphate groups, which lose approximately one water per nucleotide in the WC region. In addition, the minor groove and O2’ are partially dehydrated as well. In the non-WC region, the major groove shows a 0.5 decrease in the number of associated waters in the 2.0 M TMAO system. The results indicate that the canonical, WC region of the molecule is dehydrated more than the non-canonical region, with a loss of almost half a water molecule per nucleotide in the presence of TMAO (Table 2). The extent of dehydration is further increased in the presence of 10% TMAOP compared to a solution containing only neutral TMAO (Table S4 of the supporting information). However, the energetic cost due to dehydration is compensated by direct interaction of the charged TMAOP with PreQ1 phosphate groups (see below).
Table 2.
The average number of water molecules per nucleotide with the oxygen atom within 3.5 Å of the selected atoms of the PreQ1 RNA. Results are presented for RNA in the absence of TMAO (0 M) and in the presence of 2.0 M TMAO where 10% of the TMAO is protonated. The average values are based on an average over the last 60 ns for each simulation and the error analysis is based on random block analysis over six blocks differing by 10 ns increments.
| Selected atoms | All | Difference | |
|---|---|---|---|
| 0 M | 2.0 M | ||
| 2’-hydroxyl (O2’) | 0.9 ± 0.2 | 0.7±0.1 | −0.2 |
| Ribose (O4’) | 0.4 ± 0.1 | 0.3±0.1 | −0.1 |
| Phosphate (O1P & O2P) | 3.1±0.3 | 2.6 ± 0.2 | −0.5 |
| Minor groove (N3, O2 & N2) | 0.6 ± 0.2 | 0.4 ± 0.1 | −0.2 |
| Major groove (N7, N4, O4, O6, & N6) | 0.7 ± 0.2 | 0.5 ± 0.2 | −0.2 |
| Total | 4.5 ± 0.3 | 3.8 ± 0.3 | −0.7 |
| Selected atoms | Non-WC | Difference | |
|---|---|---|---|
| 0 M | 2.0 M | ||
| 2’-hydroxyl (O2’) | 0.9 ± 0.3 | 0.8 ± 0.2 | −0.1 |
| Ribose (O4’) | 0.4 ± 0.1 | 0.3±0.2 | −0.1 |
| Phosphate (O1P & O2P) | 3.4 ± 0.3 | 3.1±0.2 | −0.3 |
| Minor groove (N3, O2 & N2) | 0.7 ± 0.2 | 0.6 ± 0.2 | −0.1 |
| Major groove (N7, N4, O4, O6, & N6) | 1.0 ± 0.2 | 0.6 ± 0.3 | −0.4 |
| Total | 4.5 ± 0.3 | 4.0 ± 0.2 | −0.5 |
| Selected atoms | WC | Difference | |
|---|---|---|---|
| 0 M | 2.0 M | ||
| 2’-hydroxyl (O2’) | 0.9 ± 0.1 | 0.6 ± 0.1 | −0.3 |
| Ribose (O4’) | 0.3±0.2 | 0.2 ± 0.1 | −0.2 |
| Phosphate (O1P & O2P) | 3.3±0.1 | 2.3±0.2 | −1.0 |
| Minor groove (N3, O2 & N2) | 0.9 ± 0.1 | 0.5 ± 0.1 | −0.4 |
| Major groove (N7, N4, O4, O6, & N6) | 0.8 ± 0.1 | 0.7 ± 0.2 | −0.1 |
| Total | 4.6 ± 0.4 | 3.5 ± 0.4 | −1.1 |
TMAOP has favorable interactions with RNA
In order to shed light on the differences in interactions between TMAO and TMAOP with PreQ1 we calculated the number of molecules in direct contact with the RNA (Table 3) and RDFs for TMAO and TMAOP around RNA (Figure 5) in the 2.0 M TMAO/TMAOP solution. TMAOP interacts with the RNA significantly more than TMAO, with approximately 0.4 TMAOP molecules/nucleotide whereas only 0.2 neutral TMAO is present near PreQ1. Indirect interactions of both TMAO and TMAOP are evident, as judged by the maxima in the RDF being beyond 3 Å with the majority of TMAOP interactions involving the phosphate followed by the major groove; however, a two-dimensional analysis of the anionic phosphate-major groove interaction distances indicate that the major groove-TMAOP interactions are largely associated with favorable interactions with the phosphate groups (Figure S5, supporting information). Interestingly, a significant contribution to the TMAOP-RNA interactions involves TMAOP occupying the region of PreQ1 RNA occupied by the queuosine ligand (see below). The REX-CpHMD results are in agreement with those for 10% explicitly protonated TMAO (Figure S6, supporting information). These results indicate that while the destabilizing effect of TMAO includes indirect, dehydration of the RNA, direct interactions of TMAOP with the phosphates also occur such that the resulting folded state is rendered stable. In other words, the destabilizing effect of depletion of water molecules from the phosphate groups in aqueous TMAO/TMAOP solution is more than compensated by the favorable electrostatic interaction between TMAOP and RNA. It is clear that the stabilization mechanism of RNA in the presence of TMAO is more complex than for proteins, which affects secondary and tertiary structures in a similar manner [6, 31].
