Abstract
Defensins are an effector component of the innate immune system with broad antimicrobial activity. Humans express two types of defensins, α- and β-defensins, which have antiviral activity against both enveloped and non-enveloped viruses. The diversity of defensin-sensitive viral species reflects a multitude of antiviral mechanisms. These include direct defensin targeting of viral envelopes, glycoproteins, and capsids in addition to inhibition of viral fusion and post-entry neutralization. Binding and modulation of host cell surface receptors and disruption of intracellular signaling by defensins can also inhibit viral replication. In addition, defensins can function as chemokines to augment and alter adaptive immune responses, revealing an indirect antiviral mechanism. Nonetheless, many questions regarding the antiviral activities of defensins remain. Although significant mechanistic data are known for α-defensins, molecular details for β-defensin inhibition are mostly lacking. Importantly, the role of defensin antiviral activity in vivo has not been addressed due to the lack of a complete defensin knockout model. Overall, the antiviral activity of defensins is well established as are the variety of mechanisms by which defensins achieve this inhibition; however, additional research is needed to fully understand the role of defensins in viral pathogenesis.
Abbreviations: CMV, cytomegalovirus; HD, human defensin; HIV, human immunodeficiency virus; HPIV, human parainfluenza virus; HSV, herpes simplex virus; IAV, influenza A virus; RSV, respiratory syncytial virus; VSV, vesicular stomatitis virus; PIV, parainfluenza virus; HAdV, human adenovirus; HPV, human papillomavirus; BKV, BK virus; LTB4, leukotriene B4; HRV, human rhinovirus; HNP, human neutrophil peptide; DC, dendritic cell; HBD, human β-defensin; PKC, protein kinase C; SBD1, sheep β-defensin 1; MBD, murine β-defensin; PBMC, peripheral blood mononuclear cell
Keywords: virus, defensin, antimicrobial peptides, innate immunity
Graphical abstract
Highlights
-
•
Defensins are active against both enveloped and non-enveloped viruses.
-
•
Targets include viral envelopes, glycoproteins, and capsids or host cells.
-
•
Multiple antiviral mechanisms have been described.
-
•
They can also alter immune responses to affect pathogenesis.
-
•
Their role in viral pathogenesis in vivo is understudied.
Introduction and scope
Defensins are one of the most abundant classes of antimicrobial peptides in humans and have primarily been studied as effector components of the innate immune response with direct antibacterial activity. However, their antiviral properties were appreciated from the earliest functional studies of α-defensins [1], [2]. In fact, their ability to neutralize herpes simplex virus 1 (HSV-1) was one of the defining criteria in the identification and purification of the α-defensins human neutrophil peptide 1 (HNP1), HNP2, and HNP3 from human neutrophils [3]. In this review, we will summarize the known activities of α- and β-defensins against viruses, and we refer the reader to additional recent reviews on this topic for further insight [4], [5], [6], [7]. Our focus will be primarily on human defensins (HDs), and we have organized our discussion by mechanism (Fig. 1 ) to emphasize commonalities in the antiviral activities of defensins against diverse viral families.
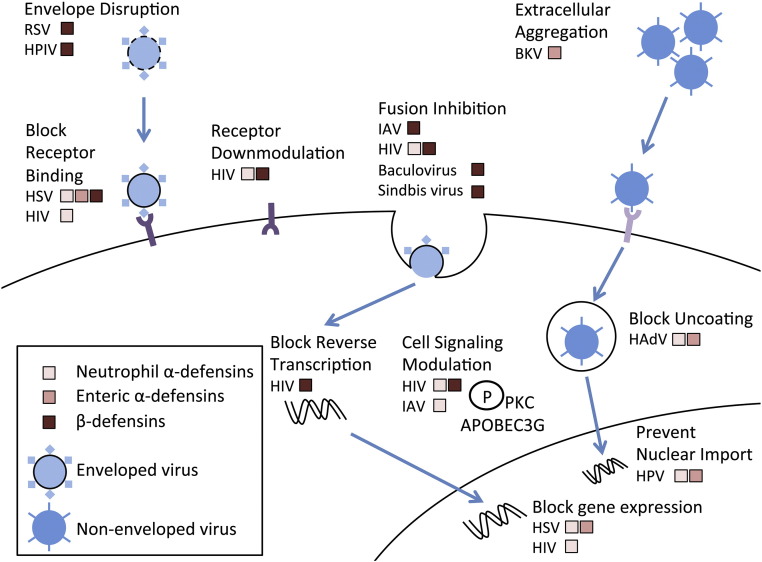
Fig. 1.
Major antiviral mechanisms of defensins. Defensins inhibit many steps in enveloped and non-enveloped viral infections. Mechanistic information is available for RSV, HPIV, HIV, HSV, IAV, BKV, HAdV, and HPV. Most mechanisms impact viral entry, but post-entry effects have been described. We have omitted defensin effects that have been reported, but their contribution to blocking infection is in doubt (e.g., aggregation of HAdV by α-defensins). Note that although the defensins relevant for each virus at each step are indicated in broad classifications (e.g., β-defensins), in most cases, not all of the defensins within these groups have been tested or have equivalent activity.
Defensin structure, expression, and physiologic concentrations
The structure, evolution, and tissue distribution of defensins have been summarized in a number of recent reviews [4], [8], [9], [10], [11], [12], [13]. Briefly, defensins are small (~ 29–42 amino acids) cationic, amphipathic peptides with a predominantly β-sheet structure stabilized by three disulfide bonds. They can be broadly classified on the basis of structure and disulfide bond organization into three groups: α-, β-, and θ-defensins. As humans lack θ-defensins, they will not be discussed in detail; however, their antiviral properties have been recently reviewed [8], [14]. Humans express 6 α-defensins and up to 31 β-defensins [15]. The α-defensins can be further subdivided into myeloid (HNP1-4) and enteric (HD5 and HD6) peptides on the basis of both expression patterns and genetic organization [11]. All of the α-defensins have been shown to multimerize into at least dimers either in solution or in crystal structures [16], [17], [18], [19]. HNP1-4 are predominantly expressed by neutrophils but can also be found in or expressed by monocyte/macrophages, natural killer cells, some T cells, B cells, and immature dendritic cells (DCs) [11], [13]. HD5 and HD6 are expressed by specialized epithelial Paneth cells of the small intestine [20], [21]. HD5 is also expressed by epithelial cells in the male and female genitourinary tracts [22], [23], [24], [25]. Human β-defensins (HBDs) are more widely expressed by epithelial cells in skin and at mucosal surfaces in contact with the environment [26]. Like the α-defensins, some β-defensins (e.g., HBD3) exist in oligomeric forms, while others, such as HBD1 and HBD2, are monomeric [27]. They are also expressed by monocytes, macrophages, and certain DCs, and a subset of β-defensins are only expressed in the male reproductive tract [26], [28]. Although there are commonalities in expression patterns of defensins in humans and other species, one important difference relevant for experimental models of infectious disease is that mice lack myeloid α-defensins [29], [30].
Quantification of physiologic defensin concentrations in vivo is complex, as defensins are present at high local concentrations within specific cell types or upon release from cells into confined anatomical niches (e.g., crypts of the small intestine) but can become diluted in extracellular fluids. For the myeloid α-defensins, Daher et al. estimated ~ 3 mM (10 mg/ml) HNPs in neutrophils, with even higher local concentrations in the azurophil granules in which they are stored [1]. For the enteric α-defensins, Ayabe et al. estimated concentrations of ≥ 3.5 mM (15–100 mg/ml) in the crypt lumen, the site of Paneth cell degranulation [31]. These concentrations are likely similar in the human small intestine, where HD5 expression exceeds that of HD6 by 6-fold [32]. In healthy patients, epithelial lining fluid of the lung contains 31–79 nM HNP1-3, nasal fluid contains ~ 2.7 μM HNP1-3, saliva contains 0.3–3 μM HNP1-3, and vaginal secretions contain ~ 1.5 μM HNPs and 0.3–14 μM HD5 [23], [33], [34], [35], [36], [37], [38]. For the β-defensins, 5–10 nM HBD2 has been measured in nasal fluid [37], [39]. However, in certain disease states, defensin levels can be highly elevated. For example, 57 μM to 2.4 mM concentrations of HNP1 have been found in epithelial lining fluid of cystic fibrosis patients [36]. Overall, the concentrations of defensins present in vivo are generally within the range that is needed for direct antiviral activity by α-defensins and generally below the concentrations required for direct antiviral activity by β-defensins.
Antiviral mechanisms through direct interactions between defensins and virus
Modes and determinants of defensin binding to viruses
There are multiple modes of defensin binding to ligands such as viral particles. First, defensins interact with lipid bilayers, which is facilitated by the presence of negatively charged phospholipids [11], [13], [40]. Second, four of the α-defensins (HNP1-3 and HD5) and HBD3 are lectins capable of binding to glycoproteins and glycolipids [41], [42], [43], [44]. Third, defensins can potentially engage in protein–protein or protein–DNA interactions. Because they are cationic and amphipathic, defensins interact with ligands through both charge–charge and hydrophobic interactions. Defensin oligomerization, particularly for α-defensins, and conformational stability imparted by disulfide bonds may further influence binding. Each of these interactions contributes to the antiviral activity of defensins, and their relative importance depends on the specific virus/defensin pair under investigation.
The property of defensins that has been most widely investigated for its contribution to antiviral activity is stabilization of the three-dimensional structural fold through the formation of disulfide bonds. Generally, destabilized or “linear” defensins are generated by substituting the conserved cysteine residues either in toto, individually, or in pairs to natural or non-natural residues that cannot form disulfide bonds such as serine or α-amino-n-butyric acid. Alternatively, wild-type defensins are reduced and chemically modified (alkylated) to prevent disulfide bond formation. All reported studies have shown that the disulfide-stabilized forms of α-defensins are required to either inhibit [HSV-1, human adenovirus (HAdV) serotype 5, influenza A virus (IAV), and human immunodeficiency virus-1 (HIV-1)] or enhance (HIV-1) virus infection [1], [2], [45], [46], [47], [48]. In two cases, the antiviral activity of β-defensins was unaffected by “linearization” [49], [50]. Given the paucity of data in this regard for β-defensins, it is unclear if this is a fundamental difference between α- and β-defensin antiviral activities. Together, these studies suggest that the effects of α-defensins on virus infection are more likely to be due to their amphipathicity or ability to multimerize, which are structurally dependent, rather than the net positive charge of the molecule that is common to both native and “linearized” forms.
