Abstract
This review focuses on economizing, prioritizing and recycling iron in Chlamydomonas, a reference organism for discovering mechanisms of acclimation to poor iron nutrition in the plant lineage. The metabolic flexibility of Chlamydomonas offers a unique opportunity to distinguish the impact of iron nutrition on photosynthetic vs. respiratory metabolism, and the contribution of sub-cellular compartments to iron storage and mobilization. Mechanisms of iron sparing include down regulation of protein abundance by transcript reduction or protein degradation. Two well studied examples of hierarchical iron allocation are the maintenance of FeSOD in the plastid and heterotrophic metabolism in acetate-grown cells at the expense of photosynthetic metabolism. The latter implicates the existence of a pathway for inter-compartment iron recycling when access to iron becomes limiting.
Introduction
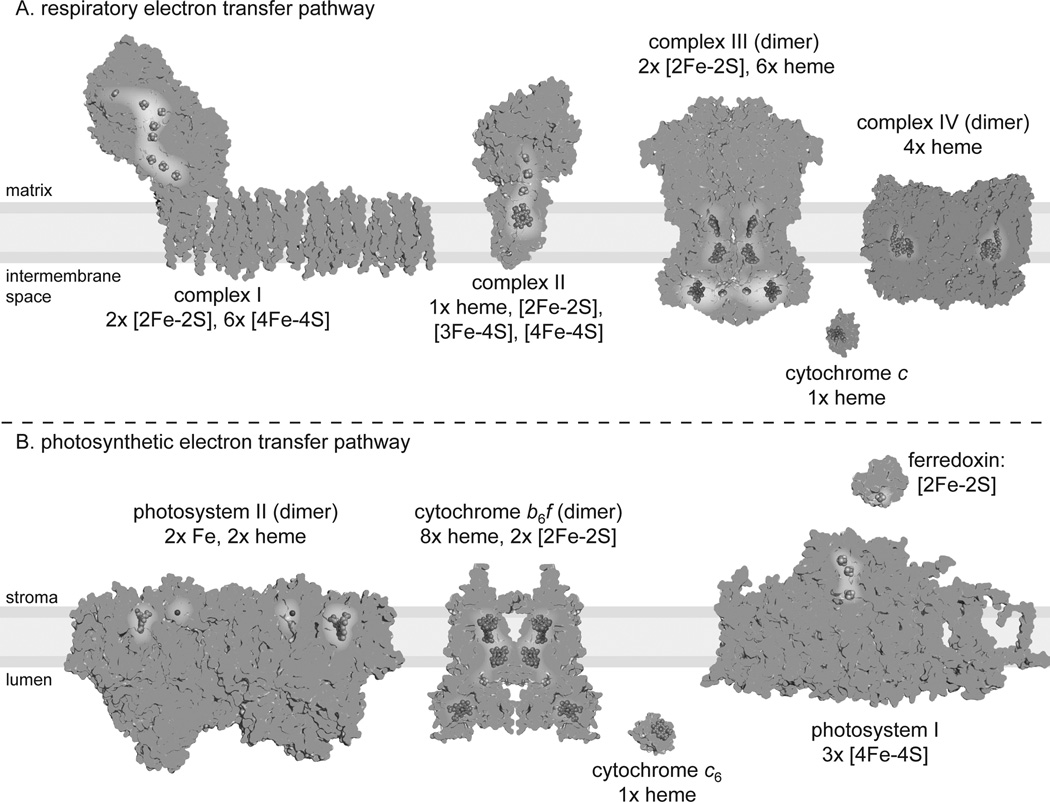
Photosynthetic organisms have a relatively high iron quota. As in other eukaryotes, all four electron transfer complexes plus cytochrome c in the mitochondrial respiratory chain require iron-sulfur clusters and/or hemes as redox cofactors in electron transfer (Figure 1A). Uniquely, an additional demand on iron assimilation is created by the photosynthetic electron transfer chain (ETC), where all proteins catalyzing so-called “linear” electron transfer from water to ferredoxin require iron to function (Figure 1B). The one exception is in the transfer of an electron between cytochrome b6f and photosystem I, which is often catalyzed by the copper-containing protein plastocyanin. However, some microbes use a heme-containing protein, cytochrome (Cyt) c6, either constitutively or conditionally in a situation of poor copper nutrition to replace plastocyanin.
Figure 1.
Iron cofactors in respiratory (A) and photosynthetic (B) electron transfer pathways. Assuming 1:1 stoichiometry and dimers for complex III and complex IV, there is a total of 53 iron atoms per respiratory electron transfer pathway. Assuming 1:1 stoichiometry and dimers for PSII and Cyt b6f, there is a total of 30 irons in the photosynthetic pathway (+1 for those algae who replace plastocyanin with Cyt c6). Nevertheless, the complexes are usually not present at 1:1 stoichiometry. For instance, in bovine heart mitochondria, the ratio of complex I: complex II: complex III: complex IV was calculated as 1.1:1.3:3:6.7 [6], which would lead to an estimate closer to 87 iron atoms per respiratory ETC. Furthermore, the abundances of the respiratory and photosynthetic ETC components are dynamic, and their stoichiometry is affected by environmental conditions [7,8]. The ratio of PSI to PSII is a common measurement of photosynthetic potential and performance, and it changes in response to light quantity, light quality, trophic status and nutrient availability. For Chlamydomonas, the PSI/PSII ratio has been estimated in the literature to vary from 0.41 to 2.02 depending on the specific culture conditions mentioned above. In the case of respiration, the complexes form associations in functional units called supercomplexes, in which the composition can differ from one supercomplex to another [9]. Therefore, it can be misleading to give a single estimate of the total contribution of the bioenergetic membranes or indeed of any given pathway to the iron quota of the cell. Illustrations are based on the following structures: complex I (PDB 3M9S; the ninth Fe-S cluster absent in eukaryotes is not shown), complex II (PDB 1ZOY), complex III (PDB 1BCC), complex IV (PDB 1OCO), cytochrome c (PDB 1CYT), PSII (PDB 3ARC; cytochrome c550 absent in land plants and green algae is not shown), Cyt b6f (PDB 1Q90), PSI (3LW5), Fd (3B2F), and Cyt c6 (1CYI).