Table 3.
The average number of TMAO/TMAOP molecules per nucleotide with the non-hydrogen atoms within 3.5 Å from the selected atoms of the PreQ1 RNA. Results are presented for RNA in the presence of 2.0 M TMAO where 10% of the TMAO is protonated. The average values are based on an average over the last 60 ns for each simulation and the error analysis is based on random block analysis over six blocks differing by 10 ns increments.
| Selected atoms | All | |
|---|---|---|
| TMAO | TMAOP | |
| 2’-hydroxyl (O2’) | 0.07 ± 0.02 | 0.05 ± 0.02 |
| Ribose (O4’) | 0.02 ± 0.03 | 0.01±0.01 |
| Phosphate (O1P & O2P) | 0.23 ± 0.06 | 0.36 ± 0.06 |
| Minor groove (N3, O2 & N2) | 0.03 ± 0.02 | 0.02 ± 0.02 |
| Major groove (N7, N4, O4, O6, & N6) | 0.04 ± 0.02 | 0.08 ± 0.01 |
| Total | 0.15 ± 0.07 | 0.30 ± 0.08 |
| Selected atoms | Non-WC | |
|---|---|---|
| TMAO | TMAOP | |
| 2’-hydroxyl (O2’) | 0.08 ± 0.02 | 0.05 ± 0.02 |
| Ribose (O4’) | 0.03 ± 0.04 | 0.03 ± 0.01 |
| Phosphate (O1P & O2P) | 0.21±0.07 | 0.39 ± 0.05 |
| Minor groove (N3, O2 & N2) | 0.06 ± 0.01 | 0.04 ± 0.02 |
| Major groove (N7, N4, O4, O6, & N6) | 0.05 ± 0.02 | 0.08 ± 0.07 |
| Total | 0.13 ± 0.07 | 0.23 ± 0.08 |
| Selected atoms | WC | |
|---|---|---|
| TMAO | TMAOP | |
| 2’-hydroxyl (O2’) | 0.06 ± 0.02 | 0.04 ± 0.02 |
| Ribose (O4’) | 0.02 ± 0.04 | 0.01±0.01 |
| Phosphate (O1P & O2P) | 0.23 ± 0.06 | 0.39 ± 0.07 |
| Minor groove (N3, O2 & N2) | 0.03 ± 0.01 | 0.02 ± 0.02 |
| Major groove (N7, N4, O4, O6, & N6) | 0.04 ± 0.04 | 0.20 ± 0.07 |
| Total | 0.16 ± 0.07 | 0.37 ± 0.09 |
Figure 5.
Radial distribution functions of TMAO versus TMAOP around the non-canonical (Non-WC) and canonical (WC) regions of the RNA molecule for the 2.0 M 10% TMAOP system. All TMAO/P non-hydrogen atoms were included in the analysis. Results are presented for the (A) 2’-hydroxyl O2’, (B) sugar O4’, (C) phosphate, (D) minor-groove side of the bases, and (E) major-groove side of the bases. [Red: protonated TMAO, Black: neutral TMAO]. The minor and major grooves were defined based on the atoms O2, N2, and N3 for the minor groove and N7, N6, O6, N4, and O4 for the major groove. Normalization was based on number of trajectory frames and the bulk density.