The capacity of defensins to function as lectins and bind selectively to sugars contributes to their antiviral properties; however, defensins also bind to host cellular and serum proteins [1], [41]. The relative affinity for viral targets versus serum components may explain why some defensins are only antiviral against particular viruses in the absence of serum. Although it has been shown that HD5 binds natural viral glycoproteins, notably, HSV-1 glycoprotein D (gD) and HIV-1 gp120, with a higher affinity than bovine serum albumin or fetuin, serum substantially attenuates the antiviral activity of HNP1 against HSV-1, even at low concentrations [1], [3], [41]. Nonetheless, this effect can be overcome at higher HNP1 concentrations or by pre-incubating the virus with HNP1 before it is added to cells. The addition of serum also abrogates the antiviral activity of HNP1-3, HBD2, and HBD3 against both X4 and R5 tropic HIV-1 [51], [52]. Serum also competes for HD5 binding to and inhibition of HAdV, which lacks viral glycoproteins [53]. Two notable exceptions to the abrogating effects of serum are inhibition of human papillomavirus (HPV) and human BK virus (BKV) infection by α-defensins, which are not blocked by 10% or 5% serum, respectively [54], [55]. The effect of serum on the modulation of HIV-1 infection by HD5 is complicated by cytotoxicity of HD5 for primary CD4+ T cells. Thus, application of HIV-1 pre-incubated with HD5 in the presence or absence of low serum (1–2%) to primary CD4+ T cells results in enhanced HIV-1 infection if the cells are subsequently cultured under standard conditions (10% fetal bovine serum and IL-2) but results in cell death if the cells are maintained under serum-deprived conditions [46], [56], [57]. Previously, HD5 was found to be pro-apoptotic for primary CD4+ T cells in the absence of serum at concentrations as low as 1.4 μM [58]. In contrast, under both serum conditions, HD5 enhances HIV-1 infection of HeLa cells expressing CD4 and CCR5 and is non-cytotoxic [46], [56]. Rapid inactivation by binding to serum components may protect host cells from damage by defensins; however, defensins likely remain potently antiviral in vivo at high local concentrations upon initial secretion and in serum-free anatomical locations (e.g., phagocytic vacuoles or the bowel lumen).
Although much of the antibacterial activity of defensins is attenuated at physiologic salt concentrations [11], this is not generally true for their antiviral activity. One instance of salt sensitivity is that HBD2 and HBD3 have attenuated anti-HIV activity in physiological salt concentrations [52], which is somewhat surprising given that even the antibacterial effects of HBD3 are generally not salt sensitive [59]. Nonetheless, the anti-HIV activity of these β-defensins under low-salt and serum-free conditions may reflect the physiological conditions of the oral cavity [60], [61]. We have shown that super-physiological concentrations of salt inhibit HD5 binding to HAdV, which implicates charge–charge interactions in HD5 binding to the viral capsid [53], [62]. In general, differential salt sensitivity may reflect variation in both the molecular interactions and the mechanisms of defensin-mediated killing or neutralization of viruses versus bacteria.
A limited number of structure–function studies have evaluated the roles of additional features of α-defensins in modulating viral infection. We have shown that the conserved salt bridge stabilizing a loop between two β-strands of HD5 is dispensable for HAdV-5 inhibition [45]. Similarly, mutation of an invariant glycine residue (Gly17) of HNP2 to glutamate severely attenuates the antibacterial activity of HNP2 but results in only a minor loss of antiviral activity against HPV-16 [54], [63]. In contrast, specific arginine residues are critical for HD5 binding to HAdV-5 and HPV-16, and this activity is not purely charge dependent, as lysine substitutions for selected arginine residues did not preserve antiviral activity [62]. Likewise, single arginine mutations at R9H or R13H of HD5 attenuate enhancement of HIV-1 infection, indicating that this property is also not simply charge dependent [64]. Furthermore, the capacity of HD5 to self-associate is critical for antiviral activity against both HAdV-5 and HPV-16 and for HIV-1 gp120 and HSV-1 gD binding [41], [62], [65], [66]. These properties were revealed through mutations that disrupt defensin activity. In the converse approach, residues in HD5 under positive selection were mutated in an effort to augment the antiviral activity of HD5 against HSV-2 [67]. One mutant (HD5 E21R) demonstrated improved anti-HSV-2 and anti-HIV-1 activity in cell culture and was both prophylactically (1 h before infection) and therapeutically (24 h after infection) protective against lethal HSV-2 challenge in a mouse model. Collectively, these studies suggest that specific features of viruses are selectively bound by defensins and that the binding interface of the defensin is sequence specific and not merely charge dependent.
The basis for selective recognition of diverse viruses by defensins is largely unknown. Defensins may interact with either the lipid bilayer or envelope glycoproteins of enveloped viruses; however, protein–protein interactions must be critical for binding to non-enveloped viruses (e.g., HPV and HAdV) and likely contribute to binding to enveloped viruses as well. For HIV-1, competition with site-specific antibodies was used to map HNP2 binding sites on gp120 [68]. For species C HAdV (HAdV-C), we have used structural studies and a chimeric approach to identify capsid determinants of HD5 binding [45], [69]. Other than for these two viruses, almost nothing is known about specific determinants of the viral particle that dictate defensin binding. Nonetheless, the fact that closely related defensins have differential antiviral (or infection-enhancing) effects indicates selectivity and may inform the design of future studies to elucidate the basis for recognition. In this regard, Table 1 summarizes the known effects of defensins on virus infection, including those defensins that have been tested but have not been found to be antiviral against specific viruses.
Table 1.
Known effects of defensins on virus infection
| Virusa | Envb | HNPc |
HDc |
HBDc |
References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | |||
| CMV | Y | I | * | * | * | * | * | * | * | * | * | [1] |
| HIV-1 | Y | I | I | I | I | E | E | NE | I | I | * | [52], [66], [83] |
| HPIV-3 | Y | * | * | * | * | * | * | NE | I | * | * | [73] |
| HSV-1/HSV-2 | Y | I | I | I | I | I | I | NE | NE | I | * | [1], [44] |
| IAV | Y | I | I | I | I | I | I | I | I | * | * | [48], [97], [98] |
| RSV | Y | * | * | * | * | * | * | NE | I | * | * | [73] |
| VSV | Y | I | * | * | * | * | * | * | * | * | * | [1] |
| VV | Y | NE | * | * | * | * | * | NE | NE | I | * | [73], [93], [129] |
| VZV | Y | * | * | * | * | * | * | * | I | * | * | [92] |
| AAV | N | I | I | * | * | * | * | * | * | * | * | [36] |
| Echovirus | N | NE | * | * | * | * | * | * | * | * | * | [1] |
| HAdV | N | |||||||||||
| HAdV-A | I | * | * | * | I | * | * | * | * | * | [[45], [58], [165],166] | |
| HAdV-B | I | * | * | * | I | * | NE | NE | * | * | ||
| HAdV-C | I | * | * | * | I | * | NE | NE | * | * | ||
| HAdV-D | E | * | * | * | E | * | NE | NE | * | * | ||
| HAdV-E | E | * | * | * | I | * | * | * | * | * | ||
| HAdV-F | E | * | * | * | NE | * | * | * | * | * | ||
| HPV | N | I | I | I | I | I | NE | NE | NE | * | * | [59] |
| PyV | N | |||||||||||
| PyV-BKV | I | * | * | * | I | * | NE | I | * | * | [60] | |
| PyV-JCV | I | * | * | * | I | * | I | I | * | * | ||
| PyV-SV40 | NE | * | * | * | I | * | NE | NE | * | * | ||
| HRV | N | * | * | * | * | * | * | * | NE | * | * | [39] |
| Reovirus | N | NE | * | * | * | * | * | * | * | * | * | [1] |
Abbreviations used: VV, vaccinia virus; VZV, varicella zoster virus; AAV, adeno-associated virus; PyV, polyomavirus; JCV, JC virus; SV40, simian virus 40.
Enveloped (Y) or non-enveloped (N) virus.
I, inhibit; E, enhanced; *, not tested; NE, no effect.
Direct virus inactivation by affecting envelope
Direct interactions between defensins and structural components of the virion, particularly the lipid bilayer of enveloped viruses, could destroy or destabilize the virus and render it non-infectious. This mechanism was proposed in the first studies of the antiviral activity of human and rabbit neutrophil α-defensins [1], [2], [3], [70]. In support of this hypothesis, HSV-1 inactivation by rabbit NP-2 or HNP1 was impaired at temperatures below 20 °C and was more effective at 40 °C than at 37 °C, implicating the fluidity of the membrane rather than binding as an important parameter [1], [2]. HNP1 has also been shown to bind directly to HSV-1 and to model membranes containing phosphatidylserine [1]. Nonetheless, the morphology of HSV-1 by electron microscopy was not altered by incubation with a neutralizing concentration of HNP1 [1]. HBD2 and HBD3 bind directly to and inactivate HIV-1, but whether this interaction affects or damages the viral envelope is unknown [52], [71], [72]. Similarly, treatment of HIV-1 with HNP1 in the absence of serum irreversibly decreases infectivity, although a physical change in particle integrity was not determined [71], [73]. More recently, the lipid bilayer of respiratory syncytial virus (RSV) exposed to neutralizing HBD2 but not to non-neutralizing HBD1 was visibly damaged when assessed by electron microscopy [74]. Although the universality of this phenotype was not quantified, the proteins of the bulk population of the virus were more buoyant in a density gradient, suggesting that most of the viral particles were disrupted. The altered morphology was correlated with a 70% reduction in virus attachment to the cell. This is perhaps the most convincing data for direct viral envelope disruption by a defensin and likely extends to human parainfluenza virus 3 (HPIV-3), which has a profile of defensin sensitivity similar to that of RSV [74].
The broad neutralization of many enveloped viruses supports the hypothesis that the lipid bilayer is the target; however, enveloped viruses are not universally susceptible and their sensitivity to α-defensins can be highly variable. For example, in one study, rabbit NP-1 and NP-2 inhibited HSV-1 up to 1000-fold; HSV-2, up to 10-fold; vesicular stomatitis virus (VSV), up to 100-fold; and IAV, up to 56-fold, but had no effect on cytomegalovirus (CMV) [2]. Similarly, HNP1 potently inhibited HSV-1 (1000-fold) and HSV-2 (100-fold) but only weakly inhibited CMV (6-fold), VSV (7-fold), and IAV (6-fold) [1], [3]. Defensin perturbation of lipid bilayers is dependent upon their composition. It is favored by negatively charged phospholipids, whereas neutral bilayers are largely inert to defensin [65], [75]. The lipid content of viral envelopes is dependent upon the subcellular location and membrane microdomains from which they bud and likely varies among viral families [76]. If direct membrane perturbation contributes to the antiviral effect of defensins, then differences in the lipid composition of the envelope may in part explain the differential susceptibility of viruses to defensins.
In contrast to disruption of enveloped viruses, increased capsid resistance to mechanical force or heat has been observed upon HD5 binding to non-enveloped HAdV-C [45], [53], [77]. This effect correlates with the inability of the HD5-bound particle to uncoat, similar to a genetic mutant of HAdV-C that is stabilized by the presence of unprocessed precursor capsid proteins [53], [78], [79]. A failure to uncoat precludes release of an internal, membrane-permeabilizing capsid protein, thereby blocking HAdV-C escape from the endosome and introduction of the viral genome into the nucleus, its replication niche [53], [79], [80], [81]. Thus, unlike enveloped viruses where destabilization of the virion impairs infectivity, an opposite, stabilizing interaction with the non-enveloped HAdV-C capsid produces a similar outcome.