Respiration requires more iron per unit (Figure 1), but the chloroplast is the major sink for iron in the cell. Roughly 80% of the cellular iron found in green leaves is located in the chloroplasts [1,2], indicating the importance in the plant lineage of the chloroplast as a site of iron utilization. Likewise, in the single-celled alga Chlamydomonas reinhardtii, the mitochondria occupy less than one-tenth of the cell volume compared to the single, cup-shaped chloroplast [3,4], again making the chloroplast the dominant iron sink.
The iron-dependent bioenergetic complexes in both the photosynthetic and respiratory ETCs are assembled in the chloroplast or mitochondrion, respectively. In the absence of an iron source (iron-chelate or heme), the complexes fail to accumulate, generally because of thermodynamic instability and protease susceptibility of individual apoproteins. It is evident that when iron availability is insufficient to maintain the cellular iron quota for a particular growth condition, there is likely to be competition between the chloroplast and the mitochondrion for this redox cofactor. How is iron delivery and utilization prioritized to one organelle over another? Within the organelle, how is the hierarchy of iron delivery / distribution to individual apoproteins maintained, and how does it change as a function of metabolism?
When the environmental supply cannot meet the cellular demand, prevailing dogma is that mechanisms for economizing, prioritizing and recycling the limiting metal nutrient are initiated [5]. This is driven by maintenance of more essential biochemical functions over dispensable ones where possible and by priming macromolecular metabolism so that upon metal resupply, proteins in key pathways are prioritized for cofactor loading. This review focuses on the extent to which these mechanisms are known to exist as part of the nutritional iron homeostasis pathway in C. reinhardtii (Figure 2).
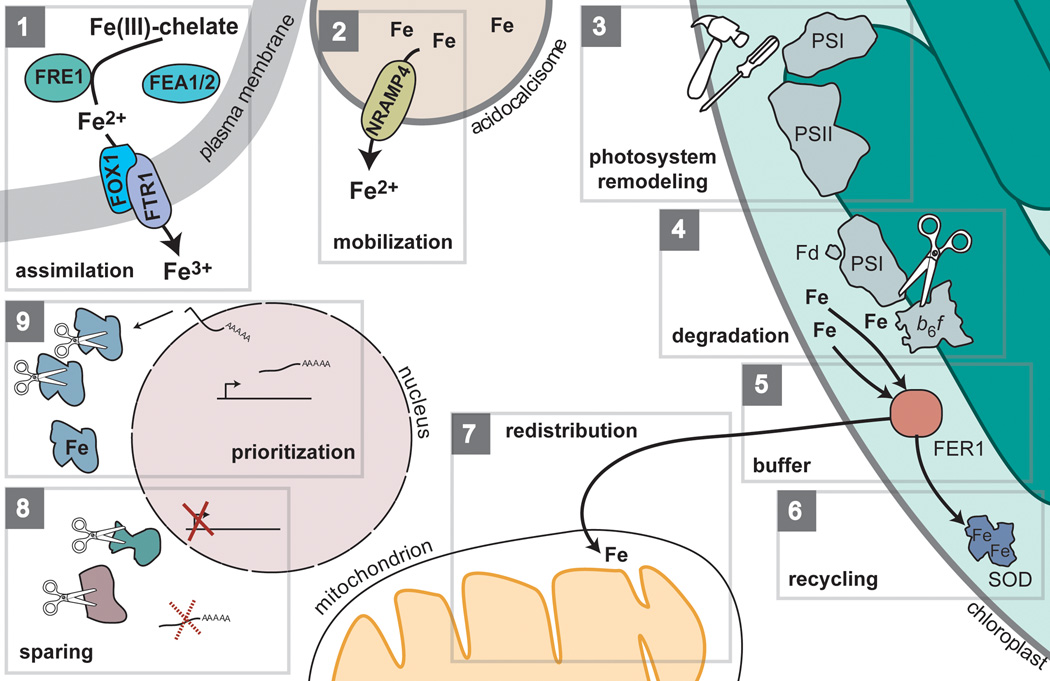
Figure 2.
