Abstract
Starvation of yeast for a single amino acid leads to derepression of enzymes in many different amino acid biosynthetic pathways. This general control is regulated by several transacting genes. Mutations in the TRA3 gene result in constitutive derepression, whereas mutations in AAS genes lead to the inability to derepress. We have isolated aas mutations as suppressors of the tra3-1 mutation. Some of these suppressors are alleles of AAS2 and others define a heretofore unidentified gene, AAS3. We have studied the regulatory behavior of strains containing both aas and tra3 mutations and strains containing the cloned AAS genes in high copy number. Either aas1- or aas2- in combination with tra3- has the Tra- phenotype, whereas aas3- in combination with tra3- has the Aas- phenotype. These interactions suggest that the AAS1 and AAS2 products act indirectly to bring about derepression by disabling the repressive effect of TRA3, whereas the AAS3 product functions more directly and is required even in the absence of the TRA3 function. When present in high copy number, the AAS3 gene complements mutations in AAS1 and AAS2, whereas AAS1 and AAS2 only complement their cognate mutations. Taken together these data suggest that AAS1 and AAS2 are negative regulators of TRA3, which in turn is a negative regulator of AAS3. AAS3 is a positive regulator, which is required for the general control response. This model of negative and positive interactions is formally identical to those proposed for the regulation of the galactose and phosphatase systems in yeast.