Extracellular aggregation
As many defensins form multimeric structures, which have been demonstrated in both crystal structures and in solution [16], [17], [18], [19], [27], [41], there is the potential that interactions between defensin peptides bound to neighboring viruses will cause virions to aggregate. This has been shown directly for IAV by HNP1 and HNP2 and for HAdV-5 and BKV by HD5 [55], [62], [82]. For IAV, it is unclear if defensin-mediated aggregation is required to block infection. Similarly, aggregation of HAdV by HD5 is not sufficient to inhibit infection, as a non-neutralizing mutant of HD5 is able to induce aggregation [62]. In contrast, aggregation and a concomitant inability to bind host cells have been shown to be the dominant mechanisms of neutralization for BKV [55]. The mechanism of inhibition of other polyomaviruses such as JCV and SV40, which are also sensitive to HD5, has not been determined [55]. In addition to multivalent binding due to defensin oligomerization, neutralization of capsid charge by defensin binding may reduce repulsion between virions. We have shown this directly for HAdV-C [62], and this effect may facilitate the aggregation of other viruses. These effects are likely interrelated, as mutants of HD5 that are impaired in self-association are incapable of both aggregating HAdV-C and fully neutralizing the capsid charge. Aggregation can impact viral infectivity by directly impeding cell binding or by causing viruses in clumps to enter fewer cells.
Blocking receptor binding
Defensin binding to viral attachment proteins could disrupt receptor interactions critical for viral entry into the cell. HNP1-3, HD5, and HBD3 bind a recombinant viral glycoprotein (gB) of both HSV-1 and HSV-2, which correlates with the ability of these defensins to inhibit HSV-1 and HSV-2 entry and adhesion [44], [83]. Lectin activity of HNP1 is critical, as deglycosylation of HSV-2 gB abrogates binding [83]. The contribution of glycoprotein binding to the antiviral mechanism of HD5 was underscored by a direct correlation between the capacity of HD5 mutants to neutralize HSV-2 and their affinity for recombinant gD [31]. HNP4 and HD6 also inhibit HSV-1 and HSV-2 infection but do not bind to viral glycoproteins [44]. Instead, HNP4 and HD6 have been shown to bind heparan sulfate, the receptor for attachment, as well as other glycosaminoglycans. HBD3 is the only defensin able to bind both host cell receptors and viral glycoproteins [44]. Those defensins that failed to bind gB or heparan sulfate, HBD1 and HBD2, were also unable to neutralize infection. Overall, blocking host cell receptors and binding to viral glycoproteins is a major mechanism by which defensins inhibit HSV-1 and HSV-2 infection.
Similarly, HNP1-4 bind HIV-1 gp41 and gp120, as well as the cell surface receptor CD4 [43], [47], [84]. The binding sites of HNP1 and HNP2 on gp120 have been mapped in antibody competition assays to the CD4 and co-receptor binding sites [68]. Conversely, the HNP1 and HNP2 binding sites on CD4 have been mapped to the gp120 binding site [68]. The mode of α-defensin binding to gp120 and gp41 may be complex, as both lectin-dependent and lectin-independent binding have been reported. In an early study, deglycosylation of gp120 reduced HNP1 binding and abolished HNP2 and HNP3 binding [43]; however, a more recent paper suggested that deglycosylation of gp120 or gp41 does not affect HNP1 binding [47]. In addition, HNP4 binding is not abolished by deglycosylation of gp120 [43], consistent with the observation that HNP4 binds more weakly to both polysaccharides and serum proteins [84]. Nonetheless, HNP4 is a more potent inhibitor of HIV-1 than are HNP1-3 [84]. In summary, like for HSV, defensins directly interfere with HIV-1 binding and attachment.
In contrast to a block in cell binding, we have observed that receptor-dependent and receptor-independent binding of HAdV-C to the cell are enhanced in the presence of an inhibitory concentration of HD5 [45], [62]. This effect may be related to neutralization of the net negative capsid charge, which promotes aggregation, in a manner comparable to enhancement of retrovirus infections by polybrene or HAdV infections by poly-cations [62], [85]. Similarly, HIV-1 binding to cells and infection is enhanced by HD5 and HD6 [46], [64]. Thus, although in several cases the net effect of defensin binding to the virus is to block cellular attachment, the opposite effect has also been observed. The balance of these activities in vivo is unclear.
Inhibition of viral fusion with or penetration of host cell lipid bilayers
To introduce their genomes into host cells, enveloped viruses must fuse their lipid bilayer with that of the host cell [86]. The fusion protein of the virus mediates this reaction by inserting a hydrophobic stretch of amino acids into the target cell membrane followed by a conformational change to a less energetic state, termed the six-helix bundle [87], [88]. The energy for lipid fusion is derived from this conformational change. HIV-1 fusion, mediated by gp41, is inhibited by HNP1 [47], [68]. Inhibition requires the disulfide-stabilized form of HNP1 and is abrogated by serum [47]. HNP1 aggregates recombinant peptides of both the carboxyl- and amino-termini of gp41 that comprise the six-helix bundle, suggesting a direct effect on formation of the post-fusion conformation of gp41 [47]. HNP1 also increases the binding and efficacy of neutralizing antibodies specific for the gp41 pre-hairpin conformation, likely due to greater antibody access to hidden neutralizing epitopes as a consequence of slowed refolding kinetics [89]. Thus, HNP1 binding directly alters HIV-1 fusion through interactions with gp41. Whether this mechanism of HIV-1 neutralization extends to other viruses has not been shown; however, a different mechanism of β- and θ-defensin blockade of enveloped virus fusion due to host cell interactions will be discussed in Antiviral Mechanisms Targeting the Cell.
Rather than fuse with the cell membrane, non-enveloped viruses must penetrate the limiting membrane of the host cell, which is generally mediated by a specific viral protein [90]. This step may occur at the plasma membrane, but it often follows a conformational change leading to uncoating of the viral capsid triggered by a drop in pH or other host factors in the endosomal pathway [91]. For HAdVs, the internal capsid protein VI is the membrane lytic factor [80], [81]. Upon binding to the viral capsid, α-defensins, including HNP1 and HD5, stabilize HAdV-C and prevent uncoating, release of protein VI, and subsequent disruption of the endosomal membrane [53], [77], [79]. To mediate this effect, α-defensins bind directly to the virus, either when mixed together in the absence of cells or when the defensin is added to virus that is pre-bound to receptors on the cell surface [45], [53], [62], [69], [79]. Thus, the defensins can recognize the virus in the complexity of host proteins, carbohydrates, and lipids that could potentially compete with the virus for defensin binding. Neutralization of HAdV by α-defensins is restricted to HAdV-B and HAdV-C, and to HAdV-A and HAdV-E to a lesser extent, whereas HAdV-D and HAdV-F infection is either unaffected or enhanced by α-defensins [45], [53]. Furthermore, resistance to HD5 neutralization can be conferred to HAdV-C through the replacement of capsid vertex proteins with those from the resistant HAdV-D [45]. The basis for enhanced HAdV-D infection is unknown. Moreover, the extent to which the antiviral mechanism against HAdV-C extends to other non-enveloped viruses has not been determined; however, HPV-16 entry is also blocked by α-defensins, possibly at an analogous step [54].
Post-entry neutralization
Infection is not completed by merely penetrating the host cell membrane. Viral transcription, protein production, assembly, and egress must all occur to complete a replicative cycle. These steps present opportunities for defensins to block viral infection, either by targeting the virus specifically or by targeting the cell. In this regard, HNP1 and HD5 are able to block HSV-1 and HSV-2 when added post-entry [44]. The defensins accumulated intracellularly in the human cervical epithelial cell line used in this study, indicating that they can still come in contact with the viruses in the cell. Hazrati et al. demonstrate that HNP2 and HD5 can bind HSV-2 DNA and speculate that this could contribute to inhibition by blocking gene expression, although a post-transcription block by an unknown mechanism was also suggested by the data [44].
Among the non-enveloped viruses sensitive to defensins, inhibition of steps after penetration of the cell membrane is not thought to be important for polyomaviruses or HAdV [53], [55]. For multiple types of papillomaviruses that are sensitive to HNP1-4 and HD5, the only step known to be inhibited is the nuclear localization of the HPV-16 genome, the last step in the virus entry pathway [54]. Unlike the case for HAdV-C neutralization, the HPV genome is exposed under conditions of HNP1 and HD5 neutralization, implying that virus uncoating occurs in the presence of the defensins [53], [54]. Thus, it is unclear if defensins block papillomavirus penetration of the host membrane or the ability of the genome to traverse the cytoplasm to the nucleus.
No mechanism but antiviral activity reported
We lack mechanistic insight for a variety of viruses that have been shown to be sensitive to defensin neutralization. Purified HBD3 and porcine β-defensin 3 were antiviral against porcine reproductive and respiratory syndrome virus [49]. HBD2 has been shown to reduce the yield of varicella zoster virus at 10 days post-infection, but not at earlier time points [92]. Transduction by recombinant adeno-associated virus is inhibited by relatively high concentrations (100 μM) of HNPs; however, ≥ 100 μM HNPs was measured in epithelial lining fluids from cystic fibrosis patients, which were also inhibitory in the same study [36]. Moreover, vaccinia virus is inhibited by HBD3 when incubated with the virus for 24 h [93]. Whether previously described or novel mechanisms contribute to the neutralization of these viruses warrants further study.
In addition to purified peptides, virus inhibition by degranulated neutrophils has been noted. When neutrophils degranulate, for example, in response to stimulation by leukotriene B4 (LTB4), they release high concentrations of defensins and other antimicrobial factors. CMV infection of human peripheral blood leukocytes containing neutrophils is weakly inhibited by LTB4 treatment [94]. A combination of α-defensins (HNP1-3), the human cathelicidin LL37, and eosinophil-derived neurotoxin mediates much of the effect. Cell-free supernatants from LTB4-stimulated neutrophils mixed with CMV were similarly inhibitory, and anti-HNP1-3 antibodies reduced the observed activity by half. These results corroborate the data on modest CMV neutralization using purified α-defensins, although the antiviral mechanism remains unknown [1], [94]. In other studies, supernatants from human neutrophils treated with LTB4 or stimulated to synthesize LTB4 strongly inhibited human coronavirus and HSV-1 and more modestly inhibited RSV and influenza B virus [95], [96]. Collectively, these studies demonstrate that naturally secreted defensins retain antiviral activity. In contrast, HNP1-3 have been shown to interact with and reduce the activity of surfactant protein D and bronchoalveolar lavage fluid containing surfactant protein D against IAV [97], [98]. These observations reinforce the importance of studying both purified peptides for their individual biological effects and more complex mixtures in which defensins and other antimicrobial factors are naturally produced, which could demonstrate synergistic or, alternatively, mutually inhibitory effects.
Antiviral mechanisms targeting the cell
Blocking fusion by cross-linking host proteins
Rather than directly targeting viral fusion proteins to block enveloped virus fusion with the host cell, HBD3 and a synthetic human θ-defensin called retrocyclin 2 have been shown to inhibit IAV fusion by cross-linking host glycoproteins [42]. Retrocyclin 2 and HBD3 binding limit the mobility of host surface proteins in the vicinity of the nascent viral fusion pore, restricting its maturation to full fusion. Inhibition is blocked by serum and by deglycosylation (PNGase treatment) of the cells, indicating that this effect is due to the lectin activity of the defensins. In this regard, a non-defensin mannan-binding lectin had a similar effect. This mechanism may be general, as inhibition is independent of direct binding to the viral hemagglutinin glycoprotein and extends to fusion reactions mediated by unrelated proteins from baculovirus and Sindbis virus. More recently for HIV-1, HNP1 treatment has also been shown to decrease the mobile fraction of CD4, CCR5, and CXCR4 receptors in the cell membrane, reflecting the same phenomenon mediated by the lectin activity of HNP1 [47].