Chlamydomonas and suboptimal iron nutrition during photoheterotrophic growth. (1) The first response is to induce the expression of the FOX1/FTR1 high-affinity iron transporter, which is composed of a copper-dependent ferroxidase (FOX1) and an iron permease (FTR1). Accompanying this transporter is a ferrireductase FRE1 and the algal-specific proteins FEA1 and FEA2, which are also proposed to participate in iron assimilation. (2) Assimilation may be combined with iron mobilization from vacuole-like compartments called acidocalcisomes. NRAMP4, whose expression is specifically induced by iron deficiency, is orthologous to the vacuole-localized Arabidopsis iron transporter NRAMP4, but its function in Chlamydomonas has not been established experimentally. (3) When, despite the operation of these two pathways, poor iron nutrition prevents cells from meeting the iron quota, the light harvesting antenna associated with photosystem I (PSI) and photosystem II (PSII) are remodeled. Specifically, the light harvesting antenna associated with PSI is disconnected at the onset of iron deficiency followed by degradation and replacement of some light-harvesting subunits [15], whereas the functional PSII antenna size increases [16]. This process is thought to reduce photooxidative stress caused by the photoreduction of dioxygen to superoxide by PSI. Superoxide destroys solvent exposed Fe-S clusters (including potentially those from PSI) releasing Fe3+. This unchelated iron can then react with hydrogen peroxide creating the highly cytotoxic hydroxyl radical, which cannot be enzymatically destroyed. (4) The loss of iron-dependent components, specifically Cyt b6f, PSI and ferredoxin, may proceed through active degradation of iron-bound proteins. The released iron may be temporarily chelated by ferritin (5) before it is recycled to other proteins (6) such as the Fe-dependent superoxide dismutase in the chloroplast. (7) Additionally, maintenance of oxygen consumption by the mitochondria points to redistribution of this iron pool for preservation of oxidative phosphorylation [16]. To further ensure optimal iron use, (8) the abundance of dispensable iron-dependent proteins is markedly reduced, possibly either through repression of expression, transcript degradation, or degradation of apoprotein. (9) The abundance of indispensible iron-dependent proteins is maintained (although sometimes at a lower abundance as in the case of the ferredoxin FDX6 [17]) by induction of gene expression and/or transcript stability.
Chlamydomonas
C. reinhardtii (referred to as Chlamydomonas) provides a unique opportunity to explore the cellular and subcellular dynamics of metal acquisition, trafficking and allocation in the plant lineage. Several paradigms in trace metal homeostasis have been established with the use of Chlamydomonas as a reference organism, including the use of “back-up” enzymes for metal-sparing and, more recently, the concept of “hot bunking” [10] or metal cofactor-recycling from one pathway to another [11,12]. Although the Chlorophyte (green algae including Chlamydomonas) and Streptophyte (non-chlorophyte green algae and land plants such as Arabidopsis) lineages are separated by at least 700 million years [13], the photosynthetic apparatus and assembly pathways are virtually identical. Nevertheless, there are several key metabolic differences. Unlike Arabidopsis, Chlamydomonas contains a light-independent chlorophyll biosynthetic pathway, can oxidize acetate, and exploits alternative bioenergetic routes such as hydrogen photoproduction and fermentation [14].
With its unique metabolic flexibility, the chloroplast is the major contributor to the iron quota, and known cellular iron homeostasis pathways and mechanisms center around this organelle. Therefore, Chlamydomonas has served as and is now well appreciated for being an excellent model for understanding the impact of poor iron nutrition (a common and globally important issue) on photosynthesis in the plant lineage.
Iron sparing
A ubiquitous method for acclimating to poor iron nutrition is iron sparing. Potential iron-binding proteins are not synthesized, synthesized at a much lower rate, or degraded before they can compete for the precious nutrient. Iron that is brought into the cell or mobilized from cellular stores can be prioritized for less dispensable iron-dependent functions, such as DNA repair/synthesis [18].
Most examples of iron sparing have come from studies with bacteria and yeast, and post-transcriptional regulation makes a large contribution to characterized microbial iron sparing strategies (reviewed in [19] and [20]). In Escherichia coli, iron sparing is accomplished with the small regulatory RNA (sRNA) RhyB, in Bacillus subtilis with the sRNA FsrA, and in Saccharomyces cerevisiae with the RNA-binding protein Cth2. The S. cerevisiae iron-sensing transcription factor, Aft1, also directly regulates the expression of some genes encoding iron dependent proteins. Cross-kingdom targets include succinate dehydrogenase (complex II), aconitase (tricarboxylic acid cycle enzyme, [4Fe-4S]) and glutamate synthase (glutamate and nitrogen metabolism, two [4Fe-4S] and one [3Fe-4S]). Although analagous sRNAs or RNA-binding proteins remain undiscovered in plants and algae, iron-responsive microRNAs have been found in Arabidopsis [21]. The role of these RNAs in the iron-deficiency response is yet to be shown.
While iron sparing is well characterized in model bacterial and fungal organisms, our understanding as to the extent this strategy occurs and how it is regulated in plants and algae is relatively deficient. Several transcriptome studies looking at the response of iron nutrition on transcript abundance are available. Among the transcripts with reduced abundance in response to poor iron nutrition are several encoding iron-dependent proteins and these are, therefore, potential targets of iron sparing. In Arabidopsis these genes encode cytochrome P450s (heme), peroxidases (heme), Rieske [Fe-S] domain-containing proteins, 2OG-Fe(II) oxygenase family proteins, and the iron-dependent superoxide dismutase, FSD1, in the plastid [22,23].
A recent genome-wide analysis of transcript abundance using RNA-Seq methodology revealed transcripts of iron-binding proteins that are reduced in abundance following poor iron supply in Chlamydomonas (Supplementary Table 1) [24]. This analysis was performed with cultures acclimated to iron-sufficient (20 µM Fe), iron-deficient (1 µM Fe) and iron-limited (0.25 µM Fe) media in the presence (photoheterotrophic) or absence of acetate (photoautotrophic). The iron deficient state is generally defined by induction of iron assimilation pathways (such as the high affinity iron transport component FOX1, Figure 2) without an impact on growth, while the iron limited state is defined by a negative impact on growth.