Discussion
Constant pH MD simulations and the pKa shift of TMAO
A central finding of the present study is the shift in the pKa of TMAO in the presence of RNA. While CpHMD is a relatively new modeling approach [13, 24, 32] its has been successfully applied to biological systems, including RNA [32, 33]. In those studies it has been used to investigate the ionizable groups on the biological macromolecule; the present study represents the first application of the method to study the ionization of an osmolyte in aqueous solution. As shown above the approach yields the correct percent protonated TMAOP in the absence of RNA and predicts a shift in the presence of RNA of up to 10% TMAOP. However, it should be noted the percent TMAOP in the experimental regimen will be a function of the concentration of the RNA. In the presented calculations the RNA concentration is approximately 8 mM, as required for computational considerations, while in experimental studies the concentration of RNA is typically in the low micromolar range [9]. Accordingly, the total amount of TMAOP will only increase slightly in the presence of experimental concentrations of RNA as compared to the absence of RNA as we assume that only TMAO in the direct vicinity of the RNA will undergo the predicted pKa shift. In addition, the TMAOP is proposed to replace cations normally found in the vicinity of RNA, such that it may be assumed that the ionizable groups on RNA [33] are not being significantly perturbed. Future experimental studies will be required to verify the presented results.
TMAOP is a surrogate cation
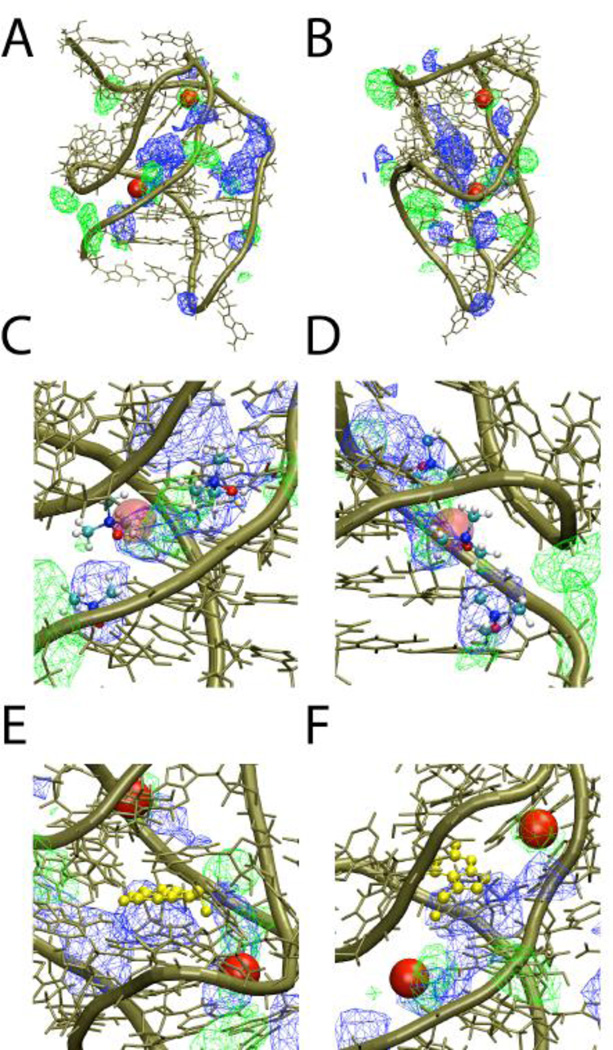
RNA is a strongly charged polyelectrolyte. Thus, cations, such as divalent Mg2+, are required to stabilize the tertiary structures of RNA [11, 34, 35]. Although it is well accepted that the most of RNA-cation interactions are non-specific several specific interactions, especially involving Mg2+, may be required to stabilize tertiary RNA structures. The PreQ1 crystal structure was obtained in the presence of Ca2+, which was shown to interact specifically with phosphates in the non-canonical loop region of the 3FU2 structure [12]. The stabilizing effect of TMAOP, a monovalent cation with low charge density, on the tertiary structure of the RNA, suggests that TMAOP could also interact specifically with the RNA in a manner similar to divalent ions albeit less strongly. To investigate this possibility, a simulation of PreQ1 in the presence of 20 mM MgCl2 was undertaken and the spatial distribution of the Mg2+ ions compared with those of TMAOP. The properties of the folded PreQ1 such as the RMSD, the Rg and the dynamics of native contact in the presence of Mg2+ are similar to those obtained in the presence of TMAOP (Figure S7). Not surprisingly, the value of Rg is lower in the presence of a high charge density Mg2+ whereas the number of native contacts is smaller compared to the TMAOP simulation. We also calculated 1D and 3D dimensional probability distributions of TMAOP and Mg2+ around the RNA (Figures S8 of the supporting information and Figure 6, respectively). The 1D probability distributions show that Mg2+ is localized adjacent to the phosphate in both the canonical and non-canonical regions of the RNA similar to the interaction involving TMAOP. The 3D probability distributions show overlap of the Mg2+ and TMAOP distributions in the vicinity of the phosphates, with TMAOP showing significant sampling towards the center of the riboswitch. Figure 6 also includes the location of two Ca2+ ions in asymmetric unit A of the 3FU2 crystal structure overlaid on the structure; two of the Mg2+ distributions show significant overlap with the ions, consistent