Modulation of cell surface receptors
HIV-1 has a complex entry pathway that utilizes several cell surface receptors and co-receptors. The relative ability of defensins to directly modulate host surface receptors important for HIV-1 infection has been debated by various groups. In peripheral blood mononuclear cells (PBMCs) and a human T cell line expressing CXCR4 and CCR5, HBD2 and HBD3 but not HBD1 reduced cell surface expression of CXCR4 but not CCR5 in 0–0.5% serum [52], [99]. A subsequent study confirmed CXCR4 downregulation in PBMCs by HBD2 and HNP1 under similar conditions [71]. In contrast, the downmodulation of CXCR4 was not observed in PBMCs treated with HBD2 in the presence of serum [61], [72]. Similarly, in primary CD4+ T cells in the presence of 10% serum, HNP1 does not alter CXCR4, CCR5, or CD4 expression on the cell surface [73]. Discrepancies among these studies are likely explained by the use of different cell types, serum concentrations, and experimental conditions. In addition, effects on receptor downregulation do not always correspond with the broad antiviral activities of the defensins against multiple HIV-1 isolates using different co-receptors [100]. Thus, the contribution of changes in cell surface receptor levels to HIV-1 infection remains unresolved.
Changes in intracellular signaling that impact infection
Many viruses regulate protein kinase C (PKC) signaling during entry and infection: HIV-1 requires phosphorylated PKC for viral fusion, transcription, integration, and assembly [73], [101], [102]. IAV requires PKC for endosomal escape and nuclear entry [103], [104]. HSV requires PKC for cell entry and nuclear egress of the viral capsid [105], [106]. As HNP1-3 are known to inhibit the activity of PKC in vitro [107], altering or inhibiting this cellular signaling pathway may be another defensin-mediated antiviral mechanism, which explains the post-entry block to infection observed for some viruses. HNP1 treatment of cells prior to or during infection with either IAV or HIV-1 reduces the levels of phosphorylated PKC [48], [73]. Treatment of CD4+ T cells with a PKC activator, bryostatin-1, partially rescued HIV-1 infection [73]. In addition, HNP1 and the PKC inhibitor Go6976 had similar inhibition kinetics [73]. Inhibition of PKC by HNP1 explains the observed block in nuclear import of the incoming pre-integration complex as well as transcription of the integrated viral genome in HIV-1-infected cells [73]. Although HSV infection is also inhibited by HNP1 through a post-entry mechanism, it is not rescued by pretreatment with bryostatin-1 [44]. In addition, cellular entry of some HAdV serotypes is also sensitive to PKC inhibition [108], [109], [110]; however, differential defensin sensitivity of chimeric HAdVs in which only certain capsid proteins are variable argues against a role for cellular targets such as PKC in HAdV neutralization by α-defensins [45]. Together, these data indicate that the PKC signaling pathway is involved in defensin-mediated neutralization of HIV and possibly IAV but is uninvolved in HSV and HAdV neutralization.
Cell signaling pathways mediated by the chemokine receptor CCR6 also play a role in defensin-mediated HIV inhibition. HBD2 is known to bind CCR6 and has been shown to induce expression of host restriction factor APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G) that has antiviral activity against HIV [100], [111]. Thus, defensins can both inhibit cellular pathways required for viral infection and activate intracellular antiviral mechanisms.
Defensin expression or secretion elicited by viral infection
In addition to their direct antiviral properties, many defensins function as cytokines or chemokines to elaborate an antimicrobial immune response, which could indirectly affect viral pathogenesis. In this regard, a number of viruses have been shown to stimulate defensin expression or secretion, whether or not the elicited defensins are directly antiviral against that virus. This has primarily been observed for β-defensins, which are often inducibly expressed, rather than the constitutively expressed α-defensins [11]. For example, HBD2 and HBD3 mRNAs were upregulated in primary bronchial epithelial cells by human rhinovirus-16 (HRV-16) infection [112]. A subsequent study confirmed the mRNA upregulation and demonstrated increased HBD2 protein production [39]. Both studies found that viral replication was required and that poly(I:C) but not single-stranded RNA could mimic viral infection, implicating a requirement for innate intracellular RNA detection pathways (RIG-I-like receptors) [113]. Consistent with this observation, multiple HRV serotypes that enter via distinct receptors and pathways had similar defensin-inducing properties [39]. Despite potent upregulation of HBD2, direct inactivation of HRV-16 was not observed [39]. IL-17A stimulation of HRV-16-infected bronchial epithelial cells synergistically induces HBD2, although the signaling pathways leading to HBD2 upregulation by these two stimuli only partially overlap [114]. When HBD2 upregulation was monitored in human volunteers following experimental HRV-16 infection, one group found elevated HBD2 message in nasal epithelial scrapings and HBD2 protein in nasal lavage following a 1-day lag from onset of symptoms [39]; however, a second group observed low HBD2 levels and an increase in HBD2 protein in sputum only on days 15–21 post-infection [115]. These discrepancies may reflect differences in the patient samples that were monitored.
For paramyxoviruses, infection of neonatal lambs with ovine parainfluenza virus 3 (PIV3) elevated sheep β-defensin 1 (SBD1) mRNA in the lung 4- to 7-fold compared to saline controls. PIV3 causes a lower respiratory infection in lambs similar to that of human PIV in children, and SBD1 is expressed and distributed similarly to HBD1 [116]. Increased inflammation and PIV3 infection due to concurrent HAdV vector infection led to further increases in SBD1 expression [117]. In contrast, bovine RSV infection of sheep did not significantly affect SBD1 expression [118]. Thus, repair and regeneration during resolution and recovery from inflammation rather than acute inflammation due to viral infection may be more associated with SBD1 upregulation [118]. The direct antiviral activity of SBD1 against these pathogens was not assessed. In studies of RSV infection of human A549 respiratory epithelial cells, both HBD2 message and protein were upregulated in an NF-κB-dependent manner [74]. Increased HBD2 expression resulted from the induction of TNF-α production upon RSV infection and replication. This pathway may be relevant in vivo, as mice challenged intranasally with RSV expressed increased amounts of murine β-defensin (MBD) 4 but not MBD3, both of which are homologs of HBD2 [15], [74]. In these studies, the elicited HBD2 was also shown to be directly antiviral by disrupting virion integrity, as discussed in Antiviral Mechanisms through Direct Interactions between Defensins and Virus. In addition, HNP1-3 levels were elevated in tracheal aspirates of infants infected with RSV during acute illness compared to convalescence [119]. By correcting for total phospholipid content in their samples, the authors suggest that increased neutrophil infiltration alone does not explain the elevated α-defensin levels; rather, increased production may also contribute [119]. These α-defensins likely contribute to the anti-RSV activity of degranulated neutrophils, but the effect of purified HNPs on RSV was not addressed in either study [95], [119].
HIV-1 infection in epithelial cells transcriptionally upregulates HBD2 and HBD3, which neutralize HIV-1, but not HBD1, which does not block HIV-1 infection [52]. In HaCaT human keratinocytes, vaccinia virus infection has been shown to stimulate the expression of HBD3, to which the virus is sensitive [93]. HSV-2 infection in keratinocytes induces HBD1 and HBD4, and HBD1 does not block HSV-2 infection [120]. Furthermore, human PBMCs, plasmacytoid DCs, and purified monocytes increase HBD1 at the RNA and protein level after HSV-1, IAV, and Sendai virus infection [121]. UV-inactivated virus is less stimulatory, indicating a role for replication. Similarly, IAV infection of mice induces expression of MBD3 and MBD4 in the nasosinus, trachea, and lungs and that of MBD1 and MBD2 in the lungs [122], [123]. Overexpression of MBD3 in MDCK cells or addition of recombinant MBD2 during infection inhibits IAV infection in cell culture [123], [124]. In summary, many studies have documented the induction of defensin expression as a consequence of viral infection, and the induced defensins may play both direct and indirect roles in viral pathogenesis.
Importance of defensins in viral pathogenesis in vivo
Most of the work demonstrating the antiviral effect of defensins has been in cell culture. Although there has been one study showing an increase in lethality from IAV infection in an MBD1 knockout mouse, those results are attributed to an increase in inflammation in the knockout mice rather than direct inhibition of viral replication [121]. Similarly, administration of rhesus θ-defensin protected mice from lethal SARS coronavirus challenge without affecting lung viral titers, likely due to a reduction in immunopathology in the treated animals [125]. In fact, there is no example of a direct role for endogenous defensins in blocking virus infection in vivo, in large part due to the lack of a complete defensin knockout animal model. Indirect evidence for the importance of defensins in vivo comes from association studies looking at defensin levels during various viral disease states. For example, patients with atopic dermatitis have reduced cathelicidin and β-defensins and are at greater risk of developing eczema vaccinatum [126], [127], [128], [129]. HIV-1 transmission across the oral epithelium of adults is rare, and the expression of HBD2 and HBD3 in adult but not in fetal oral epithelial cells may contribute to this resistance by inactivating HIV-1 as it crosses the epithelial barrier [130]. Moreover, HIV-1-positive women have lower HNP1-3 levels and lower anti-HSV-2 activity in their cervicovaginal lavage fluid compared to healthy controls [33]. Similarly, cervicovaginal lavage fluid levels of HNP1-3 correlate with the anti-HIV-1 activity of the fluid [34]. Production of HNP1-3 mRNA and protein in monocyte-derived DCs is higher in HIV-1-infected individuals compared to healthy non-infected controls [131]. Moreover, the CD4+ T cell counts in those individuals with higher levels of defensin decreased more slowly than the individuals with lower levels of defensin. α-Defensins are also detectable in the breast milk of HIV-1-infected mothers, although levels vary from approximately 0.1 to 7 nM and are positively correlated with levels of HIV-1 RNA; however, the presence of α-defensins is also positively correlated with higher maternal CD4+ T cell counts and a decrease in the risk of HIV transmission from the breastfeeding mother to her infant [132], [133]. The apparent contradiction between higher α-defensin levels correlating to higher HIV viral titers and yet lower transmission might be explained by the fact that the defensin levels were also correlated with a healthier maternal immune system, as indicated by the increased CD4+ T cell count. Alternatively, HIV-1 infection may be stimulating α-defensin expression and secretion. Thus, α-defensin levels in HIV-1-infected individuals are correlated with control of viral infection, slowed disease progression, and lower vertical transmission. Additional evidence for a contribution of defensins to HIV-1 transmission and disease progression has been reviewed elsewhere [4], [5], [61], [134], [135].