Based on this transcriptome analysis, iron sparing does not appear to be a major component of iron homeostasis during the iron-deficient state. Among the four transcripts that are at least two fold reduced in abundance under iron-deficiency in the presence of acetate is the ferredoxin FDX5. However, transcript abundance is not significantly affected by iron limitation. Among the seven transcripts reduced in abundance during iron-deficient photoautotrophic growth is a putative truncated hemoglobin (heme), which was more than ten-fold reduced. During this “pre-symptomatic” state of iron nutrition, induced iron assimilation and mobilization may obviate the need for iron sparing at the transcription level.
During the iron-limited state, in contrast, multiple examples of putative iron sparing events were found for both trophic conditions. During photoheterotrophic growth, at least 25 transcripts encoding known or putative iron-dependent proteins are reduced two-fold or more in abundance (Supplementary Table 1). The most highly decreased of these are linked to anaerobic growth, including hybrid cluster proteins (HCP4, 28-fold; HCP1 20-fold), pyruvate-ferredoxin oxidoreductase (PRF1, 15-fold), hydrogenase assembly factors (HYDG, 15-fold; HYDEF, 11- fold) and ferredoxin-sulfite reductase (SIR1, 6-fold) (Table 1). As noted already in other organisms, both aconitase and succinate dehydrogenase are down-regulated under iron-limiting photoheterotrophic conditions, and an independent metabolomics study captured the increase in the corresponding substrates [25]. Under the photoautotrophic condition, at least ten transcripts encoding known or putative iron-dependent proteins are reduced in abundance (Supplementary Table 1), including a second truncated hemoblogin. Seven transcripts encoding iron-dependent proteins are reduced in both iron-limited trophic conditions (Table1); these transcripts may represent core iron-sparing targets.
Table 1.
Select examples of RNA / protein abundance of iron-dependent proteins in response to iron and carbon status [24,26].
| Cofactor | Gene Name | Fold- change limited + acetate1 |
Fold- change limited – acetate1 |
Protein abundance (zmol/cell) replete2 |
Protein abundance (zmol/cell) limited2 |
|---|---|---|---|---|---|
| involved in anaerobic metabolism / down-reg. by Fe-limiting photoheterotrophic growth | |||||
| [4Fe-4S] | HCP4 | 0.01 | |||
| [4Fe-4S] | HCP1 | 0.04 | |||
| [4Fe-4S] | PFR1 | 0.07 | |||
| [2Fe-2S] | HYDG | 0.07 | |||
| [4Fe-4S] | HYDEF | 0.09 | |||
| [4Fe-4S] | SIR1 | 0.17 | |||
| down-reg. in both iron-limited photoheterotrophic and photo autotrophic g rowth | |||||
| [4Fe-4S] | HCP4 | 0.01 | 0.27 | ||
| [4Fe-4S] | HCP1 | 0.04 | 0.28 | ||
| [4Fe-4S] | SIR1 | 0.17 | 0.50 | ||
| heme | truncated hemoglobin | 0.03 | 0.06 | ||
| Fe, [2Fe-2S] | glutaredoxin | 0.33 | 0.35 | ||
| heme | CYP747A1 | 0.35 | 0.21 | ||
| Fe | RBD2 | 0.38 | 0.33 | ||
| correlated reduction in transcript and protein abundance | |||||
| [3Fe-4S] | GSN1 | 0.34 | 46 ± 10 | 13 ± 2 | |
| [2Fe-2S] | MitoNEET-like | 0.36 | 37 ± 15 | nd | |
| [4Fe-4S] | ACH1 | 0.49 | 379 ± 25 | 280 ± 35 | |
| [4Fe-4S] | APR1 | 0.66 | 132 ± 13 | 32 ± 14 | |
| [4Fe-4S] | HCP3 | 0.30 | 11 ± 6 | nd | |
| [2Fe-2S] | PETF | 0.61 | 87 ± 35 | nd | |
| reduction in protein abundance without significance change in transcript abundance | |||||
| heme | APX4 | 1.10 | 26 ± 3 | nd | |
| [4Fe-4S] | IDS1 | 1.07 | 61 ± 7 | nd | |
| Fe | Cre07.g320900 | 0.83 | 351 ± 68 | nd | |
| [4Fe-4S] | LEU1L | 1.12 | 159 ± 18 | 54 ± 1 | |
Fold-change of RPKMs from iron-limited cultures (0.25 µM Fe) relative to cultures grown in the replete condition (20 µM Fe).
Protein abundance was quantitated for acetate-grown cultures.
Correlated reduction in protein abundance was confirmed using a label-free, quantitative proteomics strategy for a protein similar to MitoNEET ([2Fe-2S]), the classic ferredoxin PETF ([2Fe-2S]), aconitase (4Fe-4S]), the hybrid-cluster protein HCP3 ([4Fe-4S], [4Fe-3O-3S]), glutamate synthase, NADH-dependent (GSN1, [3Fe-4S]), and an adenylylphosphosulfate reductase (APR1, [4Fe-4S]) (Table 1) [24,26].
Degradation of proteins with bound iron
Another mechanism to economize on iron may involve degradation of proteins that bind iron. However, it should be noted that in most cases a distinction has not been made between iron deficiency induced proteolysis and normal degradation coupled to unsuccessful de novo synthesis due to the absence of transcript or cofactor.