with Ca2+ replacing Mg2+ in the crystallization conditions. Figure 6C and D, which are expanded views of the images in 6A and B include representative TMAOP molecules obtained from a single snapshot of the 2.0 M TMAOP simulation. Those molecules overlap with the Mg2+ distributions with one of those molecules occupying the position of the Ca2+ ion observed in the crystal structure. These results show that TMAOP occupies regions in and around that RNA that are also sampled by Mg2+, with additional regions being occupied by the ions for which there is not significant overlap (see following paragraph). Thus, the stabilizing effect of TMAOP on the tertiary structure may be associated (at least in part) with its ability to mimic the favorable direct interactions of positively charged ions with the RNA. It should be stressed that it is more likely that TMAOP has more of the characteristics of low charge density monovalent cations. This explains the large concentration of TMAO needed to fold the A-riboswitch, which is accomplished with a few mM of Mg2+. More generally, this interpretation is in accord with experiments showing the concentration of monovalent cations has to be in the molar range to fold large ribozymes where as only a few mM concentration of Mg2+ is sufficient to fold them (see Table 1 in ref. [35]).
Figure 6.
Structure of the PreQ1 RNA (tan) including the three dimensional probability distributions of TMAOP (blue) and Mg2+ (green) from the 2.0 M 10% TMAOP and 20 mM MgCl2 simulations mapped onto the structure and the two Ca2+ ions (red sphere) identified in asymmetric unit A of the crystal structure. All non-hydrogen atoms were used to determine the - TMAOP distributions. Approximately orthogonal views (A and B) of the full RNA and (C and D) of an expanded view that includes three representative orientations of TMAOP taken from a single snapshot of the MD simulation. E and F present two orientaitons of an expanded view of the RNA focused on the crystallographic position of quenosine ligand shown in gold CPK representation alone with the TMAOP and Mg2+ distributions and the crystallographic Ca2+ ions. Normalization was based on number of trajectory frames and the bulk density.
TMAOP occupies the queuosine ligand binding site
Figure 6 reveals the presence of a large region in the central region of the PreQ1 sampled by TMAOP. Comparison of this region with the location of the queuosine ligand in the crystal structures shows these regions overlap (Figures 6 E and F). Such an overlap is not unexpected given the positive charge common to queuosine and TMAOP. What is interesting is that Mg2+ does not significantly sample this region, which is due to the strong hydration and higher charge density of the atomic dication versus the larger molecular monocation TMAOP. Interestingly, occupation of the ligand binding region by TMAOP may lead to additional stabilization of the riboswitch as evidence by the smaller RMS difference and larger number of native contacts in the presence of 10% TMAOP as compared to Mg2+ (Figure S7 of the supporting information). These results suggest a model where both TMAOP and Mg2+ (Ca2+) in part stabilize the PreQ1 RNA via direct interactions associated with their charges, with TMAOP affording additional stabilization by occupying the ligand-binding site. The results also suggest an interesting model where Mg2+ can stabilize PreQ1 in the absence of ligand while not competing for that ligand, thereby facilitating formation of the holo riboswitch. Our model predicts that the folded states of RNA can be stabilized at lower ion concentrations than usually required provided there is a modest amount of TMAOP present.