In addition to their direct antimicrobial activity, defensins are able to augment and direct the immune response to viruses in ways that impact the outcome and resolution of infection. The myriad of mechanisms by which defensins can be immunomodulatory has been reviewed elsewhere [136], [137], [138], but for the purposes of this review, we will summarize those activities that are likely relevant for viral infections. Neutrophil and enteric α-defensins have been found to selectively chemoattract different subsets of T lymphocytes, macrophages, and immature DCs [16], [139], [140], [141]. Although the chemokine receptor used by α-defensins has not been identified, the recruitment of these immune cell subsets to the site of viral infection undoubtedly contributes to the outcome of viral infections. For the β-defensins, HBD1-4 induced chemotaxis of macrophages, while HBD1 and HBD2 also induce chemotaxis of immature DCs and memory T cells [111], [142]. This recruitment has been shown to occur via direct binding and activation of the chemokine receptors CCR6 and CCR2 [111], [143]. However, a conflicting paper indicates that although HBD1-4 can all induce the chemotaxis of macrophages, they only weakly recruit DCs [142]. In addition, they found that this effect is independent of CCR6 [142]. Overall, the ability of HBDs to control immune cell chemotaxis and therefore regulate immune responses to viral infections is well established, even if there remains debate about the specific cell subsets that are recruited.
The recruitment of immune cells has been shown to be relevant in vivo, as both recombinant and naturally produced defensins augment adaptive immune responses. Intranasal inoculation of mice with ovalbumin (OVA) in combination with HNP1-3 resulted in increased OVA-specific IgG and IgM titers compared to mice that received OVA alone [144]. However, despite mucosal inoculation, no OVA-specific IgA was observed, indicating that the HNP augmentation may not allow for class switching. A subsequent study co-administered OVA intranasally with HNP1, HNP2, HBD1, or HBD2 individually and found that the predominant IgG isotypes and interleukin profiles were unique to the defensin used, highlighting the complexity of the interaction between defensins and immune cells [145]. Finally, the augmented immune response has been shown to be functionally relevant, as intraperitoneal injection of HNPs enhanced the antibody response to a syngeneic tumor challenge and increased the survival time of mice after tumor challenge [146]. Thus, although not yet shown in the context of an infectious challenge, stimulation of the adaptive immune response by defensins likely contributes to antiviral immunity.
In addition to inducing antibody secretion, direct contact with defensins can activate immune cells and induce cytokine secretion. For neutrophils, HNP1 and HBD3 suppress apoptosis via inhibition of caspase 3, which could help to prolong an immune response [147], [148]. HNP1, HNP2, and HD5 have also been shown to increase neutrophil uptake of IAV [82]. In regard to DCs, exposure to MBD2 matures and activates DCs through direct binding to TLR4, and HBD3 activates myeloid DCs and monocytes via TLR1 and TLR2 [149], [150]. HBD3 can also activate Langerhans cell-like DCs and primary human cutaneous DCs [151]. In addition, HNP1 and HBD1 promote the activation and maturation of monocyte-derived DCs and production of pro-inflammatory cytokines [152]. Defensins can elicit pro-inflammatory cytokine production from treated cells that could then stimulate DCs [8], [153], [154]. Moreover, HNP1 and HNP2 have also been shown to induce CC-chemokine expression and secretion in macrophages, which block HIV infection [155]. Therefore, some of the activation of antigen-expressing cells could be explained through an indirect mechanism.
There is also increasing evidence that defensins can be immunosuppressive. Some defensins, notably HBD1, are constitutively expressed at epithelial surfaces [156]. It has been speculated that this constitutive expression minimizes the effects of exposure to low levels of commensal and pathogenic microbes and allows for the maintenance of a non-inflammatory environment [157]. This idea is supported by the fact that HBD3 can suppress the lipopolysaccharide-induced production of pro-inflammatory cytokines by human and mouse macrophages [158], [159]. It is worth noting that the amount of HBD3 used in these studies (1 μM) is lower than the amount of HBD3 that has been shown to activate macrophages via TLRs or neutralize HIV-1 infection (4 μM) [72], [150], [158]. More recently, naturally expressed and exogenous HBD3 were also shown to reduce the innate immune response to lentiviral vector transduction of muscle cells [160]. Finally, in line with previous observations about the cathelicidin LL37, HBD2 and HBD3 can bind DNA and promote its uptake by plasmacytoid DCs leading to interferon production [161], [162]. Overall, α- and β-defensins function to augment and alter the immune responses to microbes, and the response elicited is likely dependent upon the amount of microbial stimuli and the subsequent concentration of defensin that is produced.
Conclusions and perspectives
In cell culture, both α- and β-defensins have been shown to be potently antiviral against a wide range of viral species. However, there are intriguing differences in viral neutralization when one begins to examine the interactions of individual defensins with viruses, even those from the same genus. Identification of the viral determinants that drive these differences is an area in need of further study. We now appreciate that α-defensins inhibit viruses by numerous mechanisms; however, mechanistic data involving β-defensins are mostly lacking and would add greatly to the understanding of how defensins can impact an immune response in vivo. This is especially true as β-defensins are inducibly produced by epithelial cells in response to microbial stimuli and, therefore, could provide a first line of defense against many infecting viruses. Although we have focused on antiviral mechanisms, enhancement of HIV-1 and HAdV-D, HAdV-E, and HAdV-F infection by defensins has been observed. Whether this enhancement extends to other viruses and the importance of this enhancement for infection needs to be explored. Finally, the in vivo relevance of direct defensin killing and enhancement is unclear. A lack of complete defensin knockout models hinders these studies, which need to be addressed in order to fully understand the role of defensins in viral pathogenesis. There is great interest in the potential of defensins as vaccine adjuvants and as templates for the design of novel antiviral treatments [163], [164], [165]. However, for these applications to be both safe and efficacious, a greater understanding of the complex roles of defensins in viral infection is needed.
Acknowledgements
S.S.W. and M.E.W. were supported in part by a Public Health Service, National Research Service Award, T32 GM07270, from the National Institute of General Medical Sciences. S.S.W. was supported in part by a Public Health Service, National Research Service Award, T32 AI083203, from the National Institute of Allergy and Infectious Diseases.
Edited by Eric Freed and Michael Gale
References
- 1.Daher K.A., Selsted M.E., Lehrer R.I. Direct inactivation of viruses by human granulocyte defensins. J Virol. 1986;60:1068–1074. doi: 10.1128/jvi.60.3.1068-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehrer R.I., Daher K., Ganz T., Selsted M.E. Direct inactivation of viruses by MCP-1 and MCP-2, natural peptide antibiotics from rabbit leukocytes. J Virol. 1985;54:467–472. doi: 10.1128/jvi.54.2.467-472.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganz T., Selsted M.E., Szklarek D., Harwig S.S., Daher K., Bainton D.F. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah R., Chang T.L. Vol. 1095. American Chemical Society; Washington: 2012. Defensins in viral infection; pp. 137–171. (Small wonders: peptides for disease control). [Google Scholar]
- 5.Klotman M.E., Chang T.L. Defensins in innate antiviral immunity. Nat Rev Immunol. 2006;6:447–456. doi: 10.1038/nri1860. [DOI] [PubMed] [Google Scholar]
- 6.Ding J., Chou Y.Y., Chang T.L. Defensins in viral infections. J Innate Immun. 2009;1:413–420. doi: 10.1159/000226256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gwyer Findlay E., Currie S.M., Davidson D.J. Cationic host defence peptides: potential as antiviral therapeutics. BioDrugs. 2013;27:479–493. doi: 10.1007/s40259-013-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrer R.I., Lu W. alpha-Defensins in human innate immunity. Immunol Rev. 2012;245:84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 9.Hazlett L., Wu M. Defensins in innate immunity. Cell Tissue Res. 2011;343:175–188. doi: 10.1007/s00441-010-1022-4. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer R.I. Multispecific myeloid defensins. Curr Opin Hematol. 2007;14:16–21. doi: 10.1097/00062752-200701000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Selsted M.E., Ouellette A.J. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 12.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 13.Pazgier M., Li X., Lu W., Lubkowski J. Human defensins: synthesis and structural properties. Curr Pharm Des. 2007;13:3096–3118. doi: 10.2174/138161207782110381. [DOI] [PubMed] [Google Scholar]
- 14.Penberthy W.T., Chari S., Cole A.L., Cole A.M. Retrocyclins and their activity against HIV-1. Cell Mol Life Sci. 2011;68:2231–2242. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schutte B.C., Mitros J.P., Bartlett J.A., Walters J.D., Jia H.P., Welsh M.J. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA. 2002;99:2129–2133. doi: 10.1073/pnas.042692699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill C.P., Yee J., Selsted M.E., Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 18.Pardi A., Zhang X.L., Selsted M.E., Skalicky J.J., Yip P.F. NMR studies of defensin antimicrobial peptides. 2. Three-dimensional structures of rabbit NP-2 and human HNP-1. Biochemistry. 1992;31:11357–11364. doi: 10.1021/bi00161a013. [DOI] [PubMed] [Google Scholar]
- 19.Hristova K., Selsted M.E., White S.H. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry. 1996;35:11888–11894. doi: 10.1021/bi961100d. [DOI] [PubMed] [Google Scholar]
- 20.Ouellette A.J. Paneth cell alpha-defensins in enteric innate immunity. Cell Mol Life Sci. 2011;68:2215–2229. doi: 10.1007/s00018-011-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bevins C.L., Salzman N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 22.Spencer J.D., Hains D.S., Porter E., Bevins C.L., DiRosario J., Becknell B. Human alpha defensin 5 expression in the human kidney and urinary tract. PLoS One. 2012;7:e31712. doi: 10.1371/journal.pone.0031712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quayle A.J., Porter E.M., Nussbaum A.A., Wang Y.M., Brabec C., Yip K.P. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am J Pathol. 1998;152:1247–1258. [PMC free article] [PubMed] [Google Scholar]