Two types of iron homeostasis studies are routinely performed with Chlamydomonas: iron nutrition and iron starvation. In iron nutrition studies, Chlamydomonas cultures are grown to mid-log phase, generally to a defined cell density, in medium with a known starting iron content, so that the iron supply per cell is reproducible between experiments. In the iron starvation experiment, cells grown to mid-log under replete conditions are removed from this medium, and inoculated into fresh medium lacking added iron. The cells in medium lacking iron are sampled as a function of time so that the sequence of events that occur upon progressive iron depletion can be monitored.
Each type of experiment has the potential to reveal distinct acclimation strategies, and both have been used to monitor the abundance of chloroplast proteins with various types of iron cofactors [12,27]. Moseley et al. observed that subunits of the iron-rich PSI and cytochrome b6f complexes are reduced in abundance within 24 hours of iron starvation (at T=0, cells inoculated into minus-iron acetate-containing medium at 1 × 106 cells/ml) [15]. The transcripts of these subunits, however, had roughly the same pattern of expression regardless of iron supply, leading the authors to conclude that reduction in complex abundance is not due to absence of transcript. The authors also found protease activity specific to iron-minus cells, which was responsible for degrading some light-harvesting proteins. Hence, the reduction in PSI and cytochrome b6f could be due to programmed iron-deficiency-responsive proteolysis rather than an inability to acquire cofactor. The increased abundance of both soluble and membrane-bound ferritins in Chlamydomonas under poor iron supply has been attributed to their role as iron buffers in order to protect macromolecular constituents from the reactivity of iron released from the degradation of dispensable iron-containing proteins [28,29].
A parallel genome-wide analysis of protein and transcript levels revealed several iron dependent proteins, which decrease without a drop in transcript abundance [24,26]. These include L-ascobate peroxidase (APX4, heme), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (IDS1, [4Fe-4S]), 3-isopropylmalate dehydratase (LEU1L, [4Fe-4S]), and a putative phytanoyl-CoA dioxygenase (Cre07.g320900, non-heme Fe) (Table 1). These proteins may succumb to iron-responsive degradation. Alternatively, these proteins may not be maintained because of lack of cofactor and rapid degradation of the apoprotein. In the case of 4-hydroxy-3-methylbut-2-enyl diphosphate reductase a protein homolgous to LytB, which catalyzes the last step in the methylerythritol phosphate pathway and is presumed to localize to the chloroplast, the iron-sulfur cluster may be damaged by superoxide generated in that compartment during iron limitation [30].
Preferential maintenance of indispensable iron-dependent proteins
The down-regulation of gene expression, the activation of transcript degradation and initiation of proteolysis serve to spare iron. Concurrently, a second group of iron-dependent proteins are maintained and appear to be prioritized for iron acquisition. In response to poor iron supply, this might involve increased gene expression to provide more transcripts for increased synthesis (Supplementary Table 1), increased translation of a prioritized protein, or alternatively, by a mechanism that prevents proteolysis. Note that an increase in transcript abundance may not be recapitulated at the protein level, because in the absence of the iron cofactor, the newly synthesized protein may not assemble properly and is simply degraded (as in the case of plastocyanin abundance in copper-deficient Chlamydomonas cells [31]). Nonetheless, because of normal protein turnover, the protein levels could be even lower if expression of the corresponding gene had not been increased [32].
In Chlamydomonas, it is not known if this subset of transcripts is regulated directly by iron supply. Alternatively, the lack of enzyme activity due to absence of metallation could create a feedback response from the pathway where it operates to induce gene expression. In the case of the tetrapyrrole pathway, nearly all transcripts are increased in iron limitation if acetate is present, but they are decreased if acetate is absent [24]. This result is surprising because iron-limited cells are chlorotic (lacking chlorophyll) in the presence of acetate but significantly less so in the absence of acetate. Transcript abundance for many of the genes in this pathway could be determined by the concentration of pathway intermediates or end products (which could accumulate in iron poor cells) rather than directly by iron nutrition.
In Chlamydomonas, the best characterized case of prioritization with respect to iron supply is the chloroplast-localized Fe-dependent superoxide dismutase (Fe-SOD). In contrast to several iron-dependent proteins, neither transcript abundance, polypeptide abundance nor activity of the Fe-SOD at steady state is affected by iron nutrition; the protein accumulates with bound iron even in the iron-limited situation [12]. In contrast, ferredoxin, Cyt f and a di-iron cyclase in chlorophyll biosynthesis are all reduced in abundance. This raised the question of how the FeSOD is preferentially maintained when other iron-containing proteins in the same compartment are lost. A mechanism was suggested by monitoring the fate of FeSOD in an iron starvation time course. In this situation, Fe-SOD abundance decreased rapidly, in parallel with other iron-containing proteins (Cyt f, ferredoxin and the cyclase) within 24 hours, but while the abundance of Cyt f, ferredoxin and cyclase remained low, Fe-SOD levels began to recover and were fully recovered by day five even though the RNA concentration for FeSOD was unchanged. The authors concluded that the Fe-SOD initially falls victim to lack of iron, just like other plastid iron-containing proteins, but the FeSOD is preferentially re-synthesized and preferentially metallated over other iron-dependent proteins. The identity of the factors mediating metallation and potentially enhancing the process in iron limitation is not known.