Conclusions
The enhanced stability of tertiary structures of RNA in the presence of TMAO has been attributed to the osmolyte lowering the penalty for dehydrating the phosphate moieties upon RNA folding [11]. In this picture, which is not dissimilar to the entropic stabilization mechanism proposed for proteins, TMAO destabilizes both the folded and unfolded states of RNA. Because the extent of destabilization of the unfolded state is far greater than that for the native state there is a shift in the equilibrium of the RNA toward a folded state. In addition, it was suggested that TMAO mimics the stabilizing effect of Mg2+ on a structural level, yielding similar decreases in the Rg and changes in hydroxyl radical based footprinting.
In contrast to the mechanism described above, our studies lead to a different picture for TMAO-induced stabilization of RNA tertiary structures. Our results show that the protonated form TMAOP in solution contributes to the stabilization of RNA tertiary structures. TMAOP, which interacts directly with RNA, partially mimics the specific interactions of positively charged ions such as Mg2+ and Ca2+, thus contributing to the stabilization of its structure. Further, TMAOP occupies the ligand binding site of the riboswitch, leading to further stabilization while Mg2+ does not occupy that site suggesting a model by which Mg2+ can stabilize conformations of the apo riboswitch that facilitate ligand binding and formation of the holo riboswitch. The proposed mechanism suggests that the impact of TMAO on RNA will be strongly pH dependent – a prediction that is amenable to experimental test. Another implication of our work is that in the presence of modest amounts of TMAO the metal ion concentration required for stabilization should decrease. In this context, obtaining a phase diagram of RNA structures as a function of TMAO and Mg2+ would be most interesting.
Supplementary Material
Highlights.
Trimethylamine N-oxide (TMAO) is known to stabilize RNA tertiary structure.
Constant pH MD simulations indicate RNA leads to the presence of protonated TMAO.
The presence of protonated TMAO is shown to stabilize the tertiary structure of RNA based on simulations of the PreQ1 riboswitch.
Protonated TMAO stabilizes the RNA by dehydrating the phosphodiester backbone and mimicking the behavior of ions.
Acknowledgements
Appreciation to Drs. Kenno Vanommeslaeghe and Wenbo Yu for useful discussions. Financial and computational support is acknowledged from the NIH (GM051501), the NSF (CHE 09-14033), the University of Maryland Computer-Aided Drug Design Center, and XSEDE High Performance Supercomputing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information
Figures showing the results of the REX-CpHMD and Mg2+ simulations and tables providing additional information on the TMAO/P parameters, contact definitions, and number of water or Mg2+ molecules near RNA regions.