- 24.Com E., Bourgeon F., Evrard B., Ganz T., Colleu D., Jegou B. Expression of antimicrobial defensins in the male reproductive tract of rats, mice, and humans. Biol Reprod. 2003;68:95–104. doi: 10.1095/biolreprod.102.005389. [DOI] [PubMed] [Google Scholar]
- 25.Svinarich D.M., Wolf N.A., Gomez R., Gonik B., Romero R. Detection of human defensin 5 in reproductive tissues. Am J Obstet Gynecol. 1997;176:470–475. doi: 10.1016/s0002-9378(97)70517-9. [DOI] [PubMed] [Google Scholar]
- 26.Pazgier M., Hoover D.M., Yang D., Lu W., Lubkowski J. Human beta-defensins. Cell Mol Life Sci. 2006;63:1294–1313. doi: 10.1007/s00018-005-5540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schibli D.J., Hunter H.N., Aseyev V., Starner T.D., Wiencek J.M., McCray P.B. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J Biol Chem. 2002;277:8279–8289. doi: 10.1074/jbc.M108830200. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi Y., Nagase T., Makita R., Fukuhara S., Tomita T., Tominaga T. Identification of multiple novel epididymis-specific beta-defensin isoforms in humans and mice. J Immunol. 2002;169:2516–2523. doi: 10.4049/jimmunol.169.5.2516. [DOI] [PubMed] [Google Scholar]
- 29.Shanahan M.T., Tanabe H., Ouellette A.J. Strain-specific polymorphisms in Paneth cell alpha-defensins of C57BL/6 mice and evidence of vestigial myeloid alpha-defensin pseudogenes. Infect Immun. 2011;79:459–473. doi: 10.1128/IAI.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenhauer P.B., Lehrer R.I. Mouse neutrophils lack defensins. Infect Immun. 1992;60:3446–3447. doi: 10.1128/iai.60.8.3446-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayabe T., Satchell D.P., Wilson C.L., Parks W.C., Selsted M.E., Ouellette A.J. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 32.Wehkamp J., Chu H., Shen B., Feathers R.W., Kays R.J., Lee S.K. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 2006;580:5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 33.John M., Keller M.J., Fam E.H., Cheshenko N., Hogarty K., Kasowitz A. Cervicovaginal secretions contribute to innate resistance to herpes simplex virus infection. J Infect Dis. 2005;192:1731–1740. doi: 10.1086/497168. [DOI] [PubMed] [Google Scholar]
- 34.Levinson P., Kaul R., Kimani J., Ngugi E., Moses S., MacDonald K.S. Levels of innate immune factors in genital fluids: association of alpha defensins and LL-37 with genital infections and increased HIV acquisition. AIDS. 2009;23:309–317. doi: 10.1097/QAD.0b013e328321809c. [DOI] [PubMed] [Google Scholar]
- 35.Fan S.R., Liu X.P., Liao Q.P. Human defensins and cytokines in vaginal lavage fluid of women with bacterial vaginosis. Int J Gynaecol Obstet. 2008;103:50–54. doi: 10.1016/j.ijgo.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Virella-Lowell I., Poirier A., Chesnut K.A., Brantly M., Flotte T.R. Inhibition of recombinant adeno-associated virus (rAAV) transduction by bronchial secretions from cystic fibrosis patients. Gene Ther. 2000;7:1783–1789. doi: 10.1038/sj.gt.3301268. [DOI] [PubMed] [Google Scholar]
- 37.Cole A.M., Liao H.I., Stuchlik O., Tilan J., Pohl J., Ganz T. Cationic polypeptides are required for antibacterial activity of human airway fluid. J Immunol. 2002;169:6985–6991. doi: 10.4049/jimmunol.169.12.6985. [DOI] [PubMed] [Google Scholar]
- 38.Gardner M.S., Rowland M.D., Siu A.Y., Bundy J.L., Wagener D.K., Stephenson J.L. Comprehensive defensin assay for saliva. Anal Chem. 2009;81:557–566. doi: 10.1021/ac801609r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proud D., Sanders S.P., Wiehler S. Human rhinovirus infection induces airway epithelial cell production of human beta-defensin 2 both in vitro and in vivo. J Immunol. 2004;172:4637–4645. doi: 10.4049/jimmunol.172.7.4637. [DOI] [PubMed] [Google Scholar]
- 40.Wimley W.C., Selsted M.E., White S.H. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehrer R.I., Jung G., Ruchala P., Andre S., Gabius H.J., Lu W. Multivalent binding of carbohydrates by the human alpha-defensin, HD5. J Immunol. 2009;183:480–490. doi: 10.4049/jimmunol.0900244. [DOI] [PubMed] [Google Scholar]
- 42.Leikina E., Delanoe-Ayari H., Melikov K., Cho M.S., Chen A., Waring A.J. Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nat Immunol. 2005;6:995–1001. doi: 10.1038/ni1248. [DOI] [PubMed] [Google Scholar]
- 43.Wang W., Owen S.M., Rudolph D.L., Cole A.M., Hong T., Waring A.J. Activity of alpha- and theta-defensins against primary isolates of HIV-1. J Immunol. 2004;173:515–520. doi: 10.4049/jimmunol.173.1.515. [DOI] [PubMed] [Google Scholar]
- 44.Hazrati E., Galen B., Lu W., Wang W., Ouyang Y., Keller M.J. Human alpha- and beta-defensins block multiple steps in herpes simplex virus infection. J Immunol. 2006;177:8658–8666. doi: 10.4049/jimmunol.177.12.8658. [DOI] [PubMed] [Google Scholar]
- 45.Smith J.G., Silvestry M., Lindert S., Lu W., Nemerow G.R., Stewart P.L. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 2010;6:e1000959. doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rapista A., Ding J., Benito B., Lo Y.T., Neiditch M.B., Lu W. Human defensins 5 and 6 enhance HIV-1 infectivity through promoting HIV attachment. Retrovirology. 2011;8:45. doi: 10.1186/1742-4690-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demirkhanyan L.H., Marin M., Padilla-Parra S., Zhan C., Miyauchi K., Jean-Baptiste M. Multifaceted mechanisms of HIV-1 entry inhibition by human alpha-defensin. J Biol Chem. 2012;287:28821–28838. doi: 10.1074/jbc.M112.375949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvatore M., Garcia-Sastre A., Ruchala P., Lehrer R.I., Chang T., Klotman M.E. alpha-Defensin inhibits influenza virus replication by cell-mediated mechanism(s) J Infect Dis. 2007;196:835–843. doi: 10.1086/521027. [DOI] [PubMed] [Google Scholar]
- 49.Sang Y., Ruchala P., Lehrer R.I., Ross C.R., Rowland R.R., Blecha F. Antimicrobial host defense peptides in an arteriviral infection: differential peptide expression and virus inactivation. Viral Immunol. 2009;22:235–242. doi: 10.1089/vim.2009.0005. [DOI] [PubMed] [Google Scholar]
- 50.Scudiero O., Galdiero S., Cantisani M., Di Noto R., Vitiello M., Galdiero M. Novel synthetic, salt-resistant analogs of human beta-defensins 1 and 3 endowed with enhanced antimicrobial activity. Antimicrob Agents Chemother. 2010;54:2312–2322. doi: 10.1128/AAC.01550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mackewicz C.E., Yuan J., Tran P., Diaz L., Mack E., Selsted M.E. alpha-Defensins can have anti-HIV activity but are not CD8 cell anti-HIV factors. AIDS. 2003;17:F23–F32. doi: 10.1097/00002030-200309260-00001. [DOI] [PubMed] [Google Scholar]
- 52.Quinones-Mateu M.E., Lederman M.M., Feng Z., Chakraborty B., Weber J., Rangel H.R. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS. 2003;17:F39–F48. doi: 10.1097/00002030-200311070-00001. [DOI] [PubMed] [Google Scholar]
- 53.Smith J.G., Nemerow G.R. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe. 2008;3:11–19. doi: 10.1016/j.chom.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Buck C.B., Day P.M., Thompson C.D., Lubkowski J., Lu W., Lowy D.R. Human alpha-defensins block papillomavirus infection. Proc Natl Acad Sci USA. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dugan A.S., Maginnis M.S., Jordan J.A., Gasparovic M.L., Manley K., Page R. Human alpha-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J Biol Chem. 2008;283:31125–31132. doi: 10.1074/jbc.M805902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding J., Tasker C., Valere K., Sihvonen T., Descalzi-Montoya D.B., Lu W., Chang T.L. Anti-HIV activity of human defensin 5 in primary CD4+ T cells under serum-deprived conditions is a consequence of defensin-mediated cytotoxicity. PLoS One. 2013;8:e76038. doi: 10.1371/journal.pone.0076038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furci L., Tolazzi M., Sironi F., Vassena L., Lusso P. Inhibition of HIV-1 infection by human alpha-defensin-5, a natural antimicrobial peptide expressed in the genital and intestinal mucosae. PLoS One. 2012;7:e45208. doi: 10.1371/journal.pone.0045208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu W., de Leeuw E. Pro-inflammatory and pro-apoptotic properties of human defensin 5. Biochem Biophys Res Commun. 2013;436:557–562. doi: 10.1016/j.bbrc.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harder J., Bartels J., Christophers E., Schroder J.M. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 60.Weinberg A., Quinones-Mateu M.E., Lederman M.M. Role of human beta-defensins in HIV infection. Adv Dent Res. 2006;19:42–48. doi: 10.1177/154407370601900109. [DOI] [PubMed] [Google Scholar]
- 61.Garzino-Demo A. Chemokines and defensins as HIV suppressive factors: an evolving story. Curr Pharm Des. 2007;13:163–172. doi: 10.2174/138161207779313696. [DOI] [PubMed] [Google Scholar]
- 62.Gounder A.P., Wiens M.E., Wilson S.S., Lu W., Smith J.G. Critical determinants of human alpha-defensin 5 activity against non-enveloped viruses. J Biol Chem. 2012;287:24554–24562. doi: 10.1074/jbc.M112.354068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie C., Prahl A., Ericksen B., Wu Z., Zeng P., Li X. Reconstruction of the conserved beta-bulge in mammalian defensins using d-amino acids. J Biol Chem. 2005;280:32921–32929. doi: 10.1074/jbc.M503084200. [DOI] [PubMed] [Google Scholar]
- 64.Klotman M.E., Rapista A., Teleshova N., Micsenyi A., Jarvis G.A., Lu W. Neisseria gonorrhoeae-induced human defensins 5 and 6 increase HIV infectivity: role in enhanced transmission. J Immunol. 2008;180:6176–6185. doi: 10.4049/jimmunol.180.9.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pazgier M., Wei G., Ericksen B., Jung G., Wu Z., de Leeuw E. Sometimes it takes two to tango: contributions of dimerization to functions of human alpha-defensin HNP1 peptide. J Biol Chem. 2012;287:8944–8953. doi: 10.1074/jbc.M111.332205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei G., Pazgier M., de Leeuw E., Rajabi M., Li J., Zou G. Trp-26 imparts functional versatility to human alpha-defensin HNP1. J Biol Chem. 2010;285:16275–16285. doi: 10.1074/jbc.M110.102749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang A., Chen F., Wang Y., Shen M., Xu Y., Hu J. Enhancement of antiviral activity of human alpha-defensin 5 against herpes simplex virus 2 by arginine mutagenesis at adaptive evolution sites. J Virol. 2013;87:2835–2845. doi: 10.1128/JVI.02209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furci L., Sironi F., Tolazzi M., Vassena L., Lusso P. alpha-Defensins block the early steps of HIV-1 infection: interference with the binding of gp120 to CD4. Blood. 2007;109:2928–2935. doi: 10.1182/blood-2006-05-024489. [DOI] [PubMed] [Google Scholar]
- 69.Flatt J.W., Kim R., Smith J.G., Nemerow G.R., Stewart P.L. An intrinsically disordered region of the adenovirus capsid is implicated in neutralization by human alpha defensin 5. PLoS One. 2013;8:e61571. doi: 10.1371/journal.pone.0061571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakashima H., Yamamoto N., Masuda M., Fujii N. Defensins inhibit HIV replication in vitro. AIDS. 1993;7:1129. doi: 10.1097/00002030-199308000-00019. [DOI] [PubMed] [Google Scholar]