The impact of metabolism on iron status and vice versa
The metabolic state of the cell can have a large impact on the need for iron-dependent proteins. For instance, with regards to nitrogen metabolism, nitrogen-fixation requires more iron than does nitrate assimilation, which in turn requires more iron than ammonium assimilation [27]. Consequently, the cell responds to the nitrogen source by adjusting iron uptake. Several examples are available from iron-use studies with marine cyanobacteria and diatoms. Siderophore (iron chelator involved in iron assimilation) production by the cyanobacterium Anabaena variabilis is increased when cells are grown under atmospheric nitrogen compared to with nitrate [33], while several studies have found that diatoms have a higher iron uptake rate when grown with nitrate than when grown with ammonium [34]. In addition to increased iron uptake, reallocation of iron may accommodate the synthesis of iron-rich pathways. In the cyanobacterium Crocosphaera watsonii, protein synthesis and degradation appear to be timed to recycle iron between the photosynthetic apparatus during the day and nitrogen fixation at night [10].
When iron nutrition is not optimal, however, the availability of iron can dictate which resources can be metabolized. Respiration has a higher iron demand than fermentation, and in the case of S. cerevisiae, the expression of genes encoding components of iron uptake is induced in response to the diauxic switch [35]. Accordingly, if the gene encoding the transcription factor Aft1, which is responsible for induction of iron uptake, is disrupted, the mutant is unable to grow on non-fermentable carbon sources [36].
The occurrence of two distinct metabolically active compartments, the chloroplast and the mitochondrion, both with a relatively high iron demand, presents a unique scenario in which to explore the dynamic relationship between iron nutrition and energy metabolism in the plant lineage. As briefly mentioned, the presence or absence of acetate affects the physiological outcome of poor iron nutrition in Chlamydomonas. Specifically, in the presence of acetate, iron limited cells deprioritize photosynthesis and switch mainly to heterotrophic metabolism [16]. In the absence of acetate, the photosynthetic apparatus is maintained [27]. Poor iron supply also has a significantly smaller effect on photoautotrophic cells in terms of growth rate, oxygen evolution, and chlorophyll content. The resistance of photoautotrophic cells to iron limitation is evident at the transciptome level as well; only 422 transcripts change in abundance under iron limited photoautotrophic growth, while 2050 transcripts change abundance under photoheterotrophic growth [24].
Because of the need for sequential redox reactions during iron assimilation, iron metabolism is slow, and in situations of high demand, such as during rapid exponential growth of Chlamydomonas in fully iron-replete medium, iron uptake cannot match the iron demand [12]. A simple explanation, therefore, for the resistance of photoautotrophic cells to iron limitation is the much slower growth rate, approximately half the rate as with acetate. In this way, protein synthesis and cell division is better matched to the rate of iron assimilation and distribution. This phenomenon was nicely demonstrated for the over-expression of a diiron protein stearoyl-acyl carrier protein Δ9 desaturase in E. coli, where increased production of the holoprotein was correlated with a reduced growth rate [37]. The slower growth of photoautotrophic cells may facilitate synthesis of iron-rich bioenergetic membranes even with low extracellular iron, resulting in an iron quota under conditions of poor iron nutrition that is twice that of phototoheterotrophic cells [27].
During growth on acetate, iron limitation does not significantly affect the abundance of the respiratory complexes although many iron-dependent proteins within the chloroplast are reduced in abundance [16,27]. Unlike in the photoautrophic situation, photosynthesis is dispensable when acetate is available for respiration, and iron limitation causes a switch to predominately heterotrophic metabolism. The loss of photosynthetic complexes is not solely an effect of acetate supply, because when there is adequate iron, Chlamydomonas maintains a functional photosynthetic apparatus even in the absence of light. Therefore, both iron limitation and acetate are needed for this metabolic switch. How carbon source and iron status interact to regulate bioenergetic metabolism is yet to be determined.
The ability to dispense with photosynthesis has two advantages. First, the cell can maintain relatively high growth rates with a lower iron demand. Second, iron within the chloroplast can now serve as a reservoir of iron for maintenance of the respiratory complexes, which lowers the demand for extracellular iron supply (at least temporarily). Although the loss of iron-rich complexes in the chloroplast is accompanied by increased synthesis and accumulation of ferritin, the ferritin complex from low iron cells does not appear to contain significant amounts of iron [28]. An explanation for increased abundance of ferritin but with reduced iron content is that it operates only as a transient storage instrument for iron, in other words, as a buffer during iron transit from the plastid en route to the mitochondrion for maintenance of respiration [29]. Experiments to measure the iron loading of the two ferritin forms in Chlamydomonas chloroplasts during iron-limitation induced de-greening process should address this question.
Cofactor recycling
Compared to iron-economizing strategies, which are readily evident, it is more difficult to document iron recycling. The fact that it occurs is implied in two recent works. The first example is degradation of the iron-rich photosynthetic apparatus at night concomitant with the synthesis of the iron-rich nitrogenase enzyme in C. watsonii [10], which allows the cyanobacterium to survive on a permanently reduced Fe quota. The second example is the re-synthesis of Fe-SOD subsequent to the loss of PSI, ferredoxin and cytochorme b6f in Chlamydomonas [12]. The quantitative tracking of iron from one protein to another has yet to be achieved. The mechanism responsible for degradation of dispensable iron-containing proteins has also not been explored, neither has the identity of the protease(s) nor how targets are recognized for degradation. Additionally, where these proteins are degraded is not known. A vacuole-like compartment is one possibility. It is likely that extra-plastidic degradation of iron-containing proteins enable accessibility of the released iron to other compartments (like mitochondria).
Conclusions
Although iron is a relatively abundant element in the earth’s crust, many organisms are transiently or chronically limited by access to this nutrient. In neutral to alkaline soil, iron is predominately found in poorly soluble complexes, such as ferric oxides. These complexes create a major obstacle for photosynthetic eukaryotes, which have many of the same iron demands as non-photosynthetic eukaryotes but with the added burden of supplying a relatively substantial amount of iron to the chloroplast. Estimates indicate that at least 30% of arable land is too alkaline for optimal iron uptake [38] neccesitating an understanding how plants cope with poor iron bioavailability.