References
- 1.Harries D, Rosgen J. A practical guide on how osmolytes modulate macromolecular properties. Methods in Cell Biol. 2008;84:679–735. doi: 10.1016/S0091-679X(07)84022-2. [DOI] [PubMed] [Google Scholar]
- 2.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 3.Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci USA. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yancey PH. Water stress, osmolytes and proteins. American Zoology. 2001;41:699–709. [Google Scholar]
- 5.O'Brien EP, Ziv G, Haran G, Brooks BR, Thirumalai D. Effects of denaturants and osmolytes on proteins are accurately predicted by the molecular transfer model. Proc. Natl. Acad. Sci. USA. 2008;105:13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho S, Reddy G, Straub J, Thirumalai D. Entropic stabilization of proteins by TMAO. J. Phys. Chem. B. 2011;17:13401–13407. doi: 10.1021/jp207289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou Q, Bennion BJ, Daggett V, Murphy KP. The molecular mechanism of stabilization of proteins by TMAO and its ability to counteract the effects of urea. J. Am. Chem. Soc. 2002;124:1192–1202. doi: 10.1021/ja004206b. [DOI] [PubMed] [Google Scholar]
- 8.Gluick TC, Yadav S. Trimethylamine N-oxide stabilizes RNA tertiary structure and attenuates the denaturating effects of urea. J. Am. Chem. Soc. 2003;125:4418–4419. doi: 10.1021/ja0292997. [DOI] [PubMed] [Google Scholar]
- 9.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J. Mol. Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pincus D, Hyeon C, Thirumalai D. Effects of trimethylamine N-oxide (TMAO) and crowding agents on the stability of RNA hairpins. J. Am. Chem. Soc. 2008;130:7364–7372. doi: 10.1021/ja078326w. [DOI] [PubMed] [Google Scholar]
- 11.Lambert D, Leipply D, Draper DE. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration. J. Mol. Biol. 2010;404:138–157. doi: 10.1016/j.jmb.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein DJ, Edwards TE, Ferré-D'Amaré AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nature Struc. & Mol. Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khandogin J, Brooks CL. Constant pH molecular dynamics with proton tautomerism. Biophys. J. 2005;89:141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks BR, Brooks CL, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denning EJ, Priyakumar UD, Nilsson L, MacKerell AD., Jr Impact of 2'-hydroxyl sampling on the conformational properties of RNA: update of the CHARMM all-atom additive force field for RNA. J. Comp. Chem. 2011;32:1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darden T, Perera L, Li L, Pedersen L. New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- 17.Feller SE, Pastor RW, Rojnuckarin A, Bogusz S, Brooks BR. Effect of electrostatic force truncation on interfacial and transport properties of water. J. Phys. Chem. 1996;100:17011–17020. [Google Scholar]
- 18.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 19.Nose S. A unified formulation of the constant temperature molecular-dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 20.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 21.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkane. J. Comp. Phys. 1977;23:327–341. [Google Scholar]
- 22.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD., Jr CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;4:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips J, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace JA, Shen JK, Chapter 19 - Predicting pKa Values with continuous constant pH molecular dynamics. Biothermodynamics. Part A. 2009;466:455–475. doi: 10.1016/S0076-6879(09)66019-5. [DOI] [PubMed] [Google Scholar]
- 25.Im W, Lee MS, Brooks CL. Generalized Born model with a simple smoothing function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 26.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comp. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Kang M, Peterson RD, Feigon J. Comparison of solution and crystal structures of PreQ1 riboswitch reveals calcium-induced changes in conformation and dynamics. J. Am. Chem. Soc. 2011;133:5190–5193. doi: 10.1021/ja111769g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin TY, Timasheff SN. Why do some organisms use a urea-methylamine mixture as osmolyte? Thermodynamic compensation of urea and trimethylamine N-oxide interactions with protein. Biochemistry. 1994;33:12695–12701. doi: 10.1021/bi00208a021. [DOI] [PubMed] [Google Scholar]
- 29.Youxing Q, Bolen DW. Hydrogen exchange kinetics of RNase A and the urea:TMAO paradigm. Biochemistry. 2003;42:5837–5849. doi: 10.1021/bi0206457. [DOI] [PubMed] [Google Scholar]
- 30.Singh R, Haque I, Ahmad F. Counteracting osmolyte trimethylamine N-oxide destabilizes proteins at pH below its pKa measurements of thermodynamic parameters of proteins in the presence and absence of trimethylamine N-oxide. J. Biol. Chem. 2005;280:11035–11042. doi: 10.1074/jbc.M410716200. [DOI] [PubMed] [Google Scholar]
- 31.Ferreon ACM, Moosa MM, Yann G, Deniz AA. Counteracting chemical chaperone effects on the single-molecule α-synuclein structural landscape. Proc. Natl. Acad. Sci. USA. 2012;109:17770–17771. doi: 10.1073/pnas.1201802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goh G, Knight J, Brooks CI. Constant pH molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent. J. Chem. Theory Comput. 2012;8:36–46. doi: 10.1021/ct2006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh G, Knight J, Brooks CL. pH-dependent dynamics of complex RNA macromolecules. J. Chem. Theory Comput. 2013;9:935–943. doi: 10.1021/ct300942z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leipply D, Draper DE. Dependence of RNA tertiary structural stability on Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry. 2010;49:1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heilman-Miller S, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.