- 71.Seidel A., Ye Y., de Armas L.R., Soto M., Yarosh W., Marcsisin R.A. Cyclic and acyclic defensins inhibit human immunodeficiency virus type-1 replication by different mechanisms. PLoS One. 2010;5:e9737. doi: 10.1371/journal.pone.0009737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun L., Finnegan C.M., Kish-Catalone T., Blumenthal R., Garzino-Demo P., La Terra Maggiore G.M. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol. 2005;79:14318–14329. doi: 10.1128/JVI.79.22.14318-14329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang T.L., Vargas J., DelPortillo A., Klotman M.E. Dual role of alpha-defensin-1 in anti-HIV-1 innate immunity. J Clin Invest. 2005;115:765–773. doi: 10.1172/JCI200521948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kota S., Sabbah A., Chang T.H., Harnack R., Xiang Y., Meng X. Role of human beta-defensin-2 during tumor necrosis factor-alpha/NF-kappaB-mediated innate antiviral response against human respiratory syncytial virus. J Biol Chem. 2008;283:22417–22429. doi: 10.1074/jbc.M710415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujii G., Selsted M.E., Eisenberg D. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. 1993;2:1301–1312. doi: 10.1002/pro.5560020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nayak D.P., Hui E.K. The role of lipid microdomains in virus biology. Subcell Biochem. 2004;37:443–491. doi: 10.1007/978-1-4757-5806-1_14. [DOI] [PubMed] [Google Scholar]
- 77.Snijder J., Reddy V.S., May E.R., Roos W.H., Nemerow G.R., Wuite G.J. Integrin and defensin modulate the mechanical properties of adenovirus. J Virol. 2013;87:2756–2766. doi: 10.1128/JVI.02516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Berna A.J., Ortega-Esteban A., Menendez-Conejero R., Winkler D.C., Menendez M., Steven A.C. The role of capsid maturation on adenovirus priming for sequential uncoating. J Biol Chem. 2012;287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen E.K., Nemerow G.R., Smith J.G. Direct evidence from single-cell analysis that human alpha-defensins block adenovirus uncoating to neutralize infection. J Virol. 2010;84:4041–4049. doi: 10.1128/JVI.02471-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moyer C.L., Wiethoff C.M., Maier O., Smith J.G., Nemerow G.R. Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J Virol. 2011;85:2631–2641. doi: 10.1128/JVI.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiethoff C.M., Wodrich H., Gerace L., Nemerow G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tecle T., White M.R., Gantz D., Crouch E.C., Hartshorn K.L. Human neutrophil defensins increase neutrophil uptake of influenza a virus and bacteria and modify virus-induced respiratory burst responses. J Immunol. 2007;178:8046–8052. doi: 10.4049/jimmunol.178.12.8046. [DOI] [PubMed] [Google Scholar]
- 83.Yasin B., Wang W., Pang M., Cheshenko N., Hong T., Waring A.J. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J Virol. 2004;78:5147–5156. doi: 10.1128/JVI.78.10.5147-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Z., Cocchi F., Gentles D., Ericksen B., Lubkowski J., Devico A. Human neutrophil alpha-defensin 4 inhibits HIV-1 infection in vitro. FEBS Lett. 2005;579:162–166. doi: 10.1016/j.febslet.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 85.Wang C.H., Chan L.W., Johnson R.N., Chu D.S., Shi J., Schellinger J.G. The transduction of coxsackie and adenovirus receptor-negative cells and protection against neutralizing antibodies by HPMA-co-oligolysine copolymer-coated adenovirus. Biomaterials. 2011;32:9536–9545. doi: 10.1016/j.biomaterials.2011.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harrison S.C. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weissenhorn W., Hinz A., Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;15:690–698. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klasse P.J. Vol. 14. 2012. The molecular basis of HIV entry; pp. 1183–1192. (Cell Microbiol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Demirkhanyan L., Marin M., Lu W., Melikyan G.B. Sub-inhibitory concentrations of human alpha-defensin potentiate neutralizing antibodies against HIV-1 gp41 pre-hairpin intermediates in the presence of serum. PLoS Pathog. 2013;9:e1003431. doi: 10.1371/journal.ppat.1003431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 91.Mercer J., Schelhaas M., Helenius A. Virus entry by endocytosis. Annu Rev Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 92.Crack L.R., Jones L., Malavige G.N., Patel V., Ogg G.S. Human antimicrobial peptides LL-37 and human beta-defensin-2 reduce viral replication in keratinocytes infected with varicella zoster virus. Clin Exp Dermatol. 2012;37:534–543. doi: 10.1111/j.1365-2230.2012.04305.x. [DOI] [PubMed] [Google Scholar]
- 93.Howell M.D., Streib J.E., Leung D.Y. Antiviral activity of human beta-defensin 3 against vaccinia virus. J Allergy Clin Immunol. 2007;119:1022–1025. doi: 10.1016/j.jaci.2007.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaudreault E., Gosselin J. Leukotriene B4-mediated release of antimicrobial peptides against cytomegalovirus is BLT1 dependent. Viral Immunol. 2007;20:407–420. doi: 10.1089/vim.2006.0099. [DOI] [PubMed] [Google Scholar]
- 95.Widegren H., Andersson M., Borgeat P., Flamand L., Johnston S., Greiff L. LTB4 increases nasal neutrophil activity and conditions neutrophils to exert antiviral effects. Respir Med. 2011;105:997–1006. doi: 10.1016/j.rmed.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chouinard F., Turcotte C., Guan X., Larose M.C., Poirier S., Bouchard L. 2-Arachidonoyl-glycerol- and arachidonic acid-stimulated neutrophils release antimicrobial effectors against E. coli, S. aureus, HSV-1, and RSV. J Leukoc Biol. 2013;93:267–276. doi: 10.1189/jlb.0412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hartshorn K.L., White M.R., Tecle T., Holmskov U., Crouch E.C. Innate defense against influenza A virus: activity of human neutrophil defensins and interactions of defensins with surfactant protein D. J Immunol. 2006;176:6962–6972. doi: 10.4049/jimmunol.176.11.6962. [DOI] [PubMed] [Google Scholar]
- 98.Doss M., White M.R., Tecle T., Gantz D., Crouch E.C., Jung G. Interactions of alpha-, beta-, and theta-defensins with influenza A virus and surfactant protein D. J Immunol. 2009;182:7878–7887. doi: 10.4049/jimmunol.0804049. [DOI] [PubMed] [Google Scholar]
- 99.Feng Z., Dubyak G.R., Lederman M.M., Weinberg A. Cutting edge: human beta defensin 3—a novel antagonist of the HIV-1 coreceptor CXCR4. J Immunol. 2006;177:782–786. doi: 10.4049/jimmunol.177.2.782. [DOI] [PubMed] [Google Scholar]
- 100.Lafferty M.K., Sun L., DeMasi L., Lu W., Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. 2010;115:1564–1571. doi: 10.1182/blood-2009-06-226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu G., Shen F.S., Sturch S., Aquino A., Glazer R.I., Felsted R.L. Regulation of HIV-1 gag protein subcellular targeting by protein kinase C. J Biol Chem. 1995;270:4792–4796. doi: 10.1074/jbc.270.9.4792. [DOI] [PubMed] [Google Scholar]
- 102.Contreras X., Mzoughi O., Gaston F., Peterlin M.B., Bahraoui E. Protein kinase C-delta regulates HIV-1 replication at an early post-entry step in macrophages. Retrovirology. 2012;9:37. doi: 10.1186/1742-4690-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sieczkarski S.B., Brown H.A., Whittaker G.R. Role of protein kinase C betaII in influenza virus entry via late endosomes. J Virol. 2003;77:460–469. doi: 10.1128/JVI.77.1.460-469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Root C.N., Wills E.G., McNair L.L., Whittaker G.R. Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J Gen Virol. 2000;81:2697–2705. doi: 10.1099/0022-1317-81-11-2697. [DOI] [PubMed] [Google Scholar]
- 105.Constantinescu S.N., Cernescu C.D., Popescu L.M. Effects of protein kinase C inhibitors on viral entry and infectivity. FEBS Lett. 1991;292:31–33. doi: 10.1016/0014-5793(91)80826-o. [DOI] [PubMed] [Google Scholar]
- 106.Leach N.R., Roller R.J. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology. 2010;406:127–137. doi: 10.1016/j.virol.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Charp P.A., Rice W.G., Raynor R.L., Reimund E., Kinkade J.M., Ganz T. Inhibition of protein kinase C by defensins, antibiotic peptides from human neutrophils. Biochem Pharmacol. 1988;37:951–956. doi: 10.1016/0006-2952(88)90187-6. [DOI] [PubMed] [Google Scholar]
- 108.Amstutz B., Gastaldelli M., Kalin S., Imelli N., Boucke K., Wandeler E. Subversion of CtBP1-controlled macropinocytosis by human adenovirus serotype 3. EMBO J. 2008;27:956–969. doi: 10.1038/emboj.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kalin S., Amstutz B., Gastaldelli M., Wolfrum N., Boucke K., Havenga M. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J Virol. 2010;84:5336–5350. doi: 10.1128/JVI.02494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakano M.Y., Boucke K., Suomalainen M., Stidwill R.P., Greber U.F. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J Virol. 2000;74:7085–7095. doi: 10.1128/jvi.74.15.7085-7095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang D., Chertov O., Bykovskaia S.N., Chen Q., Buffo M.J., Shogan J. beta-Defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 112.Duits L.A., Nibbering P.H., van Strijen E., Vos J.B., Mannesse-Lazeroms S.P., van Sterkenburg M.A. Rhinovirus increases human beta-defensin-2 and -3 mRNA expression in cultured bronchial epithelial cells. FEMS Immunol Med Microbiol. 2003;38:59–64. doi: 10.1016/S0928-8244(03)00106-8. [DOI] [PubMed] [Google Scholar]
- 113.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wiehler S., Proud D. Interleukin-17A modulates human airway epithelial responses to human rhinovirus infection. Am J Physiol Lung Cell Mol Physiol. 2007;293:L505–L515. doi: 10.1152/ajplung.00066.2007. [DOI] [PubMed] [Google Scholar]
- 115.Mallia P., Footitt J., Sotero R., Jepson A., Contoli M., Trujillo-Torralbo M.B. Rhinovirus infection induces degradation of antimicrobial peptides and secondary bacterial infection in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186:1117–1124. doi: 10.1164/rccm.201205-0806OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grubor B., Gallup J.M., Meyerholz D.K., Crouch E.C., Evans R.B., Brogden K.A. Enhanced surfactant protein and defensin mRNA levels and reduced viral replication during parainfluenza virus type 3 pneumonia in neonatal lambs. Clin Diagn Lab Immunol. 2004;11:599–607. doi: 10.1128/CDLI.11.3.599-607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meyerholz D.K., Grubor B., Gallup J.M., Lehmkuhl H.D., Anderson R.D., Lazic T. Adenovirus-mediated gene therapy enhances parainfluenza virus 3 infection in neonatal lambs. J Clin Microbiol. 2004;42:4780–4787. doi: 10.1128/JCM.42.10.4780-4787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kawashima K., Meyerholz D.K., Gallup J.M., Grubor B., Lazic T., Lehmkuhl H.D. Differential expression of ovine innate immune genes by preterm and neonatal lung epithelia infected with respiratory syncytial virus. Viral Immunol. 2006;19:316–323. doi: 10.1089/vim.2006.19.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thompson L., Turko I., Murad F. Mass spectrometry-based relative quantification of human neutrophil peptides 1, 2, and 3 from biological samples. Mol Immunol. 2006;43:1485–1489. doi: 10.1016/j.molimm.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 120.Shao Y., Zhang W., Dong X., Liu W., Zhang C., Zhang J. Keratinocytes play a role in the immunity to herpes simplex virus type 2 infection. Acta Virol. 2010;54:261–267. doi: 10.4149/av_2010_04_261. [DOI] [PubMed] [Google Scholar]