Because there are inherent differences within the plant lineage, a cross-examination of the responses to poor iron nutrition in phylogenetically distinct organisms provides a robust appraisal of conserved and therefore core iron-responsive mechanisms. Such an analysis comparing the poor-iron transcriptional response in Chlamydomonas, Arabidopsis, and rice revealed nine genes whose transcriptional response to iron was conserved [24]. Among these are both genes with known, such as encoding an iron nutrition–responsive ZIP family transporter (IRT), and unknown function, such as CGLD27, a protein conserved solely among cyanobacteria and plastid-containing organisms. Surprisingly, while other components of the iron-signaling pathway are not conserved, the transcriptional response of an ortholog to BRUTUS, a putative transcription factor and E3 ligase [23], is conserved. Although the Chlamydomonas ortholog has yet to be experimentally characterized, its presence in this dataset suggests that ubiquitin-mediated degradation is a core iron-responsive process within the plant lineage.
In addition to these cross-species analyses, as a single-celled reference organism for the green plant lineage, Chlamydomonas is providing novel insight into how photosynthetic eukaryotes respond to and manage suboptimal iron nutrition at the subcellular level. We are gaining insight into how the cell spares iron, prioritizes iron-dependent proteins, and partitions iron between the bioenergtic membranes of the chloroplast and the mitochondrion during an iron-limited situation. The challenge will be to decipher how iron supply and carbon source is translated into a signal that regulates these pathways. As one component, expression for a subset of genes is regulated at the transcriptional level, and iron-responsive promoter elements have been identified [24,39,40], although the transcription factor(s) mediating this regulation are currently unknown. A second level may involve non-coding RNA, as this system is prevalent in other microbes, or post-translational regulation, as several examples of iron sparing appear to involve changes in protein abundance without an effect on transcript levels.
Supplementary Material
Highlights.
Increased iron uptake is not the only mechanism to combat poor iron supply.
Iron sparing, recycling and prioritizing proteins for iron loading are key.
Chlamydomonas provides an opportunity to explore subcellular iron distribution.
Acknowledgments
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S Department of Energy (DE-FD02-04ER15529). C.E.B.-H. acknowledges support from an Individual Kirschstein National Research Service Award from the National Institutes of Health (GM100753).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacobson L. Iron in the leaves and chloroplasts of some plants in relation to their chlorophyll content. Plant Physiol. 1945;20:233–245. doi: 10.1104/pp.20.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noack K, Jiebich H. The iron-set of chloroplasts of spinach. Naturwissenschaften. 1941;29:302–302. [Google Scholar]
- 3.Hummel E, Guttmann P, Werner S, Tarek B, Schneider G, Kunz M, Frangakis AS, Westermann B. 3D Ultrastructural organization of whole Chlamydomonas reinhardtii cells studied by nanoscale soft x-ray tomography. PLoS One. 2012;7:e53293. doi: 10.1371/journal.pone.0053293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehara T, Osafune T, Hase E. Behavior of mitochondria in synchronized cells of Chlamydomonas reinhardtii (Chlorophyta) J Cell Sci. 1995;108(Pt 2):499–507. doi: 10.1242/jcs.108.2.499. [DOI] [PubMed] [Google Scholar]
- 5. Merchant SS, Helmann D. Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microb Physiol. 2012;60:91–210. doi: 10.1016/B978-0-12-398264-3.00002-4. Using examples from both prokaryotes and microbial eukaryotes, the authors provide an extensive and comprehensive review of pathways involved in acclimating to nutrient deficiency (including trace metal sparing, recycling and prioritization)
- 6.Schägger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I–V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J Biol Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 7.Chow WS, Melis A, Anderson JM. Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci U S A. 1990;87:7502–7506. doi: 10.1073/pnas.87.19.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millar AH, Whelan J, Soole KL, Day DA. Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol. 2011;62:79–104. doi: 10.1146/annurev-arplant-042110-103857. [DOI] [PubMed] [Google Scholar]
- 9.Ramírez-Aguilar SJ, Keuthe M, Rocha M, Fedyaev VV, Kramp K, Gupta KJ, Rasmusson AG, Schulze WX, van Dongen JT. The composition of plant mitochondrial supercomplexes changes with oxygen availability. J Biol Chem. 2011;286:43045–43053. doi: 10.1074/jbc.M111.252544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, Monteiro FM, Follows MJ, Valois FW, Waterbury JB. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc Natl Acad Sci U S A. 2011;108:2184–2189. doi: 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merchant S, Allen M, Kropat J, Moseley J, Long J, Tottey S, Terauchi A. Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim Biophys Acta. 2006;1763:578–594. doi: 10.1016/j.bbamcr.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 12. Page MD, Allen MD, Kropat J, Urzica EI, Karpowicz SJ, Hsieh SI, Loo JA, Merchant SS. Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii. Plant Cell. 2012 doi: 10.1105/tpc.112.098962. Based on an earlier observation that expression of a gene encoding a MnSOD is significantly induced under iron defiency, the authors discover that this MnSOD does not simply replace the plastid-localized FeSOD but rather the FeSOD is preferetnailly retained and additional expression of the MnSOD increases the capacity of iron-starved cells to cope with superoxide. The authors provide evidence for degradation and re-synthesis of the FeSOD, giving an indication that iron is preferentially supplied to the newly synthesized apoprotein.