Full text
PDF

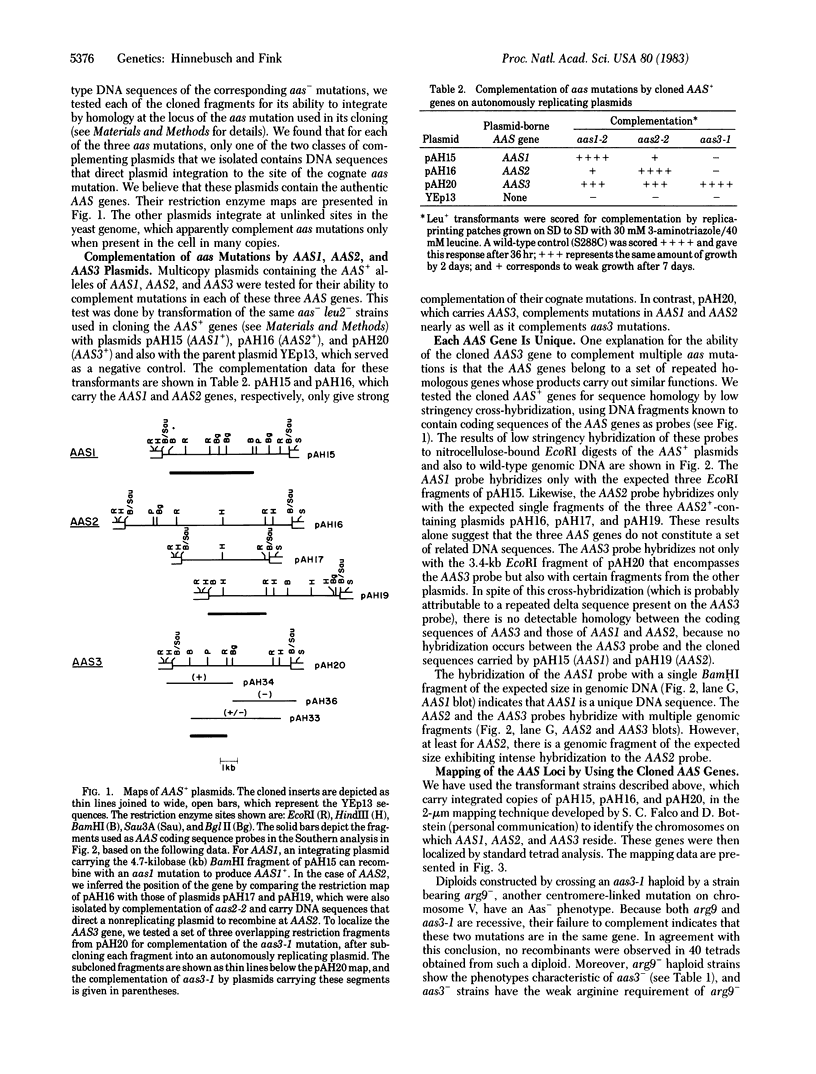
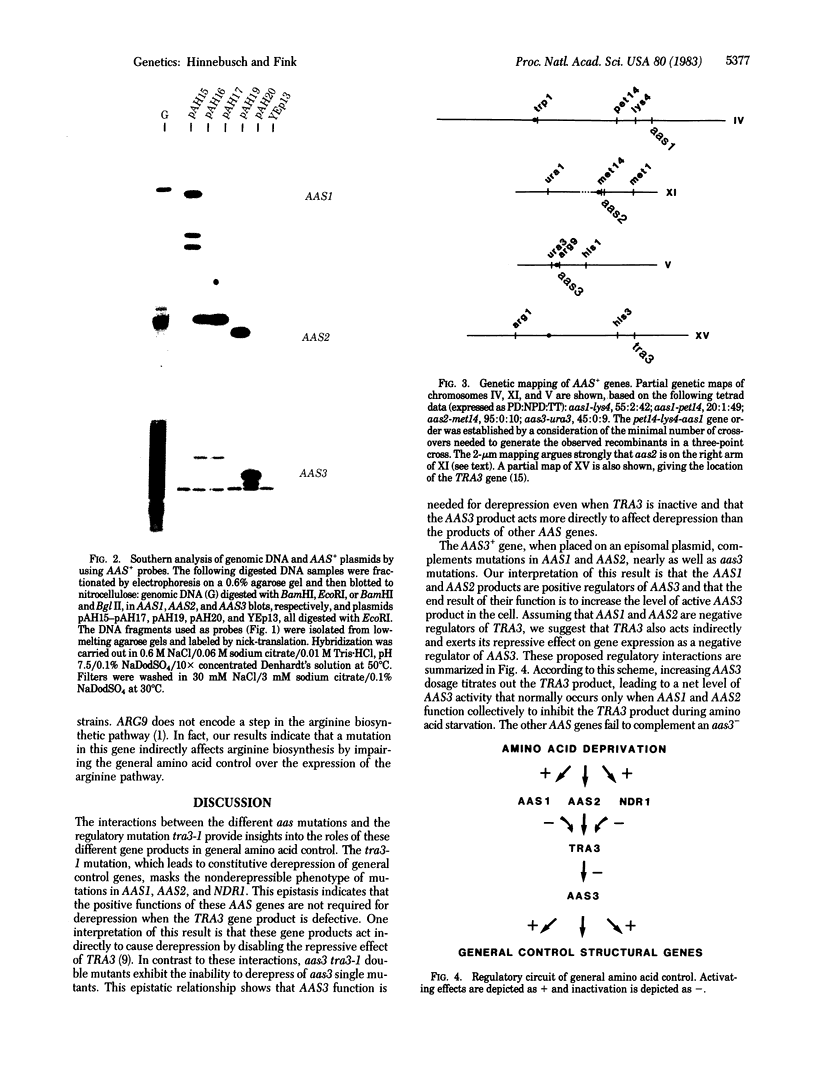

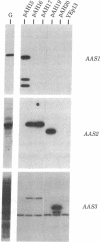
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Falco S. C., Li Y., Broach J. R., Botstein D. Genetic properties of chromosomally integrated 2 mu plasmid DNA in yeast. Cell. 1982 Jun;29(2):573–584. doi: 10.1016/0092-8674(82)90173-8. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6827–6831. doi: 10.1073/pnas.79.22.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Mol Gen Genet. 1983;189(1):148–156. doi: 10.1007/BF00326068. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Niederberger P., Miozzari G., Hütter R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jul;1(7):584–593. doi: 10.1128/mcb.1.7.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Galgoci B., Greer H. Identification of AAS genes and their regulatory role in general control of amino acid biosynthesis in yeast. Proc Natl Acad Sci U S A. 1983 May;80(9):2704–2708. doi: 10.1073/pnas.80.9.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Regulatory sites for his3 gene expression in yeast. Nature. 1982 Nov 18;300(5889):285–286. doi: 10.1038/300284a0. [DOI] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]