- 121.Ryan L.K., Dai J., Yin Z., Megjugorac N., Uhlhorn V., Yim S. Modulation of human beta-defensin-1 (hBD-1) in plasmacytoid dendritic cells (PDC), monocytes, and epithelial cells by influenza virus, Herpes simplex virus, and Sendai virus and its possible role in innate immunity. J Leukoc Biol. 2011;90:343–356. doi: 10.1189/jlb.0209079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chong K.T., Thangavel R.R., Tang X. Enhanced expression of murine beta-defensins (MBD-1, -2,- 3, and -4) in upper and lower airway mucosa of influenza virus infected mice. Virology. 2008;380:136–143. doi: 10.1016/j.virol.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 123.Jiang Y., Wang Y., Kuang Y., Wang B., Li W., Gong T. Expression of mouse beta-defensin-3 in MDCK cells and its anti-influenza-virus activity. Arch Virol. 2009;154:639–647. doi: 10.1007/s00705-009-0352-6. [DOI] [PubMed] [Google Scholar]
- 124.Gong T., Jiang Y., Wang Y., Yang D., Li W., Zhang Q. Recombinant mouse beta-defensin 2 inhibits infection by influenza A virus by blocking its entry. Arch Virol. 2010;155:491–498. doi: 10.1007/s00705-010-0608-1. [DOI] [PubMed] [Google Scholar]
- 125.Wohlford-Lenane C.L., Meyerholz D.K., Perlman S., Zhou H., Tran D., Selsted M.E. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J Virol. 2009;83:11385–11390. doi: 10.1128/JVI.01363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Howell M.D. The role of human beta defensins and cathelicidins in atopic dermatitis. Curr Opin Allergy Clin Immunol. 2007;7:413–417. doi: 10.1097/ACI.0b013e3282a64343. [DOI] [PubMed] [Google Scholar]
- 127.Howell M.D., Boguniewicz M., Pastore S., Novak N., Bieber T., Girolomoni G. Mechanism of HBD-3 deficiency in atopic dermatitis. Clin Immunol. 2006;121:332–338. doi: 10.1016/j.clim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 128.Howell M.D., Gallo R.L., Boguniewicz M., Jones J.F., Wong C., Streib J.E. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 129.Howell M.D., Jones J.F., Kisich K.O., Streib J.E., Gallo R.L., Leung D.Y. Selective killing of vaccinia virus by LL-37: implications for eczema vaccinatum. J Immunol. 2004;172:1763–1767. doi: 10.4049/jimmunol.172.3.1763. [DOI] [PubMed] [Google Scholar]
- 130.Tugizov S.M., Herrera R., Veluppillai P., Greenspan D., Soros V., Greene W.C. HIV is inactivated after transepithelial migration via adult oral epithelial cells but not fetal epithelial cells. Virology. 2011;409:211–222. doi: 10.1016/j.virol.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rodriguez-Garcia M., Climent N., Oliva H., Casanova V., Franco R., Leon A. Increased alpha-defensins 1–3 production by dendritic cells in HIV-infected individuals is associated with slower disease progression. PLoS One. 2010;5:e9436. doi: 10.1371/journal.pone.0009436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kuhn L., Trabattoni D., Kankasa C., Semrau K., Kasonde P., Lissoni F. alpha-Defensins in the prevention of HIV transmission among breastfed infants. J Acquir Immune Defic Syndr. 2005;39:138–142. [PMC free article] [PubMed] [Google Scholar]
- 133.Bosire R., John-Stewart G.C., Mabuka J.M., Wariua G., Gichuhi C., Wamalwa D. Breast milk alpha-defensins are associated with HIV type 1 RNA and CC chemokines in breast milk but not vertical HIV type 1 transmission. AIDS Res Hum Retroviruses. 2007;23:198–203. doi: 10.1089/aid.2006.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cole A.M., Cole A.L. Antimicrobial polypeptides are key anti-HIV-1 effector molecules of cervicovaginal host defense. Am J Reprod Immunol. 2008;59:27–34. doi: 10.1111/j.1600-0897.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 135.Iqbal S.M., Kaul R. Mucosal innate immunity as a determinant of HIV susceptibility. Am J Reprod Immunol. 2008;59:44–54. doi: 10.1111/j.1600-0897.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- 136.Bowdish D.M., Davidson D.J., Hancock R.E. Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol. 2006;306:27–66. doi: 10.1007/3-540-29916-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rehaume L.M., Hancock R.E. Neutrophil-derived defensins as modulators of innate immune function. Crit Rev Immunol. 2008;28:185–200. doi: 10.1615/critrevimmunol.v28.i3.10. [DOI] [PubMed] [Google Scholar]
- 138.Yang D., Biragyn A., Kwak L.W., Oppenheim J.J. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 139.Grigat J., Soruri A., Forssmann U., Riggert J., Zwirner J. Chemoattraction of macrophages, T lymphocytes, and mast cells is evolutionarily conserved within the human alpha-defensin family. J Immunol. 2007;179:3958–3965. doi: 10.4049/jimmunol.179.6.3958. [DOI] [PubMed] [Google Scholar]
- 140.Ito T., Tanabe H., Ayabe T., Ishikawa C., Inaba Y., Maemoto A. Paneth cells regulate both chemotaxis of immature dendritic cells and cytokine production from epithelial cells. Tohoku J Exp Med. 2012;227:39–48. doi: 10.1620/tjem.227.39. [DOI] [PubMed] [Google Scholar]
- 141.Yang D., Chen Q., Chertov O., Oppenheim J.J. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J Leukoc Biol. 2000;68:9–14. [PubMed] [Google Scholar]
- 142.Soruri A., Grigat J., Forssmann U., Riggert J., Zwirner J. beta-Defensins chemoattract macrophages and mast cells but not lymphocytes and dendritic cells: CCR6 is not involved. Eur J Immunol. 2007;37:2474–2486. doi: 10.1002/eji.200737292. [DOI] [PubMed] [Google Scholar]
- 143.Rohrl J., Yang D., Oppenheim J.J., Hehlgans T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J Immunol. 2010;184:6688–6694. doi: 10.4049/jimmunol.0903984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lillard J.W., Boyaka P.N., Chertov O., Oppenheim J.J., McGhee J.R. Mechanisms for induction of acquired host immunity by neutrophil peptide defensins. Proc Natl Acad Sci USA. 1999;96:651–656. doi: 10.1073/pnas.96.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brogden K.A., Heidari M., Sacco R.E., Palmquist D., Guthmiller J.M., Johnson G.K. Defensin-induced adaptive immunity in mice and its potential in preventing periodontal disease. Oral Microbiol Immunol. 2003;18:95–99. doi: 10.1034/j.1399-302x.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 146.Tani K., Murphy W.J., Chertov O., Salcedo R., Koh C.Y., Utsunomiya I. Defensins act as potent adjuvants that promote cellular and humoral immune responses in mice to a lymphoma idiotype and carrier antigens. Int Immunol. 2000;12:691–700. doi: 10.1093/intimm/12.5.691. [DOI] [PubMed] [Google Scholar]
- 147.Nagaoka I., Suzuki K., Murakami T., Niyonsaba F., Tamura H., Hirata M. Evaluation of the effect of alpha-defensin human neutrophil peptides on neutrophil apoptosis. Int J Mol Med. 2010;26:925–934. doi: 10.3892/ijmm_00000544. [DOI] [PubMed] [Google Scholar]
- 148.Nagaoka I., Niyonsaba F., Tsutsumi-Ishii Y., Tamura H., Hirata M. Evaluation of the effect of human beta-defensins on neutrophil apoptosis. Int Immunol. 2008;20:543–553. doi: 10.1093/intimm/dxn012. [DOI] [PubMed] [Google Scholar]
- 149.Biragyn A., Ruffini P.A., Leifer C.A., Klyushnenkova E., Shakhov A., Chertov O. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science. 2002;298:1025–1029. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- 150.Funderburg N., Lederman M.M., Feng Z., Drage M.G., Jadlowsky J., Harding C.V. Human-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc Natl Acad Sci USA. 2007;104:18631–18635. doi: 10.1073/PNAS.0702130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ferris L.K., Mburu Y.K., Mathers A.R., Fluharty E.R., Larregina A.T., Ferris R.L. Human beta-defensin 3 induces maturation of human langerhans cell-like dendritic cells: an antimicrobial peptide that functions as an endogenous adjuvant. J Invest Dermatol. 2013;133:460–468. doi: 10.1038/jid.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Presicce P., Giannelli S., Taddeo A., Villa M.L., Della Bella S. Human defensins activate monocyte-derived dendritic cells, promote the production of proinflammatory cytokines, and up-regulate the surface expression of CD91. J Leukoc Biol. 2009;86:941–948. doi: 10.1189/jlb.0708412. [DOI] [PubMed] [Google Scholar]
- 153.Sakamoto N., Mukae H., Fujii T., Ishii H., Yoshioka S., Kakugawa T. Differential effects of alpha- and beta-defensin on cytokine production by cultured human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L508–L513. doi: 10.1152/ajplung.00076.2004. [DOI] [PubMed] [Google Scholar]
- 154.Niyonsaba F., Ushio H., Nagaoka I., Okumura K., Ogawa H. The human beta-defensins (-1, -2, -3, -4) and cathelicidin LL-37 induce IL-18 secretion through p38 and ERK MAPK activation in primary human keratinocytes. J Immunol. 2005;175:1776–1784. doi: 10.4049/jimmunol.175.3.1776. [DOI] [PubMed] [Google Scholar]
- 155.Guo C.J., Tan N., Song L., Douglas S.D., Ho W.Z. alpha-Defensins inhibit HIV infection of macrophages through upregulation of CC-chemokines. AIDS. 2004;18:1217–1218. doi: 10.1097/00002030-200405210-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.O'Neil D.A., Porter E.M., Elewaut D., Anderson G.M., Eckmann L., Ganz T. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- 157.Semple F., Dorin J.R. beta-Defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun. 2012;4:337–348. doi: 10.1159/000336619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Semple F., Webb S., Li H.N., Patel H.B., Perretti M., Jackson I.J. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur J Immunol. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Semple F., MacPherson H., Webb S., Cox S.L., Mallin L.J., Tyrrell C. Human beta-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur J Immunol. 2011;41:3291–3300. doi: 10.1002/eji.201141648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 162.Tewary P., de la Rosa G., Sharma N., Rodriguez L.G., Tarasov S.G., Howard O.M. beta-Defensin 2 and 3 promote the uptake of self or CpG DNA, enhance IFN-alpha production by human plasmacytoid dendritic cells, and promote inflammation. J Immunol. 2013;191:865–874. doi: 10.4049/jimmunol.1201648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Eade C.R., Wood M.P., Cole A.M. Mechanisms and modifications of naturally occurring host defense peptides for anti-HIV microbicide development. Curr HIV Res. 2012;10:61–72. doi: 10.2174/157016212799304580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Verma C., Seebah S., Low S.M., Zhou L., Liu S.P., Li J. Defensins: antimicrobial peptides for therapeutic development. Biotechnol J. 2007;2:1353–1359. doi: 10.1002/biot.200700148. [DOI] [PubMed] [Google Scholar]
- 165.Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]