- 13.Becker B. Snow ball earth and the split of Streptophyta and Chlorophyta. Trends Plant Sci. 2013;18:180–183. doi: 10.1016/j.tplants.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, Spalding MH. Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol. 2007;10:190–198. doi: 10.1016/j.pbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Moseley JL, Allinger T, Herzog S, Hoerth P, Wehinger E, Merchant S, Hippler M. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 2002;21:6709–6720. doi: 10.1093/emboj/cdf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics. 2007;7:3964–3979. doi: 10.1002/pmic.200700407. [DOI] [PubMed] [Google Scholar]
- 17.Terauchi AM, Lu SF, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T, et al. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem. 2009;284:25867–25878. doi: 10.1074/jbc.M109.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanvisens N, Bañó MC, Huang M, Puig S. Regulation of ribonucleotide reductase in response to iron deficiency. Mol Cell. 2011;44:759–769. doi: 10.1016/j.molcel.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oglesby-Sherrouse AG, Murphy ER. Iron-responsive bacterial small RNAs: variations on a theme. Metallomics. 2013;5:276–286. doi: 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philpott CC, Leidgens S, Frey AG. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta. 2012;1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong WW, Yang ZM. Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol Biochem. 2010;48:153–159. doi: 10.1016/j.plaphy.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 23.Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–2236. doi: 10.1105/tpc.110.074096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urzica EI, Casero D, Yamasaki H, Hsieh SI, Adler LN, Karpowicz SJ, Blaby-Haas CE, Clarke SG, Loo JA, Pellegrini M, et al. Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell. 2012;24:3921–3948. doi: 10.1105/tpc.112.102491. The authors provide the first global analysis of RNA and protein abundance during the the three stages of iron nutrition in Chlamydomonas. Quantitation of both RNA and protein from a single experiment provides classification of proteins into 3 groups: correlated RNA and protein abudance, reduced protein abundance without a change in RNA, and opposite directions for changes in protein and RNA.
- 25.Bölling C, Fiehn O. Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol. 2005;139:1995–2005. doi: 10.1104/pp.105.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsieh SI, Castruita M, Malasarn D, Urzica E, Erde J, Page MD, Yamasaki H, Casero D, Pellegrini M, Merchant SS, et al. The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol Cell Proteomics. 2013;12:65–86. doi: 10.1074/mcp.M112.021840. The authors identify hundreds of differentially abundant proteins using label-free protein quantitation on the soluble fraction of Chlamydomonas grown under several trace metal defiency situations.
- 27.Terauchi AM, Peers G, Kobayashi MC, Niyogi KK, Merchant SS. Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth Res. 2010;105:39–49. doi: 10.1007/s11120-010-9562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long JC, Sommer F, Allen MD, Lu SF, Merchant SS. FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics. 2008;179:137–147. doi: 10.1534/genetics.107.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busch A, Rimbauld B, Naumann B, Rensch S, Hippler M. Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 2008;55:201–211. doi: 10.1111/j.1365-313X.2008.03490.x. [DOI] [PubMed] [Google Scholar]
- 30.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydrolyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 31.Li HH, Merchant S. Degradation of plastocyanin in copper-deficient Chlamydomonas reinhardtii. Evidence for a protease-susceptible conformation of the apoprotein and regulated proteolysis. J Biol Chem. 1995;270:23504–23510. doi: 10.1074/jbc.270.40.23504. [DOI] [PubMed] [Google Scholar]
- 32.Berlin CM, Schimke RT. Influence of turnover rates on the responses of enzymes to cortisone. Mol Pharmacol. 1965;1:149–156. [PubMed] [Google Scholar]
- 33.Kerry A, Laudenbach D, Trick C. Influence of iron limitation and nitrogen source on growth and siderophore production by cyanobacteria. Journal of Phycology. 1988;24:566–571. [Google Scholar]
- 34.Wang W, Dei R. Biological uptake and assimilation of iron by marine plankton: influences of macronutrients. Marine Chemistry. 2001;74:213–226. [Google Scholar]
- 35.Haurie V, Boucherie H, Sagliocco F. The Snf1 protein kinase controls the induction of genes of the iron uptake pathway at the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2003;278:45391–45396. doi: 10.1074/jbc.M307447200. [DOI] [PubMed] [Google Scholar]
- 36.Casas C, Aldea M, Espinet C, Gallego C, Gil R, Herrero E. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast. 1997;13:621–637. doi: 10.1002/(SICI)1097-0061(19970615)13:7<621::AID-YEA121>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman BJ, Broadwater JA, Johnson P, Harper J, Fox BG, Kenealy WR. Lactose fedbatch overexpression of recombinant metalloproteins in Escherichia coli BL21 (DE3): process control yielding high levels of metal-incorporated, soluble protein. Protein Expr Purif. 1995;6:646–654. doi: 10.1006/prep.1995.1085. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Barak P. Iron nutrition of plants in calcareous soils. Adv. Agron. 1982;35:217–240. [Google Scholar]
- 39.Fei X, Eriksson M, Li Y, Deng X. A novel negative Fe-deficiency-responsive element and a TGGCA-type-like FeRE control the expression of FTR1 in Chlamydomonas reinhardtii. J Biomed Biotechnol. 2010;2010:790247. doi: 10.1155/2010/790247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng X, Eriksson M. Two iron-responsive promoter elements control expression of FOX1 in Chlamydomonas reinhardtii. Eukaryot Cell. 2007;6:2163–2167. doi: 10.1128/EC.